Abstract
RGS14 is a brain scaffolding protein that integrates G protein and MAP kinase signaling pathways. Like other RGS proteins, RGS14 is a GTPase activating protein (GAP) that terminates Gαi/o signaling. Unlike other RGS proteins, RGS14 also contains a G protein regulatory (also known as GoLoco) domain that binds Gαi1/3-GDP in cells and in vitro. Here we report that Ric-8A, a non-receptor guanine nucleotide exchange factor (GEF), functionally interacts with the RGS14:Gαi1-GDP signaling complex to regulate its activation state. RGS14 and Ric-8A are recruited from the cytosol to the plasma membrane in the presence of co-expressed Gαi1 in cells, suggesting formation of a functional protein complex with Gαi1. Consistent with this idea, Ric-8A stimulates dissociation of the RGS14:Gαi1-GDP complex in cells and in vitro using purified proteins. Purified Ric-8A stimulates dissociation of the RGS14:Gαi1-GDP complex to form a stable Ric-8A:Gαi complex in the absence of GTP. In the presence of activating nucleotide, Ric-8A interacts with the RGS14:Gαi1-GDP complex to stimulate both the steady-state GTPase activity of Gαi1 and GTP binding to Gαi1. However, sufficiently high concentrations of RGS14 competitively reverse these stimulatory effects of Ric-8A on Gαi1 nucleotide binding and GTPase activity. This observation correlates with findings that show RGS14 and Ric-8A share an overlapping binding region within the last 11 amino acids of Gαi1. As further evidence that these proteins are functionally linked, native RGS14 and Ric-8A co-exist within the same hippocampal neurons. These findings demonstrate that RGS14 is a newly appreciated integrator of unconventional Ric-8A and Gαi1 signaling.
Conventional models of G protein signaling (1, 2) indicate that activated G protein-coupled receptors (GPCRs) serve as guanine nucleotide exchange factors (GEFs) towards coupled heterotrimeric (Gαβγ) G proteins. GPCR activation facilitates GDP release and subsequent GTP binding to the Gα subunit, which is followed by Gβγ dissociation from Gα-GTP. This allows free Gβγ and Gα-GTP to engage downstream effectors and linked signaling pathways. The lifetime of this signaling event is terminated by the regulators of G protein signaling (RGS) proteins, a large family of multifunctional signaling proteins that regulate the intrinsic GTPase activity of the Gα subunit and promote heterotrimer reassociation (3-5).
RGS14 is a highly unusual RGS protein that is enriched in brain (6, 7) and binds to Gαi/o and H-Ras/Raf to integrate G protein and MAP kinase signaling pathways (8). RGS14 contains a conserved RGS domain, two adjacent Ras/Rap binding domains (RBDs), and a G protein regulatory (also known as a GoLoco [GL]) domain (7, 9). Like all RGS proteins, the RGS domain of RGS14 binds directly to active Gα (specifically Gαi and Gαo) to serve as a non-selective GTPase Activating Protein (GAP) towards both of these Gα subunits (6, 7, 10). Unlike other RGS proteins, the GL domain of RGS14 binds directly to inactive Gαi1-GDP and Gαi3-GDP to inhibit guanine nucleotide binding and exchange (11-13). Furthermore, the GL domain of RGS14 forms a tight complex at the plasma membrane with inactive Gαi1 and Gαi3 independent of Gβγ(13), suggesting RGS14 serves a different role in G protein signaling compared to other RGS proteins.
Independent of conventional GPCR/G protein signaling, several unconventional G protein signaling pathways have been described recently that are involved in cell division and synaptic signaling (14-21). Ric-8A (Synembryn) is a cytosolic protein reported to bind to and act as a non-receptor GEF for Gαi1, Gαq, and Gαo proteins (22-24). Ric-8A recognizes inactive Gα-GDP proteins when they are in complex with several GL-domain containing proteins, including LGN/mPins and Activator of G protein Signaling 3 (AGS3). Like RGS14, LGN/mPins and AGS3 bind directly to inactive Gαi (22, 24), with LGN also being recruited to the plasma membrane by Gαi1 (25). However, unlike RGS14, these proteins lack an RGS domain.
Given these similarities between RGS14, LGN/mPins, and AGS3, we sought to investigate if RGS14 functionally interacts with Ric-8A to regulate unconventional G protein signaling. Here we report that RGS14 is the first example of an RGS protein that also serves as a GL protein, forming a complex with Gαi1-GDP that is regulated by Ric-8A. We show that Ric-8A interacts with RGS14 in cells and acts on the RGS14:Gαi1-GDP protein complex in vitro, thereby promoting complex dissociation to affect the activation state of Gαi1. Moreover, we demonstrate that native RGS14 and Ric-8A co-exist within the same hippocampal neurons, further supporting a functional link between these two proteins. Taken together, these findings demonstrate that RGS14 serves as a multifunctional GL protein in addition to an RGS protein. We therefore propose a working molecular model to describe how Ric-8A could regulate RGS14:Gαi1 signaling functions in cells.
Experimental Procedures
Plasmids and antibodies
The rat RGS14 cDNA used in this study (Genbank accession number U92279) was acquired as described (6). Glu-Glu (EE) tagged recombinant Gαi1 plasmid was purchased from UMR cDNA Resource Center (Rolla, Missouri). The plasmids encoding full-length RGS14 and RGS14 deletion mutants coding for amino acids 213-544 and 444-544 cloned in-frame into pcDNA3.1 (Invitrogen) were prepared as described previously (13). Oligonucleotides encoding the 8 amino acid Flag tag (Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys) were used to generate N-terminally Flag-tagged RGS14. His6-Gαi1 (N149I) derived from Escherichia coli was generated by changing bases AAC of the rat Gαi1 cDNA to ATA using the QuickChange site-directed mutagenesis kit (Stratagene), resulting in an amino acid change of N to I. Truncated His6-Gαi1 (termed Gαi1-ΔCT throughout the text) derived from E. coli was made by deleting the last 11 amino acids (IKNNLKDCGLF) of the rat Gαi1 and cloning the resulting cDNA in-frame into pET20b vector.
Anti-Flag M2 agarose beads, anti-Flag antibody, and anti-Flag HRP antibody were purchased from Sigma. Other antisera include anti-GFP antibody (Clontech), anti-His antibody (Covance), anti-Ric-8A antiserum (a gift from Dr. Greg Tall), anti-Gαi1 antibody (Santa Cruz), anti-EE antibody (BD Biosciences), anti-RGS14 antibody (Antibodies, Inc.), a rhodamine-conjugated mouse secondary IgG (Jackson), Alexa 553 goat anti-rabbit secondary IgG (Invitrogen), Alexa 546 goat anti-mouse secondary IgG (Invitrogen), Alexa 488 goat anti-rabbit secondary IgG (Invitrogen), Alexa 633 goat anti-mouse secondary IgG (Invitrogen), peroxidase-conjugated goat anti-mouse IgG antisera (Rockland Immunochemicals, Inc.), and peroxidase-conjugated goat anti-rabbit IgG antisera (Bio-Rad) .
Cell Culture
HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with sodium pyruvate and glutamate supplemented with 10% fetal bovine serum (FBS) and a mixture of 100 U/mL penicillin plus 100 μg/mL streptomycin (Sigma). Cells were incubated at 37°C with 5% CO2.
Cell transfection and anti-Flag immunoprecipitation
HeLa cells were obtained from the American Type Culture Collection (ATCC). Transfections were performed using previously described protocols with Lipofectamine 2000 (Invitrogen) (13). Cells were transiently transfected with CFP-Ric-8A and pcDNA3.1, wild-type Gαi1-EE, Flag-RGS14 (full-length), and Flag-RGS14 truncation mutants 213-544 and 444-544 either alone or in combination. Eighteen hours post-transfection, cells were lysed in buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 2 mM dithiothreitol, 10 mM MgCl2, protease inhibitor cocktail (Roche), and 1% TritonX-100. Lysates were incubated on a 4°C rotator for 1 hour, and then cleared by centrifugation at 100,000 × g for 30 mins at 4°C. Lysates were incubated with 50 μg anti-Flag M2 resin for 1.5 hours on a 4°C rotator. Resin was washed with ice-cold TBS four times and proteins were eluted by addition of Laemmli sample buffer and subsequent boiling for 5 mins. Samples were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and blotted with anti-Flag HRP, anti-GFP, and anti-EE antibodies followed by appropriate secondary antibodies. Proteins were detected by enhanced chemiluminescence.
Immunoprecipitation of pure proteins
10 μg of wild-type His6-Gαi1 (WT), His6-Gαi1 (N149I), or His6-Gαi1-ΔCT protein derived from E. coli lysates was mixed alone or with 5 μg of either purified full-length TxHis6-RGS14 or His6-YFP-Ric-8A (referred to as YFP-Ric-8A). YFP-Ric-8A was made as described (24). Proteins were diluted in buffer containing 20 mM HEPES, 150 mM NaCl, 2 mM dithiothreitol, 1 mM EDTA, and protease inhibitor cocktail. Proteins were incubated with 50 μg Protein G sepharose resin (GE Healthcare) and immunoprecipitated with either anti-RGS14 antibody or anti-Ric-8A antibody at 4°C for 3 hours. Resin was washed with ice-cold TBS four times and proteins were eluted by addition of Laemmli sample buffer and subsequent boiling for 5 mins. Samples were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and blotted with anti-His, anti-Ric-8A, and anti-Gαi1 antibodies followed by appropriate secondary antibodies. Proteins were detected by enhanced chemiluminescence.
