Abstract
The Rice Proteome Database is the first detailed database to describe the proteome of rice. The current release contains 21 reference maps based on two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) of proteins from rice tissues and subcellular compartments. These reference maps comprise 11 941 identified proteins showing tissue and subcellular localization, corresponding to 4180 separate protein entries in the database. The Rice Proteome Database contains the calculated properties of each protein such as molecular weight, isoelectric point and expression; experimentally determined properties such as amino acid sequences obtained using protein sequencers and mass spectrometry; and the results of database searches such as sequence homologies. The database is searchable by keyword, accession number, protein name, isoelectric point, molecular weight and amino acid sequence, or by selection of a spot on one of the 2D-PAGE reference maps. Cross-references are provided to tools for proteomics and to other 2D-PAGE databases, which in turn provide many links to other molecular databases. The information in the Rice Proteome Database is updated weekly, and is available on the World Wide Web at http://gene64.dna.affrc.go.jp/RPD/.
INTRODUCTION
Proteome analysis linked to genome sequence information is very useful for functional genomics. The genome and proteome of an organism do not correspond on a one-to-one basis: one gene may give rise to multiple proteins by means of alternative splicing or post-translational modification, and its expression may be temporally or spatially regulated. Since proteins are the major players in most processes of living cells, knowledge of the proteome has great relevance to the study of cells and organisms at the molecular level. Accordingly, several databases based on two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) are already available. These are ECO-2DBASE (1), HSC-2DPAGE (2), YPD (3), SIENA-2DPAGE (4,5), PHCI-2DPAGE (6,7) and SWISS-2DPAGE (8).
Rice is not only a very important agricultural resource, it is also a model plant for biological research. Several studies have dealt with the construction of proteomes for complex samples from rice, such as leaf, embryo, endosperm, root, stem, shoot and callus (9–15). Proteomics studies to date have focused mainly on changes in genome expression that are triggered by environmental factors. Examples of descriptive proteomes include the global comparison of green and etiolated rice shoots (11), an analysis of defense-associated responses in the rice leaf and leaf sheath following jasmonic acid treatment (16), an analysis of blast fungus infection of rice grown under different levels of nitrogen fertilization (17) and the characterization of proteins responsive to gibberellin in the leaf sheath of rice seedling (18).
As a complement to these more focused studies, and to facilitate future advances in rice functional genomics, we have constructed the Rice Proteome Database. The Rice Proteome Database comprises information about proteins identified on 2D-PAGE maps of protein extracts from a wide variety of tissues and subcellular compartments of rice. The proteins were identified by various techniques, including gel comparison, microsequencing using a protein sequencer, and peptide mass fingerprinting using mass spectrometry (19). The core of the Rice Proteome Database consists of a description of each of the proteins identified, including its calculated properties such as molecular weight, isoelectric point and expression level; its experimentally determined properties such as amino acid sequences, peptide masses and homologous proteins; and a 2D-PAGE image showing the location of the protein. Links are provided to tools for proteomics and to the other 2D-PAGE databases described above, which in turn provide many links to other molecular biology databases.
One major contribution of the Rice Proteome Database, in which most known rice proteins are recorded, is the wealth of new proteins on which experiments can be conducted at the biochemical and molecular levels. In addition to facilitating the identification of known proteins, sequences in the database can be used to prepare oligodeoxyribonucleotides, which are essential for cloning the corresponding cDNAs. Finally, an attempt is also made to study the physiological significance of some proteins thus identified from rice.
FORMAT AND CONTENT OF THE RICE PROTEOME DATABASE
Each entry in the Rice Proteome Database corresponds to one protein in the 2D-PAGE image data. The following three features are specific to the Rice Proteome Database.
