Abstract
Gene expression is controlled mainly by the binding of transcription factors to regulatory sequences. To generate a genomic map for regulatory sequences, the Arabidopsis thaliana genome was screened for putative transcription factor binding sites. Using publicly available data from the TRANSFAC database and from publications, alignment matrices for 23 transcription factors of 13 different factor families were used with the pattern search program Patser to determine the genomic positions of more than 2.4 × 106 putative binding sites. Due to the dense clustering of genes and the observation that regulatory sequences are not restricted to upstream regions, the prediction of binding sites was performed for the whole genome. The genomic positions and the underlying data were imported into the newly developed AthaMap database. This data can be accessed by positional information or the Arabidopsis Genome Initiative identification number. Putative binding sites are displayed in the defined region. Data on the matrices used and on the thresholds applied in these screens are given in the database. Considering the high density of sites it will be a valuable resource for generating models on gene expression regulation. The data are available at http://www.athamap.de.
INTRODUCTION
The identification of gene regulatory elements is still a major challenge for molecular biologists. Experimental methods can be complemented with bioinformatic approaches to identify transcription factors (TFs) or factor families responsible for gene expression regulation (1,2). Once a regulatory region is delineated experimentally, a bioinformatic approach may involve the use of pattern recognition programs such as MatInspector, Match or Patser to identify functional or putative TF binding sites in this region (3–5). The bases for these pattern recognition programs are alignment matrices that can be derived from random binding site selection experiments. Such experiments often determine a large array of different DNA sequences that can be bound by the factor. These data are used by pattern search programs to generate positional weight matrices that also predict novel binding sites solely on the basis of nucleotide frequencies at single matrix positions.
The recent completion of the Arabidopsis thaliana genome sequence offered a good opportunity to determine putative TF binding sites in the whole genome (6). Although most of the regulatory sequences in genes occur upstream of the transcription and translation start site, many exceptions are known (7–9). Furthermore, Arabidopsis has a high density of genes and the average length of the intergenic region is only ∼2–2.5 kb (6). Therefore, the determination of factor binding sites should not be restricted to upstream regions alone.
Resources for binding site identification used here are mostly alignment matrices derived for individual TFs and annotated in the TRANSFAC database. During the past few years the TRANSFAC database has been significantly enhanced with plant-specific data. For example, the number of plant transcription factors in the database has risen from 266–489 between the years 2000 and 2001 to currently 644 in the TRANSFAC 6.0 public database (10–12). A similar increase was achieved with the annotation of alignment matrices. This shows that a critical amount of data is available for the prediction of genomic positions of TF binding sites.
Here, we have employed alignment matrices from the TRANSFAC database and from further publications for the prediction of TF binding sites in the most recent version of the Arabidopsis genome sequence by using the matrix screening program Patser (4,12,13). The results of these screens were integrated into a genome-wide binding site map and are now available at http://www.athamap.de. This report describes the content and use of the AthaMap database resource. Tools have been developed for easy display of binding sites and for the display of the underlying data. The database will be complemented in the future with newly published binding sites and with combinatorial elements of interacting factors.
THE ATHAMAP RESOURCE
Development and content of the database
The AthaMap database structure was developed for storing positional information on putative TF binding sites, underlying data for binding site prediction, alignment matrices and additional data on TFs. The database was designed with a high degree of flexibility to facilitate future upgrades and was implemented on an MS-SQL-Server. Genomic screenings were performed using Patser (4). Software tools were programmed to import putative TF binding sites predicted by Patser into the database. This toolbox was also employed to analyse the redundancy of matches in the database (see below). Selection of TF matrices was performed in order to ensure minimal redundancy. An interactive web server interface was designed for public accessibility of the database content.