Immunofluorescence and confocal imaging
Transfected HeLa cells were fixed at room temperature for 10 mins with buffer containing 20 mM PIPES, pH 7.0, 0.5 mM EGTA, 1 mM MgCl2, 1 mM glutaraldehyde, 1 g/mL aprotinin, 0.1% TritonX-100, 2 mM taxol, and 2% paraformaldehyde. Cells were then blocked for 1 hour at room temperature in PBS containing 10% goat serum and 3% bovine serum albumin. Next, cells were incubated in this same buffer with a 1:1000 dilution of rabbit anti-Flag and/or mouse anti-EE antibodies overnight at 4°C. Cells were washed with PBS (3X) and incubated with 1:200 dilutions of Alexa 553 goat anti-rabbit and Alexa 633 goat anti-mouse secondary antibodies at room temperature for 1 hour. Cells were washed with PBS again (3X) and mounted with Vectashield mounting medium (Vector Laboratories). Confocal images were taken using a 63x oil immersion objective from a LSM510 laser scanning microscope (Zeiss). Images were processed using the Zeiss LSM image browser (version 2.801123) and Adobe Photoshop 7.0 (Adobe Systems).
Immunohistochemistry (IHC) and confocal imaging of mouse brain thin sections
To obtain brain thin sections, C57BL6 wild-type mice were perfused with saline and then with 4% paraformaldehyde. Brains were isolated, post-fixed in 4% paraformaldehyde, and then embedded in paraffin. After embedding, thin sections were cut. For IHC analysis, brain thin sections were de-paraffinized and pre-treated by microwaving in 1X citrate buffer (0.001 M citrate monohydrate in distilled water, pH 6.0). Sections were treated with 3% H2O2 and blocked with 2% goat serum in Tris-Brij buffer (0.1 M Tris-Cl, 0.1 M NaCl, 0.025 M MgCl2, and .075% Brij 35) for 15 mins. Sections were incubated with anti-Ric-8A and anti-RGS14 antibodies overnight at 4°C, and then incubated with either Alexa 546 anti-mouse and Alexa 488 anti-rabbit fluorescent secondary antibodies or anti-mouse and anti-rabbit biotinylated secondary antibodies (Vector Laboratories). Following biotinylated secondary antibody incubation, sections were incubated with avidin-biotin-peroxidase complex, and color was developed with 3, 3′-diaminobenzidine. Control sections were stained with antibody that was pre-blocked with either Ric-8A or RGS14 pure protein (10:1 ratio of protein to antibody). Confocal images were taken and processed as described above. IHC images were taken using a Nikon double-headed microscope.
Pure protein dissociation assays
Purified TxHis6-ΔRGS14 (encoding amino acids 299-544, including the RBD domains and GL domain) was created as described (6). Pre-formed ΔRGS14:Gαi1-GDP protein complex was created by mixing 85 μg pure His6-Gαi1-GDP with 25 μg pure TxHis6-ΔRGS14 at 4°C for 90 minutes. Sample was then separated over a tandem Superdex S75+S200 size-exclusion gel filtration apparatus in buffer containing 50 mM HEPES, pH 8.0, 1 mM EDTA, 150 mM NaCl, and 2 mM dithiothreitol. Elution volume containing the protein complex (500 μL of fraction corresponding to total elution volume 18000 μL – 18500 μL) was taken and mixed with 50 μM GTPγS and 10 mM MgCl2 either alone or with a 5-fold excess of YFP-Ric-8A pure protein over ΔRGS14 for 15 mins at 30°C. In other dissociation assays, pre-formed ΔRGS14:Gαi1-GDP complex was collected and mixed with a 30-fold excess of YFP-Ric-8A only, without GTPγS. After treatment, the sample was then reapplied to the gel filtration column, and resulting fractions were collected and subjected to SDS-PAGE and immunoblot analysis. Blots were probed with anti-His and anti-Ric-8A antibodies. For YFP-Ric-8A:Gαi1 complex formation, 9 μg YFP-Ric-8A was incubated with 30 μg His6-Gαi1-GDP at 4°C for 90 minutes in the buffer described above and then applied over tandem S75+S200 gel filtration columns as described above.
GTPγS binding assays
GTPγS binding studies were performed as previously described (26). Briefly, 2 μM His6-Gαi1-GDP (diluted in 20 mM HEPES and 50 mM NaCl) was incubated with 2 μM (final concentration) [35S]GTPγS (10,000 cpm/pmol) with or without amounts of TxHis6-ΔRGS14 (25 μM) and YFP-Ric-8A (either 5 μM or 125 μM) at 30°C in reaction buffer (20 mM HEPES, 100 mM NaCl, 1 mM dithiothreitol, 2 mM MgSO4, and 1 mM EDTA). Reactions were done in triplicate and stopped at the indicated time points in ice cold stop buffer (20 mM Tris, 200 mM NaCl, 2 mM MgSO4, and 1 mM GTP), quickly filtered over nitrocellulose membranes, and washed twice with wash buffer (50 mM Tris, 200 mM NaCl, and 2 mM MgSO4). Scintillation fluid (MP Biomedicals) was added to filters, and then filters were subjected to scintillation counting. The amount of [35S]GTPγS bound to the filters was quantified, and the measurements at the 0 min time point were subtracted out as background. Data are presented as mean ± S.E.M. When testing the activity of the Gαi1 mutants, the exact same protocol was performed using 2 μM Gαi1-WT, Gαi1 (N149I), and Gαi1-ΔCT alone for 0 min, 5 min, and 10 min time points.
Steady-state GTPase assays
Steady-state GTPase assays were performed as described (26, 27) at 30°C in buffer A that contained 20 mM HEPES, 100 mM NaCl, 1 mM EDTA, 2 mM MgCl2, and 0.05% Lubrol. His6-Gαi1-GDP (0.5 μM) and either full-length TxHis6-RGS14 or truncated TxHis6-ΔRGS14 (0.3 μM) were incubated for 15 min at 4°C and YFP-Ric-8A (1.5 μM) was added just before initiation of the reaction. To initiate the steady-state reaction, 0.4 μM [γ-32P]GTP (specific activity 200 cpm/pmol) in 100 uL buffer A was added. At 5 minute intervals, from 0 to 20 minutes, triplicate aliquots were removed and added to 1 mL of ice cold 5% (w/v) activated charcoal to stop the reactions. The charcoal was pelleted at 4000 × g and the clear supernatant was removed and added to scintillation vials. The resulting [32P]i released in the supernatant was measured by scintillation counting. Data are presented as mean ± S.E.M.
For steady-state GTPase experiments measuring the effects of protein concentration on response, various concentrations of full-length TxHis6-RGS14 ranging from 0 to 8.0 μM (0 nM, 10 nM, 30 nM, 100 nM, 300 nM, 1000 nM, 3000 nM, and 8000 nM) were incubated with 0.5 μM His6-Gαi1-GDP for 15 min at 4°C. YFP-Ric-8A (1.5 μM) was added just before initiation of the reaction. To initiate the steady-state reaction, 0.4 μM [γ-32P]GTP (specific activity 200 cpm/pmol) in 100 uL buffer A (see above) was added. After 10 minutes, triplicate aliquots were removed and added to 1 mL of ice cold 5% (w/v) activated charcoal to stop the reactions. The charcoal was pelleted at 4000 × g and the clear supernatant was removed and added to scintillation vials. The resulting [32P]i released in the supernatant was measured by scintillation counting. Data are presented as mean ± S.E.M.
Results
RGS14 and Ric-8A localize at the plasma membrane with Gαi1
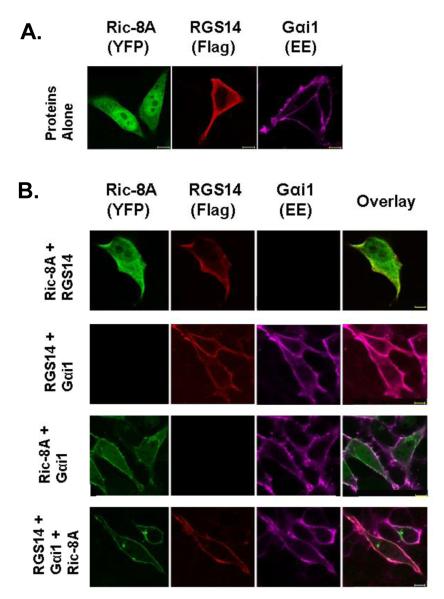
RGS14 is unusual among RGS proteins in that it contains not only an RGS domain that binds active Gαi1-GTP, but also a GL domain that binds inactive Gαi1-GDP. Therefore, we sought to determine whether RGS14 is the first example of an RGS protein that functionally interacts with Ric-8A, a reported cytosolic GEF that regulates certain GL proteins. A strong indicator of functional interaction between proteins is their capacity to co-localize together in a cellular environment. Therefore, we examined the localization of both Ric-8A and RGS14 in cells in the presence and absence of co-expressed Gαi1-GDP (Fig. 1). Flag-RGS14, YFP-Ric-8A, and wild-type Gαi1-EE were transfected alone and in combination into HeLa cells. Cells were fixed, stained with anti-Flag and anti-EE antibodies, and analyzed for immunofluorescence by confocal microscopy (Fig. 1). When expressed alone in HeLa cells, wild-type Gαi1 localizes at the plasma membrane whereas Ric-8A and RGS14 each predominately localize within the cytosol (Fig. 1A); a small amount of RGS14 is visible at the plasma membrane. When both RGS14 and Ric-8A are co-expressed, they remain mostly cytosolic (Fig. 1B – top). When either RGS14 or Ric-8A is co-expressed with wild-type Gαi1, there is a noticeable translocation of both RGS14 and Ric-8A to the plasma membrane, respectively (Fig. 1B – middle). A small portion of Ric-8A remains localized within the cytosol. Since expression of Gαi1 induces translocation of RGS14, the small amount of RGS14 visible at the plasma membrane in Fig. 1A may be due to the presence of native Gαi1 recruiting RGS14 to the membrane. When RGS14 and Ric-8A are expressed together with wild-type Gαi1 (Fig. 1B – bottom), both RGS14 and Ric-8A translocate from the cytosol to co-localize with Gαi1 at the plasma membrane. The Ric-8A that had remained cytosolic following co-expression with wild-type Gαi1 was now localized at the plasma membrane, suggesting that these three proteins may functionally interact at the plasma membrane. Taken together, it appears that the major driving force behind RGS14 and Ric-8A membrane localization is the presence of Gαi1, which is consistent with the possibility that RGS14 and Ric-8A may be acting on a common Gαi1 subunit in a functional signaling complex.
