Abstract
NEMBASE (available at http://www.nematodes.org) is a publicly available online database providing access to the sequence and associated meta-data currently being generated as part of the Edinburgh–Wellcome Trust Sanger Institute parasitic nematode EST project. NEMBASE currently holds ∼100 000 sequences from 10 different species of nematode. To facilitate ease of use, sequences have been processed to generate a non-redundant set of gene objects (‘partial genome’) for each species. Users may query the database on the basis of BLAST annotation, sequence similarity or expression profiles. NEMBASE also features an interactive Java-based tool (SimiTri) which allows the simultaneous display and analysis of the relative similarity relationships of groups of sequences to three different databases. NEMBASE is currently being expanded to include sequence data from other nematode species. Other developments include access to accurate peptide predictions, improved functional annotation and incorporation of automated processes allowing rapid analysis of nematode-specific gene families.
INTRODUCTION
The phylum Nematoda represents a highly diverse group of organisms which can be divided into five major clades (1,2). The number of species has been estimated to range from 40 000 to 10 million (3–5). However, despite the availability of the genome sequence of the free-living clade V nematode, Caenorhabditis elegans, the amount of genetic information available for other species of this medically and ecologically important phylum is very limited. In the year 2000, a consortium involving the Nematode Genomics group in Edinburgh, UK; the Pathogen Sequencing Unit of the Sanger Institute, UK; and the Genome Sequencing Center (GSC) in St Louis, MO, USA, was established to use expressed sequence tags (ESTs) as a rapid and cost-effective route to generate new sequence data for a number of human, animal and plant parasitic nematodes. ESTs are single-pass reads of cDNA fragments and as such provide a snapshot of gene expression levels of the transcriptome. By creating libraries on a sex, tissue or developmental stage-specific basis, it is possible to associate changes in transcription with the biology of the organism.
In addition to generating new sequence data, the consortium was charged with the remit of making this new sequence data accessible to the user community. Although a central repository, dbEST, exists to store ESTs (6), in order to help users gain access to this data within a biological context two complementary database resources were created (NEMBASE—developed by the Nematode Genomics group in Edinburgh and NemaGene—developed at the GSC). Here we describe NEMBASE (http://www.nematodes.org/nematodeESTs/nembase.html), which currently hosts the sequence data generated by the two UK-based groups and provides access to its associated meta-data.
CONSTRUCTION OF NEMBASE
ESTs are usually shorter than the full-length mRNAs from which they are derived (up to 700 bp) and are prone to sequencing errors. In many cases, several ESTs may be obtained from the same gene. ESTs from the same species can therefore be grouped on the basis of sequence similarity into clusters that putatively derive from one gene. The creation of a non-redundant set of gene objects helps reduce the number of sequence errors, increases the effective length of the derived transcript and allows the EST data sets for each species to be analysed in a whole-transcriptome context. Since it is unlikely that ESTs will be available for every gene, we term such collections ‘partial genomes’. In Edinburgh, we have developed an automated pipeline that rapidly processes ESTs into partial genomes (7; J. Parkinson, A. Anthony, J. Wasmuth, B. A. Hedley and M. L. Blaxter, unpublished). The process begins by: (i) collating the sequences from dbEST; (ii) clustering on the basis of BLAST similarity (8); (iii) assembling the clusters to derive consensus sequences (putative gene sequences) using phrap (P. Green, unpublished data); (iv) annotating the sequences by performing a series of BLAST analyses—BLASTN versus the non-redundant nucleotide database (GenBank/EMBL); BLASTX versus the non-redundant protein database (SwissProt/TREMBL); and BLASTN versus a database of ESTs excluding those derived from humans and mice (commonly referred to as est_others); (v) collation of the sequence and associated meta-data into a central resource using the public domain postgreSQL database solution.
This process has been applied to each of the 10 species of nematodes for which we are currently generating sequence data (see Table 1). Web front ends are constructed using PHP, Perl-CGI scripts and custom Java tools to allow results from user-specified queries to be generated on the fly. NEMBASE currently contains ∼100 000 EST sequences with an average length of 430 bp. These have in turn been processed into ∼38 000 clusters producing an average consensus length of 500 bp.