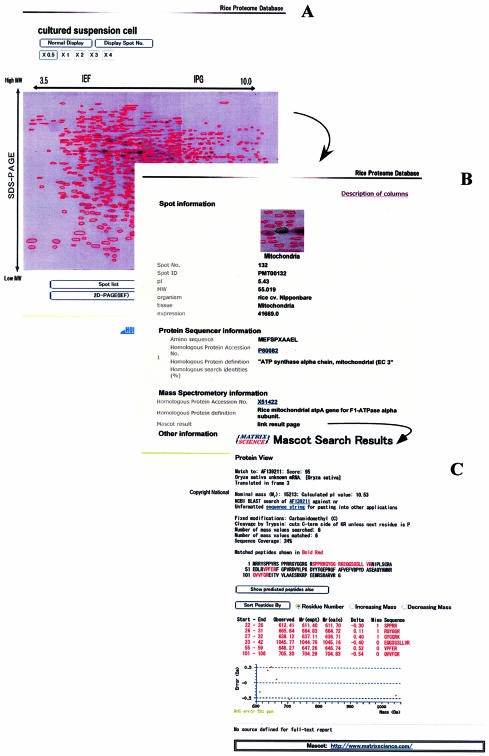
(i) The reference 2D-PAGE map shows the position of the identified entry (Fig. 1A). Spot numbers are displayed on this 2D-PAGE image. The spot list contains a table listing the number of proteins on each 2D-PAGE map in the Rice Proteome Database. Experimental protocols used for protein purification and 2D-PAGE with isoelectric focusing (IEF) and immobilized pH gradient (IPG) are shown on this page. The 2D-PAGE image is synthesized as a composite of gels run by these two different methods and the positions of individual proteins on the gels evaluated using Image Master 2D Elite software (Amersham Biosciences, Uppsala, Sweden).
Figure 1.
Example of the Rice Proteome Database compartment browser. (A) Example of a 2D-PAGE reference map in the Rice Proteome Database. This figure shows the 2D-PAGE reference map of cultured suspension cells. (B) Example of a protein entry from the Rice Proteome Database. This figure shows the spot information for PMT00132, which is spot number 132 for mitochondria in rice. (C) Example of a result from mass spectrometry on the Rice Proteome Database. This figure shows the Mascot Search results for PVM00012.
(ii) The spot information pages (Fig. 1B) provide a range of information about each protein spot, including mapping procedure and spot coordinates; calculated properties of the protein such as molecular weight, isoelectric point and expression level; experimentally determined properties, such as amino acid sequences and peptide masses obtained using protein sequencers and mass spectrometry, respectively, homologous proteins predicted by these two methods; and other information. The accession number of each homologous protein links to the NCBI site. Other information shows the accession number and the percent identity of the homologous full-length cDNA in rice, and biological information such as the known function or functions obtained via experimentation.
(iii) The Mascot Search results page displays the peptide masses derived from mass spectrometry (Fig. 1C). This page brings together the Mascot Search results such as the accession numbers of homologous proteins, scores, sequence coverage and predicted peptides. This page is also linked to the Mascot Web site (Matrix Science Ltd, London, UK).
The current release contains 21 reference maps from rice biological samples that are either tissue specific such as cultured suspension cells, endosperm, crown, seedling root, seedling leaf sheath, seedling leaf blade, stem, mature plant root, mature plant leaf sheath, mature plant leaf blade, anthers, panicle before heading, panicle after heading and panicle 1 week after flowering; or specific to a subcellular location such as cell wall, plasma membrane, vacuole membrane, Golgi membrane, mitochondrion, chloroplast and cytosol. These reference maps of proteins from various tissues and subcellular fractions have a total of 11 941 identified protein spots, corresponding to 4180 separate protein entries in the database (Table 1). The information on amino acid sequences is updated weekly.
Table 1. Content of Rice Proteome Database.
| Map | Abbreviation | Detected spot | Identified spot | No. of entries |
|---|---|---|---|---|
| Cultured suspension cells | CST | 962 | 245 | 245 |
| Endosperm | EST | 100 | 37 | 37 |
| Crown | CRT | 700 | 401 | 401 |
| Seedling root | ROY | 508 | 48 | 48 |
| Seedling leaf sheath | LSY | 431 | 145 | 145 |
| Seedling leaf blade | LBY | 679 | 235 | 135 |
| Stem | STT | 567 | 186 | 186 |
| Mature plant root | ROT | 265 | 100 | 100 |