Figure 1. RGS14 and Ric-8A are recruited to the plasma membrane by wild-type Gαi1.
Ric-8A and RGS14 translocate from the cytosol to the plasma membrane in the presence of wild-type Gαi1. Flag-RGS14, YFP-Ric-8A, and wild-type Gαi1-EE were transfected either alone (A) or in combination (B) into HeLa cells. Cells were fixed, subjected to immunofluorescence, and analyzed using confocal microscopy as described in Experimental Procedures. Scale bars represent 10 μm. Images are representative of cells observed in three separate experiments.
Ric-8A stimulates dissociation of the RGS14:Gαi1-GDP complex in cells
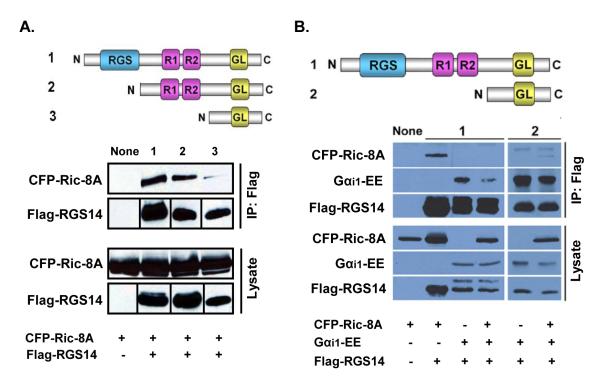
These findings prompted us to examine if RGS14, Ric-8A, and Gαi1 physically interact in cells. We previously demonstrated that RGS14 can form a stable complex with Gαi1 that can be recovered from cells by co-immunoprecipitation (13). Here we tested whether RGS14 can interact with Ric-8A in cells (Fig. 2A). HeLa cells were transfected with CFP-Ric-8A together with either full-length Flag-RGS14 or truncated forms of RGS14 that were missing either the RGS domain (construct expressing amino acids 213-544) or both the RGS domain and the tandem RBDs (construct expressing amino acids 444-544). Ric-8A was recovered together with both full-length RGS14 and RGS14 missing the RGS domain, but not with RGS14 missing the RGS domain and the tandem RBDs (Fig. 2A).
Figure 2. Ric-8A induces dissociation of the RGS14:Gαi1-GDP complex in cells.
Ric-8A induces a decrease in Gαi1 binding to RGS14 in HeLa cells. (A), CFP-Ric-8A was transfected into HeLa cells with either pcDNA3.1 (None), full-length Flag-RGS14 expressing amino acids 1-544 (1), truncated Flag-RGS14 expressing amino acids 213-544 (2), or Flag-RGS14 expressing amino acids 444-544 (3). Cells were lysed and subjected to anti-Flag immunoprecipitation, SDS-PAGE, and immunoblot. To simplify the figure, Flag-RGS14 truncation bands were cropped from their lower molecular weight positions and inserted to form one horizontal line of bands. Results are indicative of three replicate experiments. (B), Combinations of pcDNA3.1, CFP-Ric-8A, Flag-RGS14, and wild-type Gαi1-EE were transfected into HeLa cells (left-most gel). Cells were lysed and subjected to anti-Flag immunoprecipitation. Recovered proteins were subjected to SDS-PAGE and immunoblot. The right-most gel shows results from lysates transfected with combinations of pcDNA3.1, CFP-Ric-8A, wild-type Gαi1-EE, and truncated Flag-RGS14 expressing amino acids 444-544 (which does not bind Ric-8A). pcDNA3.1 was transfected in all double-transfections to bring the DNA concentration up to that of a triple-transfection (CFP-Ric-8A+Flag-RGS14+Gαi1-EE). This figure is representative of three separate experiments.
We next examined whether Ric-8A stimulates the dissociation of an RGS14:Gαi1-GDP complex in cells (Fig. 2B). CFP-Ric-8A, wild-type Gαi1-EE, full-length Flag-RGS14, or the truncated Flag-RGS14 expressing residues 444-544 were transfected alone and in combination into HeLa cells. Cell lysates were subjected to a Flag-immunoprecipitation (IP). In the absence of expressed wild-type Gαi1, Ric-8A interacts with full-length RGS14 (and does not interact non-specifically with the anti-Flag beads). In the absence of expressed Ric-8A, both full-length and truncated RGS14 strongly interact with wild-type Gαi1. However, when Ric-8A and wild-type Gαi1 are co-expressed with full-length RGS14, binding of Ric-8A to RGS14 is eliminated and binding of Gαi1 to RGS14 decreases significantly (Fig. 2B). By contrast, the truncated form of RGS14 missing the apparent Ric-8A binding region (see Fig. 2A) remains bound to Gαi1 in the presence of Ric-8A (Fig. 2B).
Purified Ric-8A stimulates dissociation of the purified RGS14:Gαi1-GDP complex in vitro
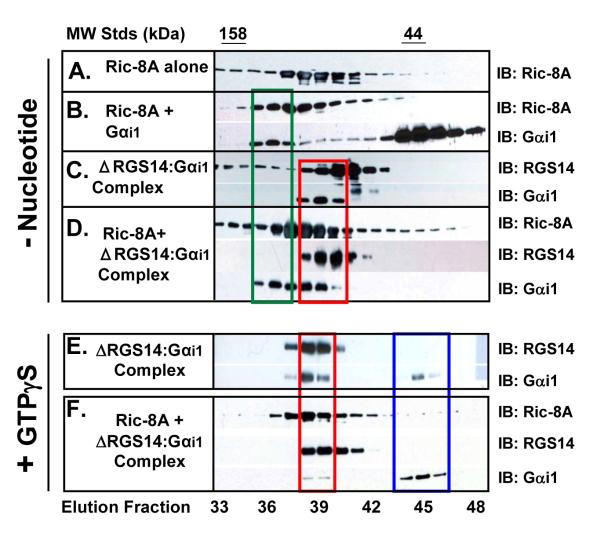
Our findings thus far (Figs. 1 and 2) are consistent with the idea that Ric-8A recognizes the RGS14:Gαi1-GDP complex and stimulates dissociation of the complex in cells, thereby causing release of Gαi1 (and possible binding of Ric-8A to free Gαi1). To test this idea directly, we examined Ric-8A interactions with RGS14 and Gαi1 using purified proteins (Fig. 3). Purified YFP-Ric-8A (24), RGS14, and Gαi1-GDP were mixed in various combinations and then subjected to size-exclusion gel chromatography to examine complex formation. Because expression of full-length recombinant RGS14 yields limiting amounts of functional full-length RGS14, we utilized a more stable truncated form of RGS14 for these studies that lacks the RGS domain (ΔRGS14) (6). As seen in Fig. S1, ΔRGS14 binds Gαi1-GDP to form a stable high molecular weight ΔRGS14:Gαi1-GDP complex detectable by size exclusion chromatography. Full-length RGS14 also forms this complex (data not shown). Ric-8A forms a stable complex with Gαi1-GDP as shown by a shift towards a higher molecular weight when compared to the Ric-8A monomer (Fig. 3, A-B). With this information, we next tested whether purified Ric-8A stimulated dissociation of the ΔRGS14:Gαi1-GDP complex. For this, we prepared a pre-formed ΔRGS14:Gαi1-GDP complex (Fig. S1). After this, we incubated pure YFP-Ric-8A with this preformed ΔRGS14:Gαi1-GDP complex in the absence of added nucleotide or in the presence of GTPγS and MgCl2. In the absence of activating nucleotide, Ric-8A induces partial dissociation of the ΔRGS14:Gαi1-GDP complex along with the formation of a new Ric-8A:Gαi1 complex (presumably nucleotide-free, (23)) (see red and green boxes in Fig. 3, B-D). However, in the presence of GTPγS/Mg2+, Ric-8A induces near-complete dissociation of the ΔRGS14:Gαi1-GDP complex, resulting in free ΔRGS14, free Gαi1-GTPγS, and free Ric-8A (Fig. 3, E-F). Gαi1 can be seen dissociating from ΔRGS14 (see red box of Fig. 3, E-F) and remaining in its monomeric form (see blue box of Fig. 3, E-F). These results clearly show that Ric-8A recognizes, binds, and induces dissociation of the ΔRGS14:Gαi1-GDP complex. We found that the purified full-length RGS14 behaved similarly to ΔRGS14 in these experiments, though our data sets were incomplete due to limiting amounts of available full-length RGS14 (data not shown).
Figure 3. Ric-8A induces dissociation of the RGS14:Gαi1-GDP complex in vitro.
Ric-8A induces dissociation of the ΔRGS14:Gαi1-GDP complex, resulting in the formation of a Ric-8A:Gαi1 complex and subsequent Gαi1-GTP. Either YFP-Ric-8A (A), YFP-Ric-8A and Gαi1-GDP (B), pre-formed ΔRGS14:Gαi1-GDP complex (C), or YFP-Ric-8A and pre-formed ΔRGS14:Gαi1-GDP complex (D) was incubated for 15 mins at 30°C without any exogenous GTP or GDP added. The reaction samples were then loaded onto tandem S75+S200 gel filtration columns and resulting products were resolved by SDS-PAGE and immunoblot. Pre-formed ΔRGS14:Gαi1-GDP complex was incubated alone (E) or with YFP-Ric-8A (F) in the presence of 50 μM GTPγS and 10 mM MgCl2 for 15 mins at 30°C. The reaction samples were then loaded onto tandem S75+S200 gel filtration columns and resulting products were resolved by SDS-PAGE and immunoblot. This figure is representative of three separate experiments for each condition.