Table 1. Sequences, clusters and number of cDNA libraries associated with each species of nematode hosted by NEMBASE.
| Cladea | Species | Description | Number of ESTs | Number of clusters | Number of libraries |
|---|---|---|---|---|---|
| I | Trichuris muris | Mouse threadworm | 2126 | 1630 | 3 |
| III | Ascaris lumbricoides | Human gut parasite | 1910 | 965 | 1 |
| III | Ascaris suum | Swine gut parasite | 29 624 | 8581 | 22 |
| III | Brugia malayi | Human lymphatic parasite | 18 741 | 8497 | 15 |
| III | Onchocerca volvulus | Human filarial parasite | 15 431 | 5226 | 9 |
| III | Toxocara canis | Canine gut parasite | 4379 | 1344 | 1 |
| V | Haemonchus contortus | Sheep gut parasite | 17 269 | 5910 | 9 |
| V | Necator americanus | Human hookworm | 4820 | 2366 | 3 |
| V | Nippostrongylus brasiliensis | Rodent gut parasite | 1250 | 842 | 3 |
| V | Teladorsagia circumcincta | Sheep gut parasite | 4379 | 2559 | 4 |
| Total | 99 929 | 37920 | 70 |
SEARCHING NEMBASE
The home page of NEMBASE describes the data presented and provides help and links to the various methods employed to mine the data.
Annotation search page
If a user is already familiar with the cluster or sequence for which they need information, they can simply type the ID into a text box. This will retrieve the associated cluster. The main feature of this page, however, is to provide users access to sequences that share homology to known genes. Using the specified keywords, a user may search the BLAST annotation to retrieve clusters of interest (e.g. list all the clusters that have homology to ‘heat shock proteins’). Queries can be limited by BLAST e-value scores. Alternatively, the user may list all the clusters associated with a species.
With the exception of retrieving clusters by the use of unique sequence or cluster IDs, the output can be specified either according to e-value scores or by abundance of ESTs associated with each cluster. In addition there is an option to provide a breakdown of library expression associated with each cluster. Output can also be generated in the form of an interactive Java applet which portrays the similarity relationships of the selected clusters to three user-selected databases using SimiTri (9). Each method of output provides a link to a page detailing information on each cluster (see below).
Sequence similarity search page
NEMBASE hosts a BLAST server which allows users to BLAST their own sequences against the sequence databases held on NEMBASE. The resulting BLAST output provides links to the clusters with significant homology to the input sequence.
Expression profile search page
Using the library expression profile table, NEMBASE offers the ability to search for clusters that contain sequences derived from specific libraries (e.g. give me all the clusters that have more than five ESTs from library X but no ESTs from library Y). Again, the SimiTri tool may be used to examine the similarity relationships of relevant clusters to three user-defined databases.
Selecting a cluster using any of the methods outlined above brings up the cluster view page (see Fig. 1), which provides detailed information associated with that cluster. This includes: number of ESTs; libraries associated with the cluster; BLAST summaries for the three databases (EMBL/SwissProtTREMBL and est_others); a list of the individual ESTs that comprise the cluster (and links out to EMBL); a schematic showing how each EST is associated with the cluster build (providing links to the detailed alignment of the ESTs to the consensus sequence and links to viewing the original sequence trace data if it is locally available); and finally a box detailing the sequence and a link that allows the user to BLAST the sequence against any of the databases hosted by NEMBASE. In addition, we are developing methods to provide accurate peptide predictions derived from the cluster consensus sequences. If available, a link from the cluster page provides the user with access to any protein information associated with that cluster. Annotation associated with the peptide predictions range from simple physical properties such as isoelectric points and molecular weights to predictions of cellular location, using PSORT (10), or the presence of protein domains, using InterPro (11).
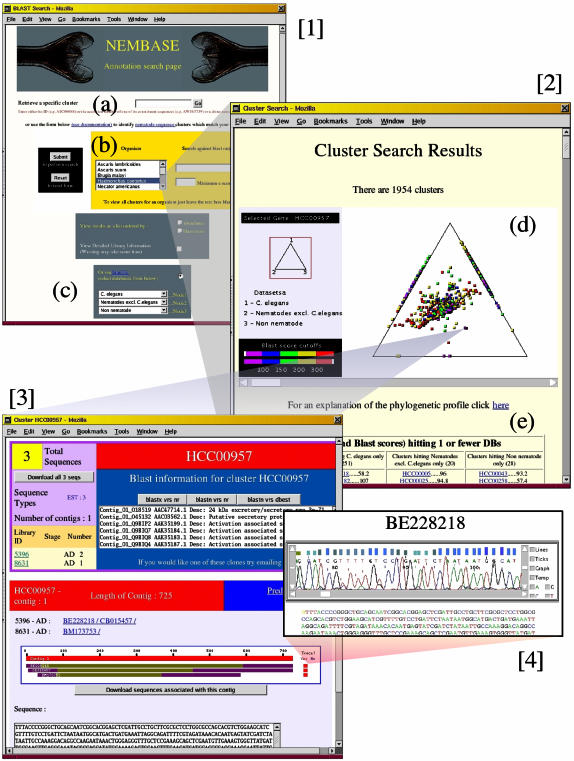
Figure 1.
