Abstract
Flytrap is a web-enabled relational database of transposable element insertions in Drosophila melanogaster. A green fluorescent protein (GFP) artificial exon carried by a transposable P-element is mobilized and inserted into a host gene intron creating a GFP fusion protein. The sequence of the tagged gene is determined by sequencing inverse-PCR products derived from genomic DNA. Flytrap contains two principle data types: micrographs of protein localization and a cellular component ontology, based on rules derived from the Gene Ontology consortium (http://www.geneontology.org), describing protein localization. Flytrap also has links to gene information contained in Flybase (http://flybase.bio.indiana.edu). The system is designed to accept submissions of micrographs and descriptions from any type of tissue (e.g. wing imaginal disk, ovary) and at any stage of development. Insertion lines can be searched using a number of queries, including Berkeley Drosophila Genome Project (BDGP) numbers and protein localization. In addition, Flytrap provides online order forms linked to each insertion line so that users may request any line generated from this project. Flytrap may be accessed from the homepage at http://flytrap.med.yale.edu.
INTRODUCTION
The Flytrap database was designed to support an ongoing genetic screen, the goal of which is to tag every gene in Drosophila melanogaster (1). In Drosophila the first implementation of this screening strategy was reported by Morin et al. (2). The strategy is designed to generate random GFP fusion proteins throughout the fly genome. The sequence of the tagged gene is determined by sequencing inverse-PCR products derived from genomic DNA. The sequence is then used to search through the entire Drosophila genome using the BLASTN algorithm (3). Since the frequency of obtaining an insertion is low, approximately 1 per 1000–2000 animals screened, an automated embryo sorter (Union BioMetrica Inc., Somerville, MA) was used to screen through up to 500 000 embryos per day. Currently there are 599 lines documented in Flytrap (Table 1). This number is expected to expand rapidly in the coming months. A similar transposon-tagging protein-trap screen has been carried out in Saccharomyces cerevisiae (4,5) and a data set is available online at http://ygac.med.yale.edu/triples/triples.htm.
Table 1. Publicly available Flytrap data sets (as of August 2003).
| Data set | Entries | Number |
|---|---|---|
| Transposon insertions | Total | 599 |
| Sequenced/defined insertion | 223 | |
| Tagged genes (annotated) | 61 | |
| Tagged genes (unannotated) | 51 | |
| Localization data | Total | 599 |
DESIGN AND IMPLEMENTATION
Flytrap was implemented using the open source MySQL database system (http://www.mysql.com). Our web server is a Macintosh G3 running OS X version 10.2.6 (Apple Computer, Cupertino, CA). The front end was implemented using the Hypertext Preprocessor (PHP) (http://www.php.net), a component of the Apache web server (http://httpd.apache.org/). The PHP script language has enabled us to embed server-side code within HTML documents. We have also incorporated several freeware libraries to generate graphical plots and histograms of localization and insertion data.
Flytrap is composed of both public and private areas. The public areas serve to generate reports on the existing data sets, and allow for data mining. Lines will be added to the public domain as they become available. Members of the Flytrap consortium may enter a password-protected area to upload data files using a web-based interface.
DATA SEARCHING AND RETRIEVAL
Users may access data within Flytrap through category-specific searches targeted at single data types (e.g. localization data, transposon insertion). The user may search by the gene designation (e.g. BDGP CG or FBgn) or the unique line identification assigned during the screen (e.g. G00005). Alternatively, expression data regarding a unique insertion may be accessed. For example, Flytrap may be queried for all tagged proteins localizing to the nucleus of somatic cells by executing a category-specific search of follicle cell localization data with ‘nucleus’ chosen as the localization. Similarly all the searches can be executed using a combination of search terms using the Boolean operators ‘and’ or ‘or’.