Ric-8A-induced dissociation of RGS14:Gαi1-GDP frees Gαi1 to bind GTP
Our findings above (Figs. 2 and 3) indicate that Ric-8A binds Gαi1 and disrupts the RGS14:Gαi1 signaling complex, thus freeing Gαi1 from the GL motif and allowing it to exchange nucleotide and bind GTP. To examine this directly, we measured the capacity of Gαi1 released from the ΔRGS14:Gαi1-GDP complex by Ric-8A to bind [35S]GTPγS (Fig. 4). In the absence of Ric-8A, Gαi1 in complex with RGS14 binds GTPγS very poorly, as expected (6, 11, 12). When Ric-8A is added in 5-fold excess of the ΔRGS14:Gαi1-GDP complex, Gαi1 readily binds GTPγS. Nucleotide binding is apparent immediately upon addition of Ric-8A, and GTPγS binding continues in a linear fashion for up to 10 min. We observe an approximate 4-fold increase in the rate of GTPγS binding to Gαi1 with addition of Ric-8A to the complex (1.04 pmol/min) compared to GTPγS binding to Gαi1 when in complex with ΔRGS14 alone (0.25 pmol/min). Pure Ric-8A protein does not bind GTPγS on its own (Fig. 4), thus the increase in nucleotide binding with Ric-8A+Gαi1-GDP is due to Ric-8A-catalyzed GTPγS binding to Gαi1. These findings show that Ric-8A stimulates dissociation of Gαi1 from RGS14, allowing Gαi1 to bind nucleotide and become activated.
Figure 4. Ric-8A-induced dissociation of the RGS14:Gαi1-GDP complex allows Gαi1 to bind GTP.
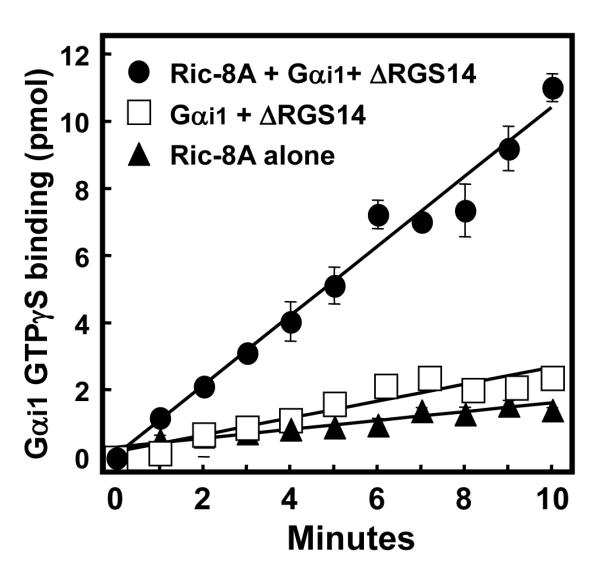
Ric-8A-stimulated dissociation of the ΔRGS14:Gαi1-GDP complex permits free Gαi1 to bind GTPγS. GTPγS binding to Gαi1 was analyzed using YFP-Ric-8A alone, pre-formed ΔRGS14:Gαi1-GDP complex, and YFP-Ric-8A plus ΔRGS14:Gαi1-GDP complex. [35S]GTPγS (2 μM; 10,000 cpm/pmol) was incubated with these protein mixtures in triplicate at 30°C. The amount of [35S]GTPγS bound to protein was quantified using scintillation counting and converted to pmol bound, with background values subtracted out. This figure is representative of three separate experiments for each condition, with data presented as mean ± S.E.M.
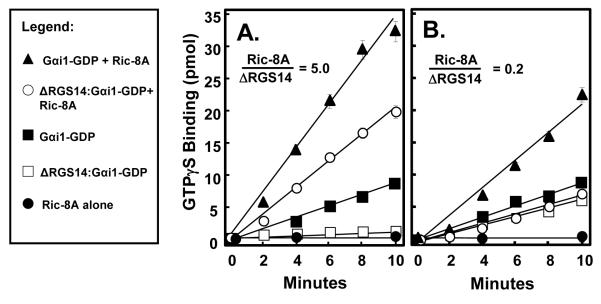
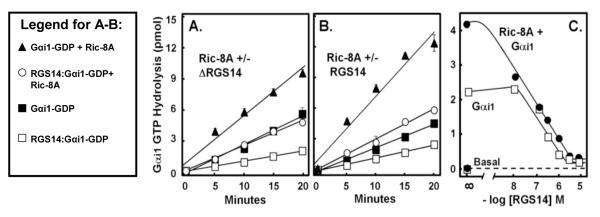
Ric-8A GEF activity towards Gαi1 is dependent on the molar ratio of Ric-8A to RGS14
Ric-8A acts as a GEF towards Gαi1 (23). Since Ric-8A is able to displace Gαi1-GDP from ΔRGS14, it appears that Ric-8A and ΔRGS14 compete for Gαi1 binding. RGS14 may affect, directly or indirectly, Ric-8A GEF activity towards Gαi1. To examine this, we measured the effects of varying the molar ratios of Ric-8A and RGS14 on the rate of nucleotide binding to Gαi1. When Ric-8A is in 5-fold molar excess of ΔRGS14, Ric-8A is able to induce dissociation of the ΔRGS14:Gαi1-GDP complex and catalyze nucleotide exchange on Gαi1 (4.10 pmol/min) in 2.3-fold excess of that observed for Gαi1 alone (1.77 pmol/min) (Fig. 5A). At these molar ratios, ΔRGS14 only partially inhibits Ric-8A GEF activity towards Gαi1 (4.10 pmol/min compared to 6.80 pmol/min with Ric-8A+Gαi1-GDP alone). By contrast, when the Ric-8A concentration is decreased so that ΔRGS14 is in 5-fold molar excess, Ric-8A no longer has any effect on Gαi1 nucleotide binding (0.74 pmol/min compared to 0.88 pmol/min for Gαi1 alone) (Fig. 5B). Pure Ric-8A protein does not bind GTPγS on its own (Fig. 5), indicating that the observed nucleotide binding is due to Ric-8A effects on Gαi1. These findings suggest that Ric-8A is neither able to force ΔRGS14:Gαi1 complex dissociation nor able to act as a GEF towards Gαi1 under these experimental conditions (Fig. 5B). Of note, the failure of Ric-8A to overcome these effects of RGS14 on Gαi1 may be due to the absence of properly modified Gαi1, since myristoylated Gαi1 has been shown to enhance the capacity of Ric-8A to act on GL:Gαi1-GDP complexes (22). We also tested whether purified full-length RGS14 containing the RGS domain behaved any differently in these assays than did ΔRGS14 missing the RGS domain. We found that the presence of the RGS domain in full-length RGS14 had no effect on Ric-8A-directed GEF activity towards Gαi1 (data not shown).
Figure 5. Ric-8A reverses RGS14 inhibition of GTPγS binding to Gαi1.
The degree of RGS14-induced inhibition of Ric-8A nucleotide exchange activity towards Gαi1 is dependent on the molar ratio of Ric-8A to RGS14. ΔRGS14:Gαi1-GDP complex was incubated with either a 5-fold excess of YFP-Ric-8A to ΔRGS14 (A) or one-fifth the concentration of YFP-Ric-8A to ΔRGS14 (B) and then mixed with [35S]GTPγS (2 μM; 10,000 cpm/pmol) at 30°C in triplicate. The amount of [35S]GTPγS bound to protein was quantified using scintillation counting. Measurements were converted to pmol [35S]GTPγS bound, with background subtracted out. This figure is representative of three separate experiments for each condition, with data presented as mean ± S.E.M.
Ric-8A stimulates an increase in the steady-state GTPase activity of Gαi1 in the presence of RGS14
All GEFs act by increasing the rate of release of GDP bound to Gα, thereby greatly reducing the rate-limiting step in guanine nucleotide exchange and steady-state hydrolysis. Thus, GEF activity is reflected both as an increase in GTPγS binding and also as an increase in steady-state GTPase activity on the target Gα (23, 28). Consistent with its reported role as a GEF, Ric-8A stimulates steady-state GTPase activity of Gαi1 (22, 23). Thus, in addition to examining Ric-8A effects on nucleotide binding (Fig. 5), we also examined its effects on Gαi1 GTPase activity and the importance of RGS14 and its RGS domain on this activity. Assays of Gαi1 steady-state GTPase activity were designed to include combinations of purified Ric-8A, Gαi1-GDP, and either truncated ΔRGS14 or full-length RGS14 (Fig. 6). ΔRGS14 inhibits the GTPase activity of Gαi1 2.8-fold (0.48 pmol/min compared to 1.37 pmol/min for Gαi1 alone) (Fig. 6A). Ric-8A overcomes this inhibition, catalyzing an increase in Gαi1 GTPase activity by 2.5-fold in the presence of ΔRGS14 (1.20 pmol/min compared to 0.48 pmol/min). However, the capacity of Ric-8A to overcome this inhibition does not exceed the intrinsic GTP hydrolysis rate of Gαi1 (1.37 pmol/min). Full-length RGS14 also inhibits the GTPase activity of Gαi1 (0.62 pmol/min compared to 1.15 pmol/min for Gαi1 alone) (Fig. 6B). Ric-8A overcomes this inhibition by 2.4-fold, however again only to the approximate rate of intrinsic Gαi1 GTP hydrolysis.
Figure 6. Ric-8A reverses RGS14 inhibition of Gαi1 steady-state GTPase activity.
