Abstract
Single molecule localization based super-resolution imaging techniques require repeated localization of many single emitters. We describe a method that uses the maximum likelihood estimator to localize multiple emitters simultaneously within a single, two-dimensional fitting sub-region, yielding an order of magnitude improvement in the tolerance of the analysis routine with regards to the single-frame active emitter density. Multiple-emitter fitting enables the overall performance of single-molecule super-resolution to be improved in one or more of several metrics that result in higher single-frame density of localized active emitters. For speed, the algorithm is implemented on Graphics Processing Unit (GPU) architecture, resulting in analysis times on the order of minutes. We show the performance of multiple emitter fitting as a function of the single-frame active emitter density. We describe the details of the algorithm that allow robust fitting, the details of the GPU implementation, and the other imaging processing steps required for the analysis of data sets.
OCIS codes: (100.6640) Superresolution, (180.2520) Fluorescence microscopy, (100.3010) Image reconstruction techniques
1. Introduction
Single molecule based super resolution (SM-SR) techniques have revolutionized fluorescence microscopy, achieving spatial resolution of approximately 20 nm, an order of magnitude improvement from conventional fluorescence microscopy that is limited by diffraction to λ/2NA or approximately 250 nm [1–5]. The SM-SR concept relies on making precise and accurate estimations of the positions of individual emitters that label the structure of interest. Resolution is then a function of both the position uncertainty and the sampling density. This concept is realized by exploiting some properties of the fluorescent probes that result in a small subset of emitters being in a fluorescent state during the acquisition of any single image. Acquired images that contain different subsets of active emitters can then be analyzed and used to generate a SR image, providing sufficient sampling density and localization precision. Initial demonstrations of SM-SR used a variety of probes including quantum dots [6, 7], photo-activatable proteins [8, 9] and organic dyes [10] and the number of probes that have been demonstrated for use in SM-SR continues to grow [4, 11].
In the case of 2D imaging, which is the focus of this work, an advantage of SM-SR over other SR techniques such as STED [12], 4Pi [13], and SSIM [14] is that it can be implemented using a relatively simple and conventional microscope such as an objective based Total Internal Reflectance Fluorescence (TIRF) microscope setup. However, the technique relies heavily on the analysis of the acquired data, primarily in making estimates of the position of on the order of 106 emitters. To simplify and speed analysis, conventional analysis approaches only attempt to localize well separated, single emitter events and data that does not fit this model is rejected. Experimental conditions must then be optimized to give a single-frame active emitter density that makes best use of the data and yet minimizes acquisition time [15].
SM-SR fitting routines that disregard events that cannot be fit to a single emitter profile result in some fraction of data being discarded. The potential loss of information is demonstrated in Fig. 1, which shows that at an active emitter density of 1 μm−2, more than 55% of 8σPSF × 8σPSF (σPSF = 127 nm) sub-regions contain 2 or more active emitters. Such nearby or overlapping emission patterns could result in a failure of the single emitter model and the data not being used in the SR image reconstruction. The distribution of the number of emitters found within these 8σPSF × 8σPSF sub-regions (σPSF = 127 nm) as a function of density is also shown in Fig. 1 and illustrates that with increasing active emitter density, isolated single-emitter events become rare and therefore a majority of the position estimates will get discarded due to an unacceptable fit to a single emitter model. It is clear that a multiple-emitter fitting approach would enable the analysis of images containing higher single-frame density of active emitters. Analysis of multiple emitters simultaneously in one sub-region does not necessarily impact the position uncertainties as visually overlapping emitters (around 100 nm between emitter centers) can be localized with similar uncertainties [16, 17]. In practice, a multi-emitter fitting model would allow one or more of several important quantities to be improved, which would result in a much better localization in cases where single frame active emitter densities are relatively high [15].
Fig. 1.