Alignment matrices from 23 TFs corresponding to 13 different TF families were used for the genomic screen for TF binding sites. Many of the factors employed for the genomic screen originate from A.thaliana. However, genomic screens were also performed with alignment matrices for factors from other plant species. The rationale behind this is the observation that binding site recognition is generally not species specific. For example, bZIP or MYB factors from different plant species recognize similar target sequences with a high conservation of a core sequence (14–16). Because Arabidopsis is frequently used to dissect the function of heterologous TFs, information on binding site locations in Arabidopsis may also be valuable for heterologous TFs (17). Furthermore, all binding sites that are displayed in the desired genomic region can be easily associated with a homologous or heterologous TF (see below).
The pattern search program Patser was employed for the identification of binding sites (4). Patser is available as a UNIX/Linux stand-alone program on the author’s web site (4) and online as part of the Regulatory Sequence Analysis Tools (18). Here, a locally installed version of Patser was used. The following command line was used to run Patser: patser-v3d -A a:t 0.325 c:g 0.175 -m matrixfile -f sequencefile -c -li -d2. Mostly the default threshold derived from the adjusted information content of the matrix was employed. In seven of 23 cases, the matrix information content was insufficient to yield specific matrix matches using the default threshold. In these cases, indicated in Table 1, a higher threshold was applied. Table 1 summarizes the number of genomic matches identified for 23 alignment matrices with Patser. Column 1 shows the designation of the factor for which an alignment matrix was available; column 2 gives the TF factor family; column 3 displays the total number of genomic positions identified and column 4 shows the TRANSFAC accession number and the reference.
Table 1. Number of putative binding sites for transcription factors in the A.thaliana genome.
| Factor | Family | Number of matches | Referencea |
|---|---|---|---|
| GAMYB | MYB | 319996 | M00345 (24) |
| CDC5 | MYB | 11449 | M00361 (25) |
| MYB.PH3[1] | MYB | 8554 | M00218 (26) |
| MYB.PH3[2] | MYB | 7581 | M00219 (26) |
| P | MYB | 186583 | M00226 (15) |
| NTMYBAS | MYB | 183549c | (27) |
| ATHB5 | HD-ZIP | 122403 | (28) |
| ATHB9 | HD-ZIP | 310 | M00417 (29) |
| AG | MADS | 46353 | M00151 (30) |
| PCF2 | TCP | 36497 | (31) |
| PCF5 | TCP | 13706 | (31) |
| ZAP1 | WRKY | 3955 | (32) |
| ANT | AP2 | 287 | (33) |
| RAV1 | AP2 | 227174 | M00344 (34) |
| TEIL | AP2 | 172861c | (35) |
| O2 | bZIP | 93011 | M00010 (36) |
| TGA1 | bZIP | 53402 | (14) |
| DOF2 | DOF | 183140c | M00353 (37) |
| LIM1 | LIM | 185915c | (38) |
| SBF1b | GTb | 195387c | M00149 (39) |
| ALFIN1 | HD-PHD | 185516c | (40) |
| HVH21 | HD-KNOTTED | 189736c | (41) |
| TBP | TBP | 30878 | (19) |
aThe matrix accession number of the TRANSFAC database (12) and the literature reference is given.
bThe factor has not been cloned. The family was predicted by binding site similarity.
cOnly the ∼180 000 highest scoring matches have been annotated into AthaMap.
The identification of TATA box binding protein (TBP) binding sites with the available matrix was a particular challenge (19). More than 200 000 putative TBP binding sites were detected by using the default threshold with Patser. Upon closer inspection it was clear that AT-rich regions are ‘hot spots’ of putative TBP binding sites. Therefore, we restricted the putative TBP binding sites in AthaMap to those sites that were detected in a region up to 400 bp upstream of the translation start point. Because the 30 878 putative TBP binding sites in AthaMap exceeds the total number of genes, some sites may still cluster in AT-rich regions.
In summary, more than 2.4 × 106 putative TF binding sites were predicted in the Arabidopsis genome.