Both full-length RGS14 and ΔRGS14 inhibit the Ric-8A-catalyzed increase in steady-state GTPase activity of Gαi1. Combinations of YFP-Ric-8A, His6-Gαi1-GDP, and either pre-formed ΔRGS14:Gαi1-GDP complex (A) or full-length RGS14:Gαi1-GDP complex (B) were used to analyze steady-state GTPase activity of Gαi1. (C), YFP-Ric-8A and Gαi1-GDP were mixed with increasing concentrations of full-length RGS14 as indicated. Protein combinations were mixed in triplicate with [γ-32P]GTP and the amount of [32P]i released in each sample was quantified using scintillation counting. Measurements were converted to pmol [γ-32P]GTP hydrolyzed, with background subtracted out. This figure is representative of three separate experiments for each condition, with data presented as mean ± S.E.M.
To examine the effects of RGS14 on Gαi1 GTPase activity more carefully, we tested a range of full-length RGS14 concentrations on Gαi1 GTPase activity in the absence or presence of Ric-8A (Fig. 6C). We found that RGS14 inhibits Ric-8A-mediated increases in Gαi1 GTPase activity in a concentration-dependent manner, with complete inhibition evident at 3 μM RGS14 (Fig. 6C). This suggests that RGS14 competes with Ric-8A for Gαi1 binding, as greater concentrations of RGS14 hinder Ric-8A from acting on Gαi1.
RGS14 and Ric-8A bind to distinct and overlapping sites of Gαi1
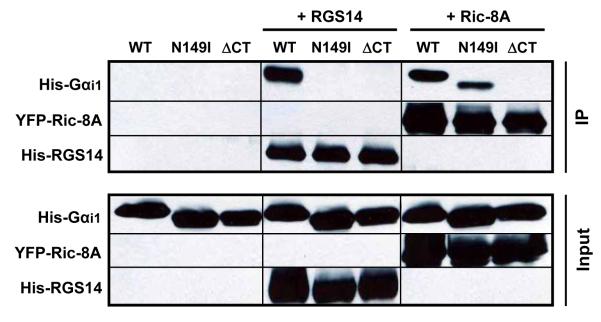
We next examined whether RGS14 and Ric-8A interact at the same or different sites of Gαi1. A recent report suggests that Ric-8A binds to the extreme C-terminus of Gαi1 since pertussis toxin disrupts Ric-8A interactions with Gαi1 (29). Based on this observation, we generated a truncation of Gαi1 (Gαi1-ΔCT) that is missing the last 11 amino acids of the protein. We also made a single point mutation in Gαi1 (N149I) that previously has been reported to block its binding to RGS14 (30, 31). GTPγS binding studies illustrate that these proteins are functional and active (0.59 pmol/min, 0.75 pmol/min, and 0.68 pmol/min for wild-type Gαi1, Gαi1 (N149I), and Gαi1-ΔCT, respectively). We examined the capacity of purified full-length TxHis6-RGS14 and YFP-Ric8A to form a stable complex with His6-Gαi1-ΔCT, His6-Gαi1 (N149I), and wild-type His6-Gαi1 derived from E. coli lysates as assessed by immunoprecipitation (Fig. 7). Both Ric-8A and RGS14 bind wild-type Gαi1, as expected (Fig. 7). Ric-8A interacts with Gαi1 (N149I) whereas RGS14 does not, indicating a distinct site of interaction for the two proteins on Gαi1 (Fig. 7). In contrast, Ric-8A fails to bind Gαi1-ΔCT, which is consistent with a recent report (29) showing that pertussis toxin-mediated ADP-ribosylation of a cysteine (C351) within this deleted region of Gαi1 blocks its functional interactions with Ric-8A. Surprisingly, RGS14 also fails to bind to this truncated form of Gαi1, suggesting an overlapping binding region that is shared by Ric-8A and RGS14 within the last 11 amino acids of Gαi1 (Fig. 7). These findings show that RGS14 and Ric-8A bind to both distinct sites and overlapping regions of Gαi1.
Figure 7. RGS14 and Ric-8A bind to distinct and overlapping regions of Gαi1.
RGS14 binds Gαi1 distinct from Ric-8A at residue N149, whereas both RGS14 and Ric-8A share an overlapping binding region at the extreme C-terminus of Gαi1. Wild-type His6-Gαi1 (WT), His6-Gαi1 (N149I) (N149I), and His6-Gαi1-ΔCT (ΔCT) proteins derived from E. coli were mixed alone or with either purified full-length TxHis6-RGS14 or purified YFP-Ric-8A. Protein mixtures were subjected to either anti-RGS14 or anti-Ric-8A immunoprecipitation, SDS-PAGE, and immunoblot. Results are indicative of three replicate experiments.
Ric-8A and RGS14 co-exist within the same hippocampal neurons
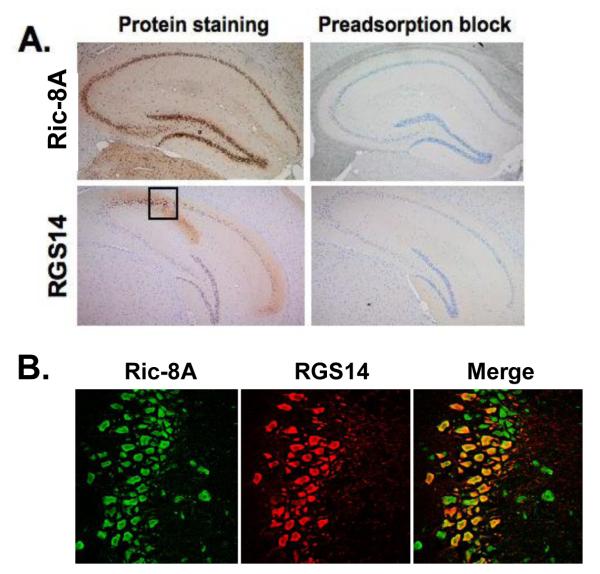
Thus far, our findings provide evidence that Ric-8A can functionally regulate the RGS14:Gαi1 complex. For this to be physiologically relevant, we would expect native RGS14 and Ric-8A to exist within the same cells. Since both RGS14 and Ric-8A are natively expressed in brain (6, 7, 32, 33), we studied the localization patterns of each of these proteins within brain using IHC staining techniques and confocal microscopy of fixed tissue (Fig. 8). Consistent with our recent observations (34), we find that RGS14 is present in hippocampus, but with a protein expression pattern that is largely restricted to neurons and neurites of the CA2 and CA1 sub-regions (Fig. 8A). We find that Ric-8A protein is also highly expressed in neurons of the CA2 and CA1 regions of the hippocampus (Fig. 8A). Staining of RGS14 and Ric-8A with anti-RGS14 and anti-Ric-8A antibodies is blocked by pre-adsorption of the antibodies with pure RGS14 and Ric-8A proteins, respectively (Fig. 8A, right panels). Most importantly, Ric-8A and RGS14 co-localize within the same CA2 hippocampal neurons as visualized by confocal imaging (corresponding to the area shown in the black box in Fig. 8A) (Fig. 8B). Ric-8A and RGS14 co-localize mainly to the cytosol of the soma of these neurons. These results further support the idea that Ric-8A and RGS14 are functionally linked within hippocampal neurons to regulate their functions.
Figure 8. RGS14 and Ric-8A co-exist and co-localize within the same hippocampal neurons.
RGS14 and Ric-8A co-localize within neurons of the hippocampus, specifically in the CA2 region of the hippocampus. (A), Mouse brain thin sections were subjected to immunohistochemistry and stained for RGS14 and Ric-8A. Control sections were incubated with antibody that was pre-adsorbed with RGS14 and Ric-8A pure protein (1:10 ratio of antibody to protein) (right panels). (B), Mouse brain thin sections were labeled with RGS14 and Ric-8A antibodies, followed by fluorescently-conjugated secondary IgG. Sections were analyzed by confocal microscopy as described in Experimental Procedures.
Discussion
RGS14 is a complex signaling protein that contains an RGS domain, tandem Ras/Rap binding domains, and a GL domain. Previous studies have focused largely on the presumed function of RGS14 as a regulator of GPCR-G protein signaling (6, 7, 10, 35, 36). However, findings here and elsewhere (8, 12, 13, 30) strongly suggest that RGS14 serves as a scaffold that integrates unconventional G protein signaling events rather than as a conventional RGS protein. In support of this idea, we show that RGS14 functionally interacts with Ric-8A, a defined regulator of unconventional G protein signaling pathways (22-24). Our key findings indicate the following: 1) RGS14 and Ric-8A co-localize at the plasma membrane with wild-type Gαi1; 2) RGS14 and Ric-8A interact with each other in cells; 3) Ric-8A stimulates dissociation of the RGS14:Gαi1-GDP complex in cells and in vitro; 4) Ric-8A serves as a GEF to facilitate nucleotide exchange (e.g. GTPγS binding) on the Gαi1 that it liberates from RGS14; 5) the capacity of Ric-8A to overcome the inhibitory effects of RGS14 on Gαi1 nucleotide exchange and GTPase activity depends on the molar ratio of RGS14 relative to Ric-8A; 6) RGS14 and Ric-8A bind to both distinct and overlapping regions of Gαi1; and 7) native RGS14 and Ric-8A co-exist within the same hippocampal neurons.
Our findings indicate that Ric-8A can functionally regulate the activation state of the RGS14:Gαi1-GDP signaling complex, which may potentially play a role in hippocampal signaling functions since RGS14 expression is highly restricted to this brain region. In this regard, RGS14 shows structural and mechanistic parallels with two other brain proteins, LGN (mPins) and AGS3. Like RGS14, these proteins contain GL domains that form stable complexes with Gαi1-GDP, and LGN has been shown to be recruited to the plasma membrane in cells to form an LGN:Gαi1-GDP complex (22, 24, 25). Similar to its effects on RGS14, Ric-8A also recognizes and induces dissociation of both the AGS3:Gαi1-GDP and LGN:Gαi1-GDP complexes, subsequently facilitating GTP binding to free Gαi1 (22, 24). As is the case with RGS14, excess amounts of both LGN and AGS3 have been shown to inhibit Ric-8A effects on Gαi1, suggesting competition between these GL proteins and Ric-8A for Gαi1 binding (22, 24). Taken together, our findings strongly suggest that RGS14 acts as a GL protein as well as an RGS protein.