| Mature plant leaf sheath | LST | 509 | 115 | 115 |
| Mature plant leaf blade | LBT | 718 | 350 | 350 |
| Anthers | ANT | 1080 | 365 | 365 |
| Panicle before heading | BHT | 704 | 441 | 441 |
| Panicle after heading | AHT | 559 | 361 | 361 |
| Panicle 1 week after flowering | AFT | 1073 | 324 | 324 |
| Cell wall | PCW | 513 | 111 | 111 |
| Plasma membrane | PPM | 464 | 159 | 90 |
| Vacuolar membrane | PVM | 141 | 74 | 43 |
| Golgi membrane | PGM | 361 | 187 | 44 |
| Mitochondria | PMT | 672 | 369 | 121 |
| Chloroplast | PCP | 252 | 159 | 66 |
| Cytosolic fraction | PCF | 683 | 352 | 325 |
| Total | 11 941 | 4764 | 4180 |
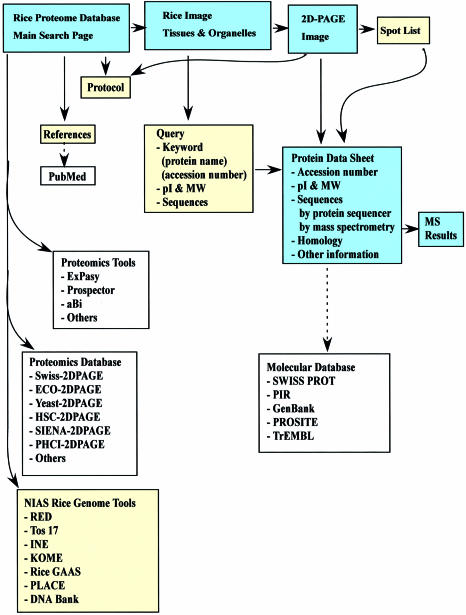
The Rice Proteome Database has links to the NIAS Rice genome tools which are the Rice Expression Database (RED), the Rice full-length cDNA Database (KOME), the Rice Genome Integrated map Database (INE), the Rice Mutant Panel Database (Tos17), the Rice Genome Annotation Database (RiceGAAS), and DNA Bank. The Rice Proteome Database also links to many useful proteomics tools and other proteomics databases (Fig. 2).
Figure 2.
Overview of the Rice Proteome Database.
HOW TO USE THE RICE PROTEOME DATABASE
The Rice Proteome Database can be reached on the world wide web through the Rice Proteome Database home page at http://gene64.dna.affrc.go.jp/RPD/. The Rice Proteome Database home page and the contents of the Rice Proteome Database are maintained by the authors (qxzhsk@bank.dna.affrc.go.jp and skomatsu@affrc.go.jp). The Rice Proteome Database home page provides introductory material on the Rice Proteome Database. A Rice Proteome Database entry may be obtained from the server in one of four ways.
(i) By selecting a spot on one of the 2D-PAGE reference maps. The Rice Proteome Database contains information on proteins identified from several tissues and organelles on 2D-PAGE reference maps. These 2D-PAGE maps can be reached by clicking the individual tissues/organelles denoted by red boxes. Only spots with sequence data are highlighted and labeled ‘Annotation Data Available’.
(ii) By ‘protein keyword’ or ‘protein database accession identifiers’ using the protein name or accession number. The Rice Proteome Database can be searched using proteins as keywords.
(iii) By isoelectric point and molecular weight for any protein. The Rice Proteome Database can be searched with a range of isoelectric points and molecular weights.
(iv) By similarity search with the user’s amino acid sequences. The query sequence can be searched using the homology search tools BLASTP and BLASTX for the presence of amino acid sequences identical or similar to previously reported amino acid sequences in the Rice Proteome Database.
FUTURE PROSPECTS
Information on post-translational modifications such as phosphorylation, glycosylation and other modifications, obtained experimentally by immunoblot, will be added to the Rice Proteome Database at the end of March 2004. The experimentally determined 2D-PAGE image results for rice embryo, nucleus and other new samples will also be added, and the number of identified proteins will be increased.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Mr Itaru Usami at Infocom Co., and Dr Arun Sharma, Dr Guangxiao Yang and Dr Muhammed Khan at NIAS for their technical assistance in database preparation, and the members of the Rice Proteome Analysis Center for their assistance in protein identification. This work was supported by a grant from the Rice Genome Project, Ministry of Agriculture, Forestry and Fisheries of Japan.