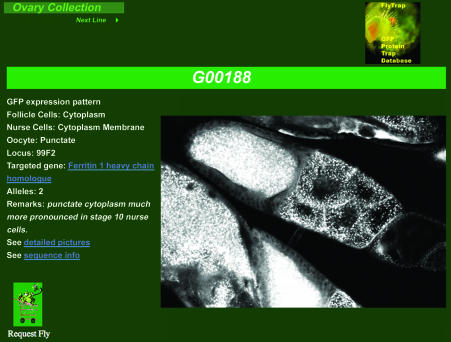
The results are presented in a tabular format and may be downloaded as a tab-delimited text file. Category-specific reports may be sorted by clicking on data fields (e.g. Gene Trapped, Cytology) to group results in preferred hierarchies. To further enhance the utility of Flytrap, all trapped genes are linked to a complete Flybase (6) report to give the user a comprehensive explanation of the gene that is trapped. Each line identifier may be clicked on to generate a corresponding detailed report for that line (Figure 1). The designation of the line (i.e. G00005 versus ZCL2071) indicates that the lines were derived at different stages, and in some cases different locations, during the screen.
Figure 1.
Example of a detailed record generated by Flytrap. Transposon-tagged proteins were visualized directly by GFP fluorescence. Over 1000 images of subcellular staining patterns have been recorded and may be viewed through Flytrap. The detailed record includes information about the line, including the subcellular localization in different cell types, the cytological position of the insertion, the identity of the targeted gene, the number of insertions obtained in the screen (Alleles), remarks about the line, links to additional pictures and links to supporting DNA sequence that was used to identify the targeted gene. Additionally, a user can click on the Request Fly link to add the line to a shopping cart. After adding any number of lines a user can check out and have the lines delivered via postal or overnight services.
The detailed report for each line indicates whether additional tissues have been examined. An icon will appear at the top of the screen describing which tissue has been examined and by clicking on the icon the user will open up an additional screen detailing the images and observations made in a given tissue. From the detailed report the user may also choose to add the line to a ‘shopping-cart’. After selecting all the desired lines, the user can ‘check out’ and have the line(s) delivered by the USPS at no cost to the user, or by an overnight carrier paid by the user.
SIGNIFICANCE
In the ever-expanding realm of genome-sized data sets, it is increasingly important that data sets adhere to common rules established by genomic consortia. By adopting open source applications (e.g. MySQL, PHP and Apache) to maintain data sets sharing a common lexicon, free exchange of data will continue to push forward progress in our understanding of large-scale data sets. Free access to expression data in Flytrap combined with the access to fly stocks will greatly facilitate rapid progress in research.
SUPPLEMENTARY MATERIAL
A tab-delimited text file detailing the current Flytrap data set is available as Supplementary Material at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank members, past and present, of the Cooley lab for helpful discussions. We are grateful to William Chia and Xavier Morin for fruitful and ongoing collaboration. Additionally we would like to thank Kevin White for invaluable discussions on the implementation of a MySQL database. We would also like to thank Jeff Axelrod and Barbara Wakimoto for contributing wing disk and testis images, respectively. We would also like to acknowledge Alain Debec for inspiring the layout of the details page. This work was supported by grants to L.C. from the NIH (GM43301, GM52702).
REFERENCES
- 1.Spradling A.C., Stern,D., Beaton,A., Rhem,E.J., Laverty,T., Mozden,N., Misra,S. and Rubin,G.M. (1999) The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics, 153, 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morin X., Daneman,R., Zavortink,M. and Chia,W. (2001) A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl Acad. Sci. USA, 98, 15050–15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 4.Kumar A., Cheung,K.H., Ross-Macdonald,P., Coelho,P.S., Miller,P. and Snyder,M. (2000) TRIPLES: a database of gene function in Saccharomyces cerevisiae. Nucleic Acids Res., 28, 81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar A., Cheung,K.H., Tosches,N., Masiar,P., Liu,Y., Miller,P. and Snyder,M. (2002) The TRIPLES database: a community resource for yeast molecular biology. Nucleic Acids Res., 30, 73–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fly Base Consortium (2003) The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res., 31, 172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.