AthaMap contains a low level of binding site redundancy
In several cases alignment matrices from different TFs of the same TF family were used in the screens. Therefore, it is interesting to estimate the level of redundancy within the genomic positions. This is of particular importance for members of the MYB factor family represented by six different matrices (Table 1). To determine the level of redundancy, the genomic positions detected by all MYB matrices were investigated for colocalization in the genome using a software tool developed in the lab (L. Bülow and R. Hehl, unpublished). This tool detects identical matches for two different matrices. MYB.PH3 recognizes two different sets of binding sites (MYB.PH3[1] and MYB.PH3[2], Table 1) that were both annotated to the TRANSFAC database (12,20). Based on the matrix consensus sequence it was expected that CDC5 would not detect the same binding sites as GAMYB, MYB.PH3[1], MYB.PH3[2] and P (data not shown). GAMYB, MYB.PH3[1], MYB.PH3[2] and P recognize a consensus sequence with a characteristic AAC trinucleotide sequence, which is not conserved in the matrix for CDC5. In accordance with this expectation, mostly GAMYB, MYB.PH3 and P show a certain level of redundancy in binding site identification. Table 2 shows that 34 863 of the genomic matches detected with the GAMYB (319 996) and NTMYBAS (183 549) matrices are identical. Similarly, of the 8554 matches detected with MYB.PH3[1], 2166, 2929 and 215 are also identified with GAMYB, NTMYBAS and P, respectively. CDC5 does not detect the same positions as were identified with GAMYB, MYB.PH3[1], MYB.PH3[2] and P (Table 2). From these values we deduce a high number of unique positions and estimate that the level of redundancy in AthaMap is relatively low. Only those matrices that harbour a certain degree of sequence similarity match identical positions in the genome. Furthermore, it is advantageous to represent TF binding sites for different matrices within the same TF family because the specificity of DNA binding is then determined mainly by the nucleotides flanking the core region.
Table 2. Number of putative binding sites in the AthaMap database detected by the same MYB transcription factor matrix.
| GAMYB | CDC5 | MYB.PH3[1] | MYB.PH3[2] | NTMYBAS | P | |
|---|---|---|---|---|---|---|
| GAMYB | 319996 | 0 | 2166 | 2580 | 34863 | 51616 |
| CDC5 | 11449 | 0 | 0 | 159 | 0 | |
| MYB.PH3[1] | 8554 | 17 | 2929 | 215 | ||
| MYB.PH3[2] | 7581 | 219 | 461 | |||
| NTMYBAS | 183549 | 28911 | ||||
| P | 186583 |
Accessing AthaMap
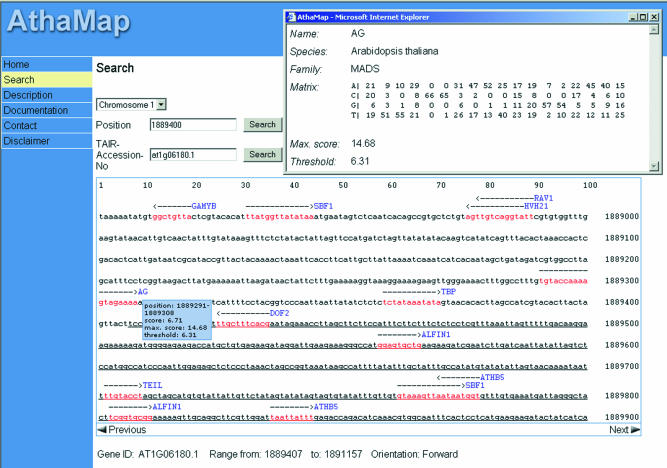
AthaMap is accessible at http://www.athamap.de. AthaMap provides two different search modes for the user. The chromosomal regions of interest can be retrieved either by submitting the Arabidopsis Genome Initiative identification or TAIR accession number (13) or by entering the genomic position. AthaMap displays the DNA sequence 500 bp upstream and downstream of the putative gene start or submitted position, respectively, with all identified TF binding sites highlighted in red. The arrow above the sequence adjacent to the factor’s name indicates the length of the putative binding site and its orientation. To facilitate navigation within the sequence, buttons were placed in the lower corners of the sequence display window which can be used to scroll 500 bp in either direction. Transcribed and translated gene regions are underlined and the respective genes are identified below the sequence window with gene name and gene features like start, stop and orientation (Fig. 1). The designations were taken from TAIR annotation tables (13).