RGS14 and Ric-8A co-localize with Gαi1-GDP at the plasma membrane in cells
Our cellular localization findings (Fig. 1) suggest that Ric-8A, RGS14, and Gαi1 may functionally interact at the plasma membrane in cells. Since both Ric-8A and RGS14 directly bind to inactive Gαi1 in cells (6, 11, 12, 23), we examined the subcellular localization of both Ric-8A and RGS14 in the presence of wild-type Gαi1. While a majority of Ric-8A is recruited to the plasma membrane in the presence of wild-type Gαi1, almost all Ric-8A is recruited to the plasma membrane when expressed with both wild-type Gαi1 and RGS14 (Fig. 1). The fact that Ric-8A and RGS14 co-localize at the same time with Gαi1 at the plasma membrane supports the possibility that these proteins functionally interact together through sequential formations/dissociations of RGS14:Gαi1 and Ric-8A:Gαi1 complexes, and perhaps through formation of a transient ternary RGS14:Gαi1-GDP:Ric-8A complex. Our data throughout support both the idea of the formation of RGS14:Gαi1 and Ric-8A:Gαi1 complexes and the concept that Gαi1 is exchanged between RGS14 and Ric-8A before dissociation as free Gαi1-GTP.
Ric-8A induces dissociation of the RGS14:Gαi1-GDP complex and subsequently facilitates nucleotide exchange on Gαi1
Mechanistically, our results show that Ric-8A interacts with the RGS14:Gαi1 complex to regulate its activation state. In the absence of nucleotide, Ric-8A forces Gαi1 dissociation from RGS14 to form a stable (and presumably nucleotide free (23)) Ric-8A:Gαi1 complex. In the presence of GTPγS, Ric-8A-induced dissociation of RGS14:Gαi1 allows Ric-8A to act as a GEF towards free Gαi1, which results in a rapid uncoupling of the Ric-8A:Gαi1 complex and formation of free Gαi1-GTPγS. Our findings are consistent with previous reports describing Ric-8A regulation of other GL:Gαi1-GDP complexes both in the presence and absence of exogenous GTP (22, 24). While these intermediate ternary biochemical complexes can be isolated under controlled experimental conditions, the lifetime of an RGS14:Gαi1-GDP:Ric-8A complex in cells is likely very transient (24). This is reflected by our failure to observe a stable heterotrimeric RGS14:Gαi1-GDP:Ric-8A complex in cells or as purified proteins; in both cases, Ric-8A seems to displace Gαi1 from RGS14 (Figs. 2 and 3). However, such a transition complex must exist since Gαi1 transfer occurs from RGS14 to Ric-8A (Fig. 3). We observed Ric-8A/RGS14 complex formation in cells (Fig. 2), but failed to observe this with purified proteins (Fig. 3, and data not shown). Reasons for the discrepancy between these two findings are unclear. We do not observe a stable Ric-8A/RGS14 complex when native RGS14 is co-immunoprecipitated from mouse brain (data not shown), though this does not definitively rule out such a complex. One possibility is that our observed cellular interactions are due to post-translation modifications (e.g. fatty acylation, phosphorylation) on either protein that promote a favorable conformation for binding. Alternatively, an intermediary protein may facilitate an interaction which may be independent of any Ric-8A effects on the RGS14:Gαi1-GDP complex (as is the case with Frmpd1 and AGS3 (37)). Recovered Ric-8A bound to RGS14 (Fig. 2) may also be the result of native Gαi1 bridging the two proteins together, however our dissociation data (Fig. 2B) does not support this idea. Such an intermediary protein bringing Ric-8A and RGS14 together may facilitate RGS14 to “switch” from regulating G protein signaling to regulating H-Ras/Raf-1-madiated MAP kinase signaling (8) (or other unknown signaling pathways). The role of Ric-8A in this context remains to be studied.
Ric-8A accelerates nucleotide exchange and GTPase activity of Gαi1 following RGS14:Gαi1-GDP dissociation
We observe that Ric-8A accelerates both GTPγS binding to and the steady-state GTPase activity of Gαi1 in the presence of RGS14, however these Ric-8A effects can be reversed by increasing concentrations of RGS14 (Figs. 4-6); this was the case for both full-length RGS14 and truncated RGS14 missing the RGS domain (ΔRGS14). Even with a dominant GDI function, Ric-8A is able to overcome ΔRGS14 inhibition of GTPγS binding to Gαi1, stimulating over a 20-fold increase in Gαi1 nucleotide binding when introduced to the ΔRGS14:Gαi1-GDP complex (Fig. 5A). A five-fold excess of ΔRGS14 to Ric-8A completely inhibits this Ric-8A-induced GTPγS binding, indicating that ΔRGS14 maintains Gαi1 in an inactive state. Full-length RGS14 appears to be as effective as ΔRGS14 at inhibiting Gαi1-directed steady-state GTP hydrolysis, both alone and in the presence of Ric-8A (Fig. 6). The presence of the RGS domain and its GAP activity might be expected to enhance GTP hydrolysis. However, it is likely that nucleotide exchange, and not GTP hydrolysis, is rate-limiting under the experimental conditions used. In this case, the GAP activity of the RGS domain would not be apparent in this in vitro assay, but is necessarily important in the context of cellular signaling.
Like we observe with the GTPγS binding assay, Ric-8A is able to overcome RGS14 inhibition of steady-state Gαi1 GTPase activity, catalyzing a 2.4-fold increase in Gαi1 steady-state GTPase activity when introduced to the RGS14:Gαi1-GDP complex (Fig. 6B). Again, increasing concentrations of RGS14 inhibit Ric-8A effects on Gαi1 GTP hydrolysis (Fig. 6C). Since the GEF activity of Ric-8A serves to enhance GDP release and increase the velocity of and/or eliminate the rate-limiting step in nucleotide exchange and hydrolysis, enhanced RGS14 binding to Gαi1-GDP would result in increased GDI activity reflected as an inhibition of GTPγS binding and steady-state GTPase activity that is more difficult for Ric-8A to overcome (as we observe). Therefore, RGS14 may bind Gαi1-GDP and hinder Ric-8A (by competitive or non-competitive inhibition) from binding and catalyzing Gαi1-directed GTP binding and hydrolysis.
Ric-8A and RGS14 bind Gαi1 at both distinct and overlapping sites
In studies designed to identify sites(s) of RGS14 and Ric-8A interactions on Gαi1 (Fig. 7), we found that RGS14 and Ric-8A compete for an overlapping binding site on the extreme C-terminus of Gαi1. Whereas residue N149 of Gαi1 has been shown to interact with the GL domain of RGS14 (30), identified binding sites on Gαi1 for Ric-8A were previously unknown. A recent study suggests that Ric-8A binds to the extreme C-terminus of Gαi1 since pertussis toxin-stimulated modification of C351 within this region inhibits Ric-8A activation of Gαi1 in cells (29). By comparing the binding properties of Gαi1 (N149I) (which does not bind RGS14 (31)) and Gαi1-ΔCT (missing the last 11 amino acids including C351), we determined that Ric-8A and RGS14 share distinct and overlapping binding regions on Gαi1 (Fig. 7). The presence of an overlapping binding region correlates with our other data (Figs. 5 and 6) that shows increasing concentrations of RGS14 block Ric-8A GEF activity towards Gαi1. Taken together, these findings are consistent with the idea that RGS14 and Ric-8A compete for the exact same or very proximal residues within the extreme C-terminal 11 amino acids of Gαi1. Since RGS14 binds N149 of Gαi1 and Ric-8A does not, it is also possible that RGS14 and Ric-8A are acting on distinct and overlapping regions of Gαi1 at the same time. RGS14 may interact with Gαi1 at residue N149 to carry out additional functions and/or to affect Ric-8A:Gαi1 interactions by allosteric modulation. These findings are the first to show any binding site for Ric-8A on Gαi1, and also the first to show a second binding region on Gαi1 for RGS14. Solved co-crystal structures of the RGS14:Gαi1 and the Ric-8A:Gαi1 complexes will be necessary to precisely define the binding interfaces between these proteins.
Working model for how Ric-8A regulates the RGS14:Gai1-GDP signaling complex
Since RGS14 was first identified as a Rap binding protein that contains an RGS domain (7, 9), much of the previous work on this protein has focused on its presumed role as an RGS protein that modulates GPCR/G protein signaling (6, 7, 10, 36). However, our findings here combined with findings elsewhere (8, 12, 13, 30) suggest that RGS14 may serve as a GL protein that integrates unconventional Ric-8A/G protein signaling with Ras/Raf/MAP kinase signaling (7, 8, 12). These findings provide a framework for a working model (Fig. S2) to describe how these proteins and the functionally opposed RGS and GL domains work together to bind and modulate the functions of Ric-8A, inactive Gαi-GDP, and active Gαi-GTP. Our proposed model highlights the GL domain as the first point of contact between Gαi and RGS14 rather than the RGS domain. In its basal resting state, RGS14 exists in a stable complex with Gαi1-GDP at the cell membrane. We postulate that following a signaling event (as yet undefined), Ric-8A recognizes the RGS14:Gαi1-GDP complex to stimulate nucleotide exchange and GTP binding to Gαi1, which then promotes dissociation of RGS14 (because the GL domain does not bind Gα-GTP). Of note, a role for a GPCR in this activation step cannot be ruled out. Once free from Gαi1, RGS14 would be available to act on other downstream binding partners (e.g. active H-Ras and Raf kinases to modulate MAP kinase signaling) (7, 8, 12). In this model, we envision that the lifetime of this newly-formed RGS14 signaling complex is limited by the RGS domain, which acts on nearby Gαi1-GTP to restore Gαi1-GDP and to promote reformation of the Gαi1-GDP:GL-RGS14 complex. This event is coupled with dissociation of RGS14 from its binding partners and a return to the basal resting state. An attractive feature of this model is that the structural configuration of RGS14 that incorporates both the RGS domain and GL domain into the same protein could serve to spatially restrict the function of the RGS domain towards the pre-bound Gα, thus eliminating the need for strict intrinsic RGS/Gα selectivity (i.e. even though the RGS domain is capable of acting on other Gα, it will only act on the one that is nearby). This idea is consistent with earlier observations that the RGS domain is a non-selective GAP for Gαi/o (6, 7, 10)), while the GL domain is specific for Gαi1 and Gαi3 (11-13). This proposed activation/deactivation cycle (Fig. S2) is entirely consistent with our findings here and with previous findings (8, 13, 22, 24), and future studies will examine untested steps in this model.