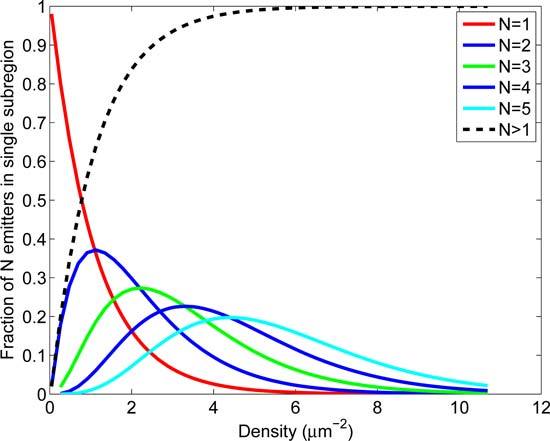
Proximity of emitters as a function of emitter density. The probabilities of finding N=1–5 emitters within a 8σPSF×8σPSF square sub-region (σPSF = 127 nm) at different densities were calculated for a uniformly distributed population of emitters and plotted as a function of density. As the emitter density increases beyond 1 μm−2, the fraction of subregions containing single emitters reduces dramatically (red line), emphasizing the need for fitting algorithms that can accommodate multiple emitters within a single sub-region.
Here, we describe an analysis method that uses the Maximum Likelihood Estimator (MLE) in order to perform simultaneous position estimates of multiple emitters within a small sub-region. In contrast to other techniques that use deflation methods, whereby the best single fluorophore fit is made to the image and the analysis proceeds with the residuum image that is calculated by subtracting the single fluorophore fit model [18–21], all emitter positions within the subregion are estimated simultaneously. The sub-region data is fit to models assuming N emitters, where N is varied from N=1, to N = Nmax using a process that we will subsequently refer to as Multi-emitter Fitting Analysis (MFA). Based on the log-likelihood, a chi square distributed test statistic is used to either choose one model, or reject all fitting models. In this manuscript, we describe a procedure that allows robust application of the MFA, including model selection criteria, uncertainty calculations, and other procedures for analyzing a SM-SR data set and image reconstruction.
2. Theoretical basis for the multi-emitter fitting algorithm
2.1. Multiple emitter extension to the pixelized single emitter model
The impulse response of a microscope to a point source of light is defined as the point spread function (PSF) and in the 2D case, can be well approximated by the Gaussian function [22,23]:
| (1) |
where σ0 represents the standard deviation of the Gaussian.
Given the pixelization that occurs from a CCD based detector system, this continuous distribution can be modified to represent the expected photon count in pixels on the camera. For an individual pixel k located at a position {x,y} and assumed to rectangular, the expected number of photons in that pixel, which are emitted from a point object in focus, can be calculated by integrating Eq. (1) across the pixel assuming a square shaped pixel. This pixelized single emitter profile is given as:
| (2) |
where μk(x, y) is the expected photon count for a given pixel ‘k’, I0 is the total emitted photon counts expected, b0 is the background and ΔEx(x, y) and ΔEy(x, y) are:
| (3a) |
| (3b) |
where x0 and y0 are emitter positions.
This model can be extended to account for emission from multiple emitters by assuming each emitter contributes independently to the expected photon counts at a given pixel k. The expected photon count for pixel k, μk(x, y) generated by N emitters can then be calculated by summing over the total number of emitters N and is defined as:
| (4) |
2.2. Maximum likelihood estimator
To estimate the emitter positions, we maximize the likelihood function [24]:
| (5) |
where the likelihood of the parameters θ given the data D is modeled as a photon counting process for each pixel, with the expected counts given by the multi-emitter model μk defined in Eq. (4) and the observed counts dk. The maximum likelihood estimator (MLE) is used to estimate the emitter positions {xi, yi}....{xN, yN} and the background fluorescence rate b0, giving θ̂ = {b0, x1, y1,...xN, yN}T. To ensure robust estimation, we find that it is necessary to confine the intensity parameter Ii = I0 in Eq. (4), where I0 is obtained from independent measurements.
Maximization of Eq. (5) can be performed using the Newton-Raphson method (NR) to iteratively maximize the log-likelihood. The iterative step for parameter θi can be written for a Poisson noise model as follows [25]:
| (6) |
All derivatives of μ(θ) are identical in form to those from the single-emitter model and are given in our previous work [25].
3. The analysis procedure
Our fitting routine operates independently on each image of a time series. First, a series of image filters are applied to each frame to find points of interests and then each frame is partitioned into an array of sub-regions around these points. In each sub-region, the positions of N proposed emitters in a model of N = 1 to Nmax are found sequentially where the N emitter model uses position information from the N – 1 emitter model. We generate the p-value from a test statistic based on the log-likelihood ratio (LLR) to compare fits for each model. The model with the highest p-value is selected and the associated uncertainties and fits are determined based on a modified Fisher information matrix. The process is repeated for all frames and a reconstructed image is generated from the estimates by placing bivariate Gaussian shapes at the estimated locations using estimator uncertainties to build the bi-variate covariance matrix. Below we outline these steps in further detail.