Screenshots from a typical search strategy on NEMBASE. [1] Annotation search page: on this page users may either retrieve a cluster by entering its ID or the ID of one of its constituent sequences (a) or select a species and enter some text to search for keywords in the BLAST annotation associated with clusters from that species (b). Output may be viewed in terms of relative abundance, BLAST score or using the SimiTri Java tool (c). [2] SimiTri output page: selecting the SimiTri output option creates the embedded Java applet (d); individual clusters are represented by coloured tiles on the graphic. The relative position of the tiles indicates the clusters’ relative similarity to the three selected data sets. Clusters with similarity to only one or no data set are listed below the applet (e). Clicking on a tile whilst holding the control key held down, or selecting a cluster from the list below the applet launches the detailed cluster page [3]. This provides information on the number and source of the constituent sequences, summaries of BLAST annotation and further links to e.g. raw trace chromatograms [4] associated with the sequences.
FUTURE OF NEMBASE
NEMBASE is currently in the process of expansion to include sequence data generated at the GSC from an additional 20 species of nematodes. By summer 2004 it is expected that the current 330 000 ESTs derived from non-C.elegans species will be incorporated within NEMBASE. Further tools are being developed to help exploit these data including improved methods of functional annotation and peptide prediction, automated processes allowing rapid analysis of nematode-specific gene families and graphical tools to help relate these new data to the C.elegans genome.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank the Wellcome Trust for their support of this project.
REFERENCES
- 1.Blaxter M.L., De Ley,P., Garey,J.R., Liu,L.X., Scheldeman,P., Vierstraete,A., Vanfleteren,J.R., Mackey,L.Y., Dorris,M., Frisse,L.M. et al. (1998) A molecular evolutionary framework for the phylum Nematoda. Nature, 392, 71–75. [DOI] [PubMed] [Google Scholar]
- 2.Dorris M., De Ley,P. and Blaxter,M.L. (1999) Molecular analysis of nematode diversity and the evolution of parasitism. Parasitol. Today, 15, 188–193. [DOI] [PubMed] [Google Scholar]
- 3.Maggenti A.R. (1983) Nematode higher classification as influenced by species and family concepts. In Stone,A.R., Platt,H.M. and Khalil,L.F. (eds), Concepts in Nematode Systematics. Academic Press, London, pp. 25–40. [Google Scholar]
- 4.Lambshead P.J.D. (1993) Recent developments in marine benthic biodiversity research. Oceanis, 19, 5–24. [Google Scholar]
- 5.Platt H.M. (1994) Foreword. In Lorenzen,S. (ed.), The Phylogenetic Systematics of Free-living Nematodes. The Ray Society, London, pp. i–ii. [Google Scholar]
- 6.Boguski M.S., Lowe,T.M. and Tolstoshev,C.M. (1993) dbEST—database for ‘expressed sequence tags’. Nature Genet., 4, 332–333. [DOI] [PubMed] [Google Scholar]
- 7.Parkinson J., Guiliano,D.G. and Blaxter,M. (2002) Making sense of EST sequences by CLOBBing them. BMC Bioinformatics, 3, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul S.F., Gish,W., Miller,W., Myers,M.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 9.Parkinson J. and Blaxter,M. (2003) SimiTri—visualizing similarity relationships for groups of sequences. Bioinformatics, 19, 390–395. [DOI] [PubMed] [Google Scholar]
- 10.Nakai K. and Kanehisa,M. (1992) A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics, 14, 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apweiler R., Attwood,T.K., Bairoch,A., Bateman,A., Birney,E., Biswas,M., Bucher,P., Cerutti,L., Corpet,F., Croning,M.D.R. et al. (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res., 29, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]