RGS14 and Ric-8A are brain proteins important for hippocampal functions
We find that native RGS14 and Ric-8A co-exist and co-localize within the same neurons of the CA2 and CA1 sub-regions of the hippocampus (Fig. 8). These findings highlight the likelihood for functional interplay between Ric-8A and RGS14 in hippocampal signaling pathways. Our findings here and those in previous reports (33, 38) indicate that Ric-8A is widely expressed in brain, including but not limited to those hippocampal neurons that contain RGS14. Thus, Ric-8A must also serve roles in addition to regulation of the RGS14:Gαi1-GDP signaling complex. In this regard, LGN/mPins, AGS3, and other proteins that contain GL domains are also highly enriched in various brain regions (39-41). Furthermore, we observe via size-exclusion chromatography that most of the Ric-8A in soluble brain lysates exists as an uncomplexed monomer (data not shown). Therefore, it is possible that Ric-8A acts as a master regulator of multiple GL:Gαi-GDP signaling complexes involved with brain signaling. Consistent with this idea, both LGN/mPins and AGS3 have each been reported to serve important roles in synaptic plasticity in brain (15, 17, 39, 42). Genetic deletion of Ric-8A is reported to alter hippocampal learning behavior (32). Of particular relevance to these reports and our findings here, we observe that RGS14 is expressed almost exclusively in CA2 neurons of mouse hippocampus and that genetic deletion of RGS14 in mouse brain results in animals with a targeted enhancement of hippocampal-based learning and memory and synaptic plasticity in CA2 neurons (34). These studies, combined with our results here and other reports showing that the RGS14 binding partners H-Ras, Rap2 and Raf-1 are also important for hippocampal learning and memory (43-49) strongly suggest that RGS14 is a newly appreciated multifunctional GL and RGS protein that integrates unconventional Ric-8A/Gαi and MAP kinase signaling pathways important for hippocampal cognitive processing.
Supplementary Material
Acknowledgment
We would like to thank the Neuropathology and Histochemistry Core of the Emory University Neuroscience NINDS Core Facilities. This core helped isolate the mouse brains, slice them into thin sections, and embed the sections in paraffin. This work was supported in part by the Neuropathology and Histochemistry Core of the Emory Neuroscience NINDS Core Facilities grant (P30NS055077).
This work was supported by National Institutes of Health grants (R01NS049195 and R01NS037112 to J.R.H., GM088242 to G.T.T., and Pharmacological Sciences Training Grant T32 GM008602 to C.P.V.) and a PhRMA Foundation Predoctoral Pharmacology/Toxicology Fellowship to C.P.V.
Abbreviations
- RGS
Regulator of G protein Signaling
- MAP
Mitogen-activated protein
- GAP
GTPase Activating Protein
- GEF
Guanine nucleotide exchange factor
- Ric-8A
Resistance to inhibitors of cholinesterase-8A
- GPCR
G Protein – Coupled Receptor
- RBD
Ras/Raf binding domain
- GL
GoLoco
- AGS
Activator of G protein Signaling
- HRP
Horseradish peroxidase
- GFP
Green Fluorescent Protein
- CFP
Cyan Fluorescent Protein
- YFP
Yellow Fluorescent Protein
- IgG
Immunoglobulin G
- EGTA
Ethylene glycol tetraacetic acid
- EDTA
Ethylenediaminetetraacetic acid
- Tx
Thioredoxin
- PIPES
piperazine-N,N’-bis(2-ethanesulfonic acid)
- GTPγS
Guanosine 5′- O - thiotriphosphate
- CA1/CA2
Cornu Ammonis 1/Cornu Ammonis 2
- GDI
Guanine nucleotide dissociation inhibitor
- His6
Hexahistidine
Footnotes
Supporting Information Available The material shown in Figure S1 shows that a pure protein RGS14:Gαi1-GDP complex can be consistently isolated using size-exclusion gel filtration. The complex is made by mixing purified RGS14 (missing the RGS domain) and purified Gαi1-GDP at 4°C and then applying the protein mixture to a tandem S200+S75 Superdex size-exclusion gel filtration apparatus. This complex was used in the gel filtration work presented in Figure 3.
The material shown in Figure S2 illustrates our working model involving Ric-8A, RGS14, and Gαi1-GDP. Ric-8A recognizes the RGS14:Gαi1-GDP complex, and subsequently induces its dissociation and promotes nucleotide exchange on Gαi1. RGS14 is now free to potentially interact with other binding partners. Due to the close proximity of activated Gαi1, the RGS domain of RGS14 induces GTP hydrolysis on the Gα subunit. The RGS14 GL domain then rebinds Gαi1-GDP to complete one round of this cycle. All this material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Gilman AG. G proteins: transducers of receptor-generated signals. Annu. Rev. Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 2.Hamm HE. The many faces of G protein signaling. J. Biol. Chem. 1998;273:669–672. doi: 10.1074/jbc.273.2.669. [DOI] [PubMed] [Google Scholar]
- 3.De Vries L, Zheng B, Fischer T, Elenko E, Farquhar MG. The regulator of G protein signaling family. Annu. Rev. Pharmacol. Toxicol. 2000;40:235–271. doi: 10.1146/annurev.pharmtox.40.1.235. [DOI] [PubMed] [Google Scholar]
- 4.Hollinger S, Hepler JR. Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol. Rev. 2002;54:527–559. doi: 10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- 5.Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu. Rev. Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- 6.Hollinger S, Taylor JB, Goldman EH, Hepler JR. RGS14 is a bifunctional regulator of Galphai/o activity that exists in multiple populations in brain. J. Neurochem. 2001;79:941–949. doi: 10.1046/j.1471-4159.2001.00629.x. [DOI] [PubMed] [Google Scholar]
- 7.Traver S, Bidot C, Spassky N, Baltauss T, De Tand MF, Thomas JL, Zalc B, Janoueix-Lerosey I, Gunzburg JD. RGS14 is a novel Rap effector that preferentially regulates the GTPase activity of Gαo. Biochem. J. 2000;350:19–29. [PMC free article] [PubMed] [Google Scholar]
- 8.Shu F-j., Ramineni S, Hepler JR. RGS14 is a multifunctional scaffold that integrates G protein and Ras/Raf MAPkinase signalling pathways. Cell. Signal. 2010;22:366–376. doi: 10.1016/j.cellsig.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snow BE, Antonio L, Suggs S, Gutstein HB, Siderovski DP. Molecular cloning and expression analysis of rat Rgs12 and Rgs14. Biochem. Biophys. Res. Commun. 1997;233:770–777. doi: 10.1006/bbrc.1997.6537. [DOI] [PubMed] [Google Scholar]
- 10.Cho H, Kozasa T, Takekoshi K, De Gunzburg J, Kehrl JH. RGS14, a GTPase-activating protein for Giα, attenuates Giα- and G13α-mediated signaling pathways. Mol. Pharmacol. 2000;58:569–576. doi: 10.1124/mol.58.3.569. [DOI] [PubMed] [Google Scholar]
- 11.Kimple RJ, De Vries L, Tronchère H, Behe CI, Morris RA, Farquhar MG, Siderovski DP. RGS12 and RGS14 GoLoco motifs are Gαi interaction sites with guanine nucleotide dissociation inhibitor activity. J. Biol. Chem. 2001;276:29275–29281. doi: 10.1074/jbc.M103208200. [DOI] [PubMed] [Google Scholar]
- 12.Mittal V, Linder ME. The RGS14 GoLoco domain discriminates among Gαi isoforms. J. Biol. Chem. 2004;279:46772–46778. doi: 10.1074/jbc.M407409200. [DOI] [PubMed] [Google Scholar]
- 13.Shu F.-j., Ramineni S, Amyot W, Hepler JR. Selective interactions between Giα1 and Giα3 and the GoLoco/GPR domain of RGS14 influence its dynamic subcellular localization. Cell. Signal. 2007;19:163–176. doi: 10.1016/j.cellsig.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Colombo K, Grill SW, Kimple RJ, Willard FS, Siderovski DP, Gonczy P. Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science. 2003;300:1957–1961. doi: 10.1126/science.1084146. [DOI] [PubMed] [Google Scholar]
- 15.Groves B, Gong Q, Xu Z, Huntsman C, Nguyen C, Li D, Ma D. A specific role of AGS3 in the surface expression of plasma membrane proteins. Proc. Natl. Acad. Sci. USA. 2007;104:18103–18108. doi: 10.1073/pnas.0709282104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hampoelz B, Knoblich JA. Heterotrimeric G proteins: new tricks for an old dog. Cell. 2004;119:453–456. doi: 10.1016/j.cell.2004.10.025. [DOI] [PubMed] [Google Scholar]
- 17.Sans N, Wang PY, Du Q, Petralia RS, Wang Y-X, Nakka S, Blumer JB, Macara IG, Wenthold RJ. mPins modulates PSD-95 and SAP102 trafficking and influences NMDA receptor surface expression. Nat. Cell Biol. 2005;7:1179–1190. doi: 10.1038/ncb1325. [DOI] [PubMed] [Google Scholar]
- 18.Sato M, Blumer JB, Simon V, Lanier SM. Accessory proteins for G proteins: partners in signaling. Annu. Rev. Pharmacol. Toxicol. 2006;46:151–187. doi: 10.1146/annurev.pharmtox.46.120604.141115. [DOI] [PubMed] [Google Scholar]