3.1. Image pre-processing and segmentation
For each data set, all frames are analyzed independently. Experimentally acquired images are first offset and gain corrected to convert pixel intensity values to photon counts. To aid subregion selection, a two step image filtering process is carried out to reduce Poisson noise and background and to identify potential emitter locations. The first filtering step is calculated from the original image I, as follows:
| (7) |
where uniform[image,q] represents a uniform filter process with a square kernel size q operating on the 2-D matrix image. The uniform filter acts as a smoothing filter by reassigning the value of each pixel to the average pixel value within the square kernel centered at the pixel position. The analysis is not strongly dependent on the smoothing filter so the uniform filter is chosen for speed. Subtraction means a pixel-wise subtraction between results obtained for each filter process. The second filtering step is performed on the first filtered image A1 as follows:
| (8) |
where max[image,q] represents a maximum filter process used to obtain local maximum values within a square kernel size q. Through this process, all pixels within a kernel take the maximum value within the kernel. These two filtered images A1 and A2 are then compared pixel-wise to identify regions of interest:
| (9) |
Through this process, pixels with local maximum intensities in the uniformly filtered image A1 are identified in A3. Sub-regions of size 6σPSF × 6σPSF that are centered at pixels where A3 = 1 are selected for further analysis.
3.2. Multi-emitter fitting analysis (MFA)
Each sub-region is analyzed using a Multi-emitter Fitting Analysis as depicted in Fig. 2. The analysis proceeds sequentially from a N = 1 model to a N = Nmax model. For the N = 1 model, the center of mass of the sub-region is used as the initial position estimate. For the N ≠ 1, multi-emitter models, the N – 1 position estimates found in the previous step are used as N – 1 of the initial position estimates. The remaining initial position estimate is found by calculating the residuum image generated by a subtraction of the N – 1 model (Eq. (4)) from the data in the sub-region. If the value of the maximum intensity pixel in the residuum image is low enough to assume that all emitters in the sub-region have been found, the analysis does not proceed further. Otherwise, from the residuum image, the last initial estimate is calculated from the position of the pixel with the maximum count value, giving {xdef, ydef} and then is adjusted in a “Push&Pull” process to {xadj, yadj} = {xdef ± σPSF/2, ydef ± σPSF/2}. If {xdef, ydef} is within σPSF of the edge of the sub-region, that position is likely to correspond an emitter outside of the region, and the sign of the adjustment is such to move the adjusted position further away from the center of the sub-region. Otherwise, the sign of the adjustment is such to move the adjusted position towards the center of mass of the N – 1 position estimates. This compensates for the effect that in a N – 1 model of an underlying N emitter system, the estimated positions of N-1 emitters are displaced such that after deflation, the position of the maximum value pixel is biased away from the actual position of that emitter. This effect is illustrated in Fig. 2(b). We found that the ”Push&Pull” adjustment of only one of the initial position estimates is sufficient to allow robust convergence. The initial estimates are then updated using a fixed number of iterations of Eq. (6). After obtaining estimates for each model, models with location estimates outside the fitting boundary, which is a 8σPSF × 8σPSF square region concentric with image sub-region (red box Fig. 2(b)–2(e)), are discarded. Models with positions estimates within the fitting boundary but outside the data sub-region (black region between red and yellow box in Fig. 2(b)–2(e)) are allowed since emitters located in this region will affect the data sub-region. The position and background estimates, along with their log-likelihood, are saved for each remaining model for a further model selection process.
Fig. 2.