Figure 1.
A screenshot of an AthaMap database search result. The region between nucleotides 1 888 900 and 1 889 900 on chromosome 1 of A.thaliana is displayed. Putative TF binding sites in the sequence are indicated in red. Information on the AG binding site is given in the small boxes. All features are described in the text.
The AthaMap documentation contains a complete list of the matrices used for the genomic screenings, the number of matches detected with Patser, the score thresholds for each matrix used by Patser, the maximum score and the sequences from which the matrix was derived, including the corresponding publication. Furthermore, the documentation of the database provides the TRANSFAC accession number of the matrix. This will enable the user of AthaMap to find more recent information about the respective factor in the regularly updated TRANSFAC database (12). All of the matrices used will eventually be annotated in TRANSFAC.
Figure 1 shows a screen shot of AthaMap displaying the upstream and part of the transcribed region (underlined) of locus At1g06180.1 on chromosome 1. The information provided for the putative MADS box binding site identified with the Arabidopsis AG matrix (Table 1) is shown. The name of the factor, factor family, species, matrix, maximum score of the matrix and the threshold used by Patser is displayed in a pop-up window by clicking on the factor’s name. The tool tip box appears by moving the mouse over the arrow and identifies the position of the site, the score of this particular site and the maximum score and threshold used by Patser. The score of the match can be compared with the maximum score displayed in the same window. A high score close to the maximum score represents a high-quality binding site. A low score close to the threshold represents a low-quality binding site. These values enable the user to judge the quality of a match or putative binding site by comparing the particular score of a match with the threshold and maximum score determined by Patser.
Other resources for identifying cis-regulatory sequences in A.thaliana
An online resource designated Regulatory Sequence Analysis (RSA) tools is available for the detection of TF binding sites in the Arabidopsis and in other genomes (18). For detection of TF binding sites or cis regulatory sequences in plant genes, the databases Place, PlantCare and TRANSFAC can be employed (12,21,22). These three databases require an input sequence in which putative binding sites should be detected while the genomic detection of binding sites with the RSA tools require that a matrix or a binding site consensus sequence is provided.
AthaMap is one of only two databases that display putative binding sites directly in the genome of A.thaliana and the only database that displays TF binding sites in the whole genomic sequence including coding and complete intergenic regions. The other Arabidopsis genome database on cis regulatory sequences, AtcisDB (A.thaliana cis-regulatory database), contains the 5′ regulatory sequences of 29 388 annotated Arabidopsis genes (23). The main differences between AtcisDB and AthaMap are the restriction to 5′ sequences (AtcisDB) versus the complete genome (AthaMap) and the accessibility of the underlying data. AtcisDB contains only known cis-acting elements while AthaMap also identifies novel putative cis-acting TF binding sites. This demonstrates that AthaMap and AtcisDB are complementary. Furthermore, AthaMap provides a high level of transparency, which means that the process of binding site detection in AthaMap can be reproduced by the user. All parameters for binding site detection are identified and each site is associated with a TF.
AVAILABILITY
The AthaMap resources are freely available for non- commercial users at http://www.athamap.de. The database will be updated on a regular basis. All updates and changes will be announced on the AthaMap home page.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank the members of the Intergenomics network at the Technical University, Braunschweig, for many helpful discussions. This work was supported by the German Ministry of Education and Research (BMBF grant no. 031U110C/031U210C) and was carried out as part of the Intergenomics network, Braunschweig. Further support was provided by the Forschungsschwerpunkt Agrarbiotechnologie des Landes Niedersachsen (VW-Vorab).