- 19.Schade MA, Reynolds NK, Dollins CM, Miller KG. Mutations that rescue the paralysis of C. elegans ric-8 (synembryn) mutants activate the Gαs pathway and define a third major branch of the synaptic signaling network. Genetics. 2004;169:631–649. doi: 10.1534/genetics.104.032334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willard FS, Kimple RJ, Siderovski DP. Return of the GDI: the GoLoco motif in cell division. Annu. Rev. Biochem. 2004;73:925–951. doi: 10.1146/annurev.biochem.73.011303.073756. [DOI] [PubMed] [Google Scholar]
- 21.Hess HA, Röper J-C, Grill SW, Koelle MR. RGS-7 completes a receptor-independent heterotrimeric G protein cycle to asymmetrically regulate mitotic spindle positioning in C. elegans. Cell. 2004;119:209–218. doi: 10.1016/j.cell.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 22.Tall GG, Gilman AG. Resistance to inhibitors of cholinesterase 8A catalyzes release of Gαi-GTP and nuclear mitotic apparatus protein (NuMA) from NuMA/LGN/Gαi-GDP complexes. Proc. Natl. Acad. Sci. USA. 2005;102:16584–16589. doi: 10.1073/pnas.0508306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tall GG, Krumins AM, Gilman AG. Mammalian Ric-8A (synembryn) is a heterotrimeric Gα protein guanine nucleotide exchange factor. J. Biol. Chem. 2003;278:8356–8362. doi: 10.1074/jbc.M211862200. [DOI] [PubMed] [Google Scholar]
- 24.Thomas CJ, Tall GG, Adhikari A, Sprang SR. Ric-8A catalyzes guanine nucleotide exchange on Gαi1 bound to the GPR/GoLoco exchange inhibitor AGS3. J. Biol. Chem. 2008;283:23150–23160. doi: 10.1074/jbc.M802422200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du Q, Macara IG. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 2004;119:503–516. doi: 10.1016/j.cell.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 26.Tall GG, Gilman AG. Purification and functional analysis of Ric-8A: a guanine nucleotide exchange factor for G-protein α subunits. Methods Enzymol. 2004;390:377–388. doi: 10.1016/S0076-6879(04)90023-7. [DOI] [PubMed] [Google Scholar]
- 27.Krumins AM, Gilman AG. Assay of RGS protein activity in vitro using purified components. Methods Enzymol. 2002;344:673–685. doi: 10.1016/s0076-6879(02)44748-9. [DOI] [PubMed] [Google Scholar]
- 28.Siderovski DP, Willard FS. The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int. J. Biol. Sci. 2005;1:51–66. doi: 10.7150/ijbs.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodard GE, Huang N-N, Cho H, Miki T, Tall GG, Kehrl JH. Ric-8A and Giα recruit LGN, NuMA, and Dynein to the cell cortex to help orient the mitotic spindle. Mol. Cell. Biol. 30:3519–3530. doi: 10.1128/MCB.00394-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimple RJ, Kimple ME, Betts L, Sondek J, Siderovski DP. Structural determinants for GoLoco-induced inhibition of nucleotide release by Gα subunits. Nature. 2002;416:878–881. doi: 10.1038/416878a. [DOI] [PubMed] [Google Scholar]
- 31.Willard FS, Zheng Z, Guo J, Digby GJ, Kimple AJ, Conley JM, Johnston CA, Bosch D, Willard MD, Watts VJ, Lambert NA, Ikeda SR, Du Q, Siderovski DP. A point mutation to Gαi selectively blocks GoLoco motif binding. J. Biol. Chem. 2008;283:36698–36710. doi: 10.1074/jbc.M804936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tõnissoo T, Kõks S, Meier R, Raud S, Plaas M, Vasar E, Karis A. Heterozygous mice with Ric-8 mutation exhibit impaired spatial memory and decreased anxiety. Behav. Brain Res. 2006;167:42–48. doi: 10.1016/j.bbr.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 33.Tõnissoo T, Meier R, Talts K, Plaas M, Karis A. Expression of ric-8 (synembryn) gene in the nervous system of developing and adult mouse. Gene Expr. Patterns. 2003;3:591–594. doi: 10.1016/s1567-133x(03)00119-4. [DOI] [PubMed] [Google Scholar]
- 34.Lee SE, Simons SB, Heldt SA, Zhao M, Schroeder JP, Vellano CP, Cowan DP, Ramineni S, Yates CK, Feng Y, Smith Y, Sweatt JD, Weinshenker D, Ressler KJ, Dudek SM, Hepler JR. RGS14 is a natural suppressor of both synaptic plasticity in CA2 neurons and hippocampal-based learning and memory. Proc. Natl. Acad. Sci. USA. 2010;107:16994–16998. doi: 10.1073/pnas.1005362107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hepler JR, Cladman W, Ramineni S, Hollinger S, Chidiac P. Novel activity of RGS14 on Goα and Giα nucleotide binding and hydrolysis distinct from its RGS domain and GDI activity. Biochemistry. 2005;44:5495–5502. doi: 10.1021/bi048359d. [DOI] [PubMed] [Google Scholar]
- 36.Rodríguez-Muñoz M, de la Torre-Madrid E, Gaitán G, Sánchez-Blázquez P, Garzón J. RGS14 prevents morphine from internalizing Mu-opioid receptors in periaqueductal gray neurons. Cell. Signal. 2007;19:2558–2571. doi: 10.1016/j.cellsig.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 37.An N, Blumer JB, Bernard ML, Lanier SM. The PDZ and band 4.1 containing protein Frmpd1 regulates the subcellular location of Activator of G-protein Signaling 3 and its interaction with G-proteins. J. Biol. Chem. 2008;283:24718–24728. doi: 10.1074/jbc.M803497200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S-C, Lai H-L, Chiu Y-T, Ou R, Huang C-L, Chern Y. Regulation of type V adenylate cyclase by Ric8a, a guanine nucleotide exchange factor. Biochem. J. 2007;406:383–388. doi: 10.1042/BJ20070512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blumer JB, Chandler LJ, Lanier SM. Expression analysis and subcellular distribution of the two G-protein regulators AGS3 and LGN indicate distinct functionality. J. Biol. Chem. 2002;277:15897–15903. doi: 10.1074/jbc.M112185200. [DOI] [PubMed] [Google Scholar]
- 40.Pizzinat N, Takesono A, Lanier SM. Identification of a truncated form of the G-protein regulator AGS3 in heart that lacks the tetratricopeptide repeat domains. J. Biol. Chem. 2001;276:16601–16610. doi: 10.1074/jbc.M007573200. [DOI] [PubMed] [Google Scholar]
- 41.Takesono A, Cismowski MJ, Ribas C, Bernard M, Chung P, Hazard S, Duzic E, Lanier SM. Receptor-independent activators of heterotrimeric G-protein signaling pathways. J. Biol. Chem. 1999;274:33202–33205. doi: 10.1074/jbc.274.47.33202. [DOI] [PubMed] [Google Scholar]
- 42.Wiser O, Qian X, Ehlers M, Ja WW, Roberts RW, Reuveny E, Jan YN, Jan LY. Modulation of basal and receptor-induced GIRK potassium channel activity and neuronal excitability by the mammalian PINS homolog LGN. Neuron. 2006;50:561–573. doi: 10.1016/j.neuron.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 43.Costa RM, Federov NB, Kogan JH, Murphy GG, Stern J, Ohno M, Kucherlapati R, Jacks T, Silva AJ. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature. 2002;415:526–530. doi: 10.1038/nature711. [DOI] [PubMed] [Google Scholar]
- 44.Fu Z, Lee SH, Simonetta A, Hansen J, Sheng M, Pak DTS. Differential roles of Rap1 and Rap2 small GTPases in neurite retraction and synapse elimination in hippocampal spiny neurons. J. Neurochem. 2007;100:118–131. doi: 10.1111/j.1471-4159.2006.04195.x. [DOI] [PubMed] [Google Scholar]
- 45.Kushner SA, Elgersma Y, Murphy GG, Jaarsma D, van Woerden GM, Hojjati MR, Cui Y, LeBoutillier JC, Marrone DF, Choi ES, De Zeeuw CI, Petit TL, Pozzo-Miller L, Silva AJ. Modulation of presynaptic plasticity and learning by the H-ras/extracellular signal-regulated kinase/synapsin I signaling pathway. J. Neurosci. 2005;25:9721–9734. doi: 10.1523/JNEUROSCI.2836-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manabe T, Aiba A, Yamada A, Ichise T, Sakagami H, Kondo H, Katsuki M. Regulation of long-term potentiation by H-Ras through NMDA receptor phosphorylation. J. Neurosci. 2000;20:2504–2511. doi: 10.1523/JNEUROSCI.20-07-02504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryu J, Futai K, Feliu M, Weinberg R, Sheng M. Constitutively active Rap2 transgenic mice display fewer dendritic spines, reduced extracellular signal-regulated kinase signaling, enhanced long-term depression, and impaired spatial learning and fear extinction. J. Neurosci. 2008;28:8178–8188. doi: 10.1523/JNEUROSCI.1944-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sweatt JD. Mitogen-activated protein kinases in synaptic plasticity and memory. Curr. Opin. Neurobiol. 2004;14:311–317. doi: 10.1016/j.conb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 49.Wu G-Y, Deisseroth K, Tsien RW. Spaced stimuli stabilize MAPK pathway activation and its effects on dendritic morphology. Nat. Neurosci. 2001;4:151–158. doi: 10.1038/83976. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.