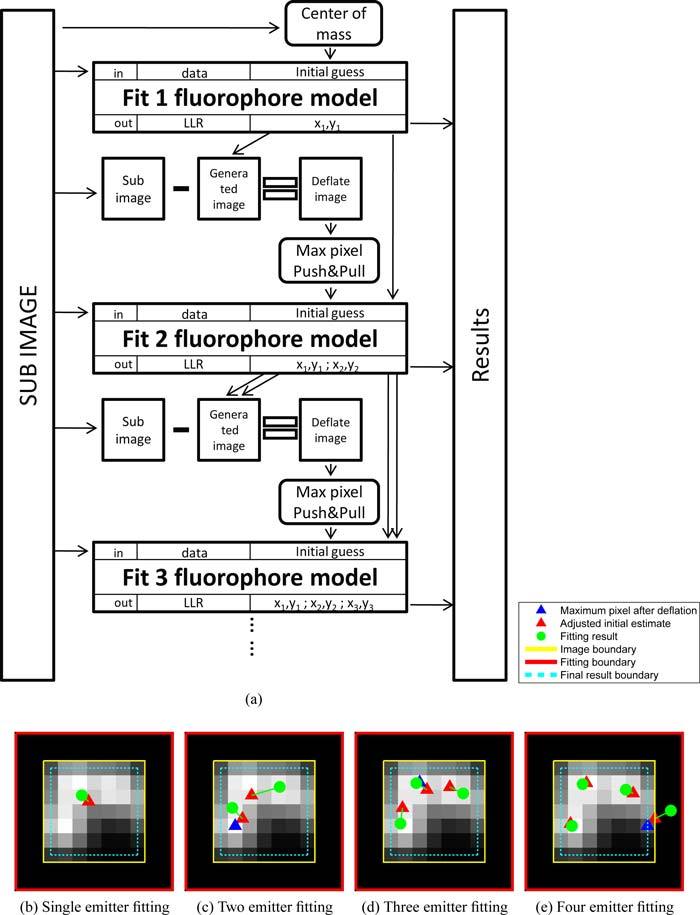
Illustration of execution steps in the multi-emitter estimation task. (a) Fitting algorithm flowchart. For a given sub-region, MFA is performed sequentially from the N = 1 emitter model to either the Nmax emitter model or is terminated if the maximum pixel counts in the residuum image is lower than 10 counts. (b) through (e): Demonstration of the results from each estimation task from the 1 emitter model through the 4 emitter model. The 5 emitter model fitting is not performed by the algorithm, because of the low photon counts in the deflated image.
3.3. Model selection
To compare between models, we used a test statistic based on the log-likelihood ratio (LLR) as an indicator for the quality of fit. The LLR is shown in Eq. (10) and approximates a chi square distribution with K – (2N + 1) degrees of freedom, where K is the number of pixels in the sub-region and N is the number of emitters in the model.
| (10) |
where D represents the sub-region data, θ̂ are the MLE estimates and L(D|D) gives the upper limit of likelihood of the data set with Poisson noise (when μk = dk). The model is accepted if it has the maximum chi-square p-value of all models and passes the p-value threshold set by user. Considering that the variance of intensities in real or realistically simulated data would broaden the LLR distribution and thus result in a smaller p-value, typically a small p-value of 10−3 to 10−6 is used as the threshold in our analysis and is still sufficient to reject incorrect models and the un-converged fit. After obtaining the uncertainty for the position estimates, emitters with estimated positions near (within σPSF/2) or outside of the sub-region boundary are discarded. The parameters describing the remaining emitters are passed to the image reconstruction process.
3.4. Precision of the estimated parameters
For unbiased estimators, the Cramer-Rao Lower Bound (CRLB), given as where is the Fisher information matrix, is often used to calculate the precision of estimated parameters [16, 25, 26]. However, as known from the analysis of Gaussian mixture models [27], the Fisher information matrix is singular at {xi, yi} = {xj, yj}, and near this singular point, can not be used to correctly calculate estimator precision. We implemented a phenomenological correction to the Fisher information matrix by modifying the off diagonal terms that give rise to the singularity. Given our parameter set θ = {b0, x1, y1,...xN, yN}T, the corrections are given by:
| (11) |
A is given by , where σi and σj are the intermediate precision calculations obtained from F(θ) assuming A = 0. F(θ), which we designate the modified Fisher Information matrix, replaces the original Fisher Information matrix in our precision calculation process, is non-singular at {xi, yi} = {xj, yj} and quickly converges to I(θ) once far from the point of singularity. Thus it provides reasonable precision estimates in the regions both near and far from the point of singularity.
3.5. Filtering and SR image reconstruction
After obtaining estimates and their uncertainties, a rejection process is performed to remove repeated localizations that can occur due to overlapped sub-regions. An emitter estimate is removed if there is another estimate with a smaller uncertainty within a distance of the previous emitter’s localization uncertainty coming from the the same image frame but a different sub-region. Another filtering process is performed to remove the estimates with position uncertainties greater than the resolution threshold and would therefore not contribute to the desired resolution in