REFERENCES
- 1.VanBogelen R.A., Abshire,K.Z., Moldover,B., Olson,E.R. and Neidhardt,F.C. (1997) Escherichia coli proteome analysis using the gene–protein database. Electrophoresis, 18, 1243–1251. [DOI] [PubMed] [Google Scholar]
- 2.Dunn M.J., Corbett,J.M. and Wheeler,C.H. (1997) HSC-2DPAGE and the two-dimensional gel electrophoresis database of dog heart proteins. Electrophoresis, 18, 2795–2802. [DOI] [PubMed] [Google Scholar]
- 3.Payne W.E. and Garrels,J.I. (1997) Yeast protein database (YPD): a database for the complete proteome of Saccharomyces cerevisiae. Nucleic Acids Res., 25, 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bini L., Heid,H., Liberatori,S., Geier,G., Pallini,V. and Zwilling,R. (1997) Two-dimensional gel electrophoresis of Caenorhabditis elegans homogenates and identification of protein spots by microsequencing. Electrophoresis, 18, 557–562. [DOI] [PubMed] [Google Scholar]
- 5.Bini L., Magi,B., Marzocchi,B., Arcuri,F., Tripodi,S., Cintorino,M., Sanchez,J.-C., Frutiger,S., Hughes,G.J., Pallini,V. et al. (1997) Protein expression profiles in human breast ductal carcinoma and histologically normal tissue. Electrophoresis, 18, 2832–2841. [DOI] [PubMed] [Google Scholar]
- 6.Shaw A.C., Larsen,M.R., Roepstorff,P., Holm,A., Christiansen,G. and Birkelund,S. (1999) Mapping and identification of HeLa cell proteins separated by immobilized pH-gradient two-dimensional gel electrophoresis and construction of a two-dimensional polyacrylamide gel electrophoresis database. Electrophoresis, 20, 977–983. [DOI] [PubMed] [Google Scholar]
- 7.Shaw A.C., Larsen,M.R., Roepstorff,P., Justesen,J., Christiansen,G. and Birkelund,S. (1999) Mapping and identification of interferon gamma regulated HeLa cell proteins separated by immobilized pH gradient two-dimensional gel electrophoresis. Electrophoresis, 20, 984–993. [DOI] [PubMed] [Google Scholar]
- 8.Hoogland C., Sanchez,J.-C., Tonella,L., Binz,R.-A., Bairoch,A., Hochstrasser,D.F. and Appel,R.D. (2000) The 1999 SWISS-2DPAGE database update. Nucleic Acids Res., 28, 286–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komatsu S., Kajiwara,H. and Hirano,H. (1993) A rice protein library: a data-file of rice proteins separated by two-dimensional electrophoresis. Theor. Appl. Genet., 86, 935–942. [DOI] [PubMed] [Google Scholar]
- 10.Zhong B., Karibe,H., Komatsu,S., Ichimura,H., Nagamura Y., Sasaki,T. and Hirano,H. (1997) Screening of rice genes from a cDNA catalog based on the sequence data-file of proteins separated by two-dimensional electrophoresis. Breed. Sci., 47, 245–251. [Google Scholar]
- 11.Komatsu S., Muhammad,A. and Rakwal,R. (1999) Separation and characterization of proteins from green and etiolated shoots of rice: towards a rice proteome. Electrophoresis, 20, 630–636. [DOI] [PubMed] [Google Scholar]
- 12.Komatsu S., Rakwal,R. and Li,Z. (1999) Separation and characterization of proteins in rice suspension cultured cells. Plant Cell Tissue Organ Cult., 55, 183–192. [Google Scholar]
- 13.Tsugita A., Kawakami,T., Uchimiya,Y., Kamo,M., Miyatake,N. and Nozu,Y. (1994) Separation and characterization of rice proteins. Electrophoresis, 15, 708–720. [DOI] [PubMed] [Google Scholar]
- 14.Shen S., Matsubae,M., Takao,T., Tanaka,N. and Komatsu,S. (2002) A proteomics analysis of leaf sheath from rice. J. Biochem., 132, 613–620. [DOI] [PubMed] [Google Scholar]
- 15.Koller A., Washburn,M.P., Lange,B.M. andon,N.L., Deciu,C., Haynes,P.A., Hays,L., Schieltz,D., Ulaszek,R., Wei,J. et al. (2002) Proteomic survey of metabolic pathway in rice. Proc. Natl Acad. Sci. USA, 99, 11969–11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rakwal R. and Komatsu,S. (2000) Role of jasmonate in the rice self-defense mechanism using proteome analysis. Electrophoresis, 21, 2492–2500. [DOI] [PubMed] [Google Scholar]
- 17.Konishi H., Ishiguro,K. and Komatsu S. (2001) A proteomics approach towards understanding blast fungus infection of rice grown under different levels of nitrogen fertilization. Proteomics, 1, 1162–1171. [DOI] [PubMed] [Google Scholar]
- 18.Shen S., Sharma,A. and Komatsu,S. (2003) Characterization of proteins responsive to gibberellin in the leaf-sheath of rice (Oryza sativa L.) seedling using proteome analysis. Biol. Pharm. Bull., 26, 129–136. [DOI] [PubMed] [Google Scholar]
- 19.Komatsu S., Konishi,H., Shen,S. and Yang,G. (2003) Rice proteomics: A step toward functional analysis of the rice genome. Mol. Cell. Proteomics, 2, 2–10. [DOI] [PubMed] [Google Scholar]