REFERENCES
- 1.Hehl R., Steffens,N.O. and Wingender,E. (2004) Isolation and analysis of gene regulatory sequences. In Klee,H. and Christou,P. (eds), Handbook of Plant Biotechnology. Wiley and Sons Ltd, in press. [Google Scholar]
- 2.Hehl R. and Wingender,E. (2001) Database-assisted promoter analysis. Trends Plant Sci., 6, 251–255. [DOI] [PubMed] [Google Scholar]
- 3.Quandt K., Frech,K., Karas,H., Wingender,E. and Werner,T. (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res., 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hertz G.Z. and Stormo,G.D. (1999) Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics, 15, 563–577. [DOI] [PubMed] [Google Scholar]
- 5.Kel A.E., Gößling,E., Reuter,I., Cheremushkin,E., Kel-Margoulis,O.V. and Wingender,E. (2003) MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res., 31, 3576–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Arabidopsis Genome Initiative. (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 7.Dean C., Favreau,M., Bond-Nutter,D., Bedbrook,J. and Dunsmuir,P. (1989) Sequences downstream of translation start regulate quantitative expression of two petunia rbcS genes. Plant Cell, 1, 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sieburth L.E. and Meyerowitz,E.M. (1997) Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell, 9, 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong R.L., Hamaguchi,L., Busch,M.A. and Weigel,D. (2003) Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell, 15, 1296–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wingender E., Chen,X., Hehl,R., Karas,H., Liebich,I., Matys,V., Meinhardt,T., Prüß,M., Reuter,I. and Schacherer,F. (2000) TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res., 28, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wingender E., Chen,X., Fricke,E., Geffers,R., Hehl,R., Liebich,I., Krull,M., Matys,V., Michael,H., Ohnhäuser,R. et al. (2001) The TRANSFAC system on gene expression regulation. Nucleic Acids Res., 29, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matys V., Fricke,E., Geffers,R., Gossling,E., Haubrock,M., Hehl,R., Hornischer,K., Karas,D., Kel,A.E., Kel-Margoulis,O.V. et al. (2003) TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res., 31, 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhee S.Y., Beavis,W., Berardini,T.Z., Chen,G., Dixon,D., Doyle,A., Garcia-Hernandez,M., Huala,E., Lander,G., Montoya,M. et al. (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res., 31, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schindler U., Beckmann,H. and Cashmore,A.R. (1992) TGA1 and G-box binding factors: two distinct classes of Arabidopsis leucine zipper proteins compete for the G-box-like element TGACGTGG. Plant Cell, 4, 1309–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grotewold E., Drummond,B.J., Bowen,B. and Peterson,T. (1994) The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell, 76, 543–553. [DOI] [PubMed] [Google Scholar]
- 16.Hoeren F.U., Dolferus,R., Wu,Y., Peacock,W.J. and Dennis,E.S. (1998) Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol gehydrogenase gene (ADH1) by low oxygen. Genetics, 149, 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J.C., Lee,S.H., Cheong,Y.H., Yoo,C.M., Lee,S.I., Chun,H.J., Yun,D.J., Hong,J.C., Lee,S.Y., Lim,C.O. et al. (2001) A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. Plant J., 25, 247–259. [DOI] [PubMed] [Google Scholar]
- 18.van Helden J. (2003) Regulatory Sequence Analysis Tools. Nucleic Acids Res., 31, 3593–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shahmuradov I.A., Gammerman,A.J., Hancock,J.M., Bramley,P.M. and Solovyev,V.V. (2003) PlantProm: a database of plant promoter sequences. Nucleic Acids Res., 31, 114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solano R., Nieto,C. and Paz-Ares,J. (1995) MYB.Ph3 transcription factor from Petunia hybrida induces similar DNA-bending/distortions on its two types of binding site. Plant J., 8, 673–682. [DOI] [PubMed] [Google Scholar]
- 21.Higo K., Ugawa,Y., Iwamoto,M. and Korenaga,T. (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res., 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lescot M., Dehais,P., Thijs,G., Marchal,K., Moreau,Y., Van de Peer,Y., Rouze,P. and Rombauts,S. (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res., 30, 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davuluri R.V., Sun,H., Palaniswamy,S.K., Matthews,N., Molina,C., Kurtz,M. and Grotewold,E. (2003) AGRIS: Arabidopsis Gene Regulatory Information Server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics, 4, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gubler F., Raventos,D., Keys,M., Watts,R., Mundy,J. and Jacobsen,J.V. (1999) Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J., 17, 1–9. [DOI] [PubMed] [Google Scholar]
- 25.Hirayama T. and Shinozaki,K. (1996) A cdc5+ homolog of a higher plant, Arabidopsis thaliana. Proc. Natl Acad. Sci. USA, 93, 13371–13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solano R., Nieto,C., Avila,J., Canas,L., Diaz,I. and Paz-Ares,J. (1995) Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYB.Ph3) from Petunia hybrida. EMBO J., 14, 1773–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang S., Sweetman,J.P., Amirsadeghi,S., Barghchi,M., Huttly,A.K., Chung,W.I. and Twell,D. (2001) Novel anther-specific myb genes from tobacco as putative regulators of phenylalanine ammonia-lyase expression. Plant Physiol., 126, 1738–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johannesson H., Wang,Y. and Engström,P. (2001) DNA-binding and dimerization preferences of Arabidopsis homeodomain-leucine zipper transcription factors in vitro. Plant Mol. Biol., 45, 63–73. [DOI] [PubMed] [Google Scholar]
- 29.Sessa G., Steindler,C., Morelli,G. and Ruberti,I. (1998) The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol. Biol., 38, 609–622. [DOI] [PubMed] [Google Scholar]
- 30.Huang H., Mizukami,Y., Hu,Y. and Ma,H. (1993) Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res., 21, 4769–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kosugi S. and Ohashi,Y. (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J., 30, 337–348. [DOI] [PubMed] [Google Scholar]
- 32.de Pater S., Greco,V., Pham,K., Memelink,J. and Kijne,J. (1996) Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Res., 24, 4624–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nole-Wilson S. and Krizek,B.A. (2000) DNA binding properties of the Arabidopsis floral development protein AINTEGUMENTA. Nucleic Acids Res., 28, 4076–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kagaya Y., Ohmiya,K. and Hattori,T. (1999) RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res., 27, 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kosugi S. and Ohashi,Y. (2000) Cloning and DNA-binding properties of a tobacco Ethylene-Insensitive3 (EIN3) homolog. Nucleic Acids Res., 28, 960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt R.J., Ketudat,M., Aukerman,M.J. and Hoschek,G. (1992) Opaque-2 is a transcriptional activator that recognizes a specific target site in 22-kD zein genes. Plant Cell, 4, 689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yanagisawa S. and Schmidt,R.J. (1999) Diversity and similarity among recognition sequences of Dof transcription factors. Plant J., 17, 209–214. [DOI] [PubMed] [Google Scholar]
- 38.Kawaoka A., Kaothien,P., Yoshida,K., Endo,S., Yamada,K. and Ebinuma,H. (2000) Functional analysis of tobacco LIM protein Ntlim1 involved in lignin biosynthesis. Plant J., 22, 289–301. [DOI] [PubMed] [Google Scholar]
- 39.Lawton M.A., Dean,S.M., Dron,M., Kooter,J.M., Kragh,K.M., Harrison,M.J., Yu,L., Tanguay,L., Dixon,R.A. and Lamb,C.J. (1991) Silencer region of a chalcone synthase promoter contains multiple binding sites for a factor, SBF-1, closely related to GT-1. Plant Mol. Biol., 16, 235–249. [DOI] [PubMed] [Google Scholar]
- 40.Bastola D.R., Pethe,V.V. and Winicov,I. (1998) Alfin1, a novel zinc-finger protein in alfalfa roots that binds to promoter elements in the salt-inducible MsPRP2 gene. Plant Mol. Biol., 38, 1123–1135. [DOI] [PubMed] [Google Scholar]
- 41.Krusell L., Rasmussen,I. and Gausing,K. (1997) DNA binding sites recognised in vitro by a knotted class 1 homeodomain protein encoded by the hooded gene, k, in barley (Hordeum vulgare). FEBS Lett., 408, 25–29. [DOI] [PubMed] [Google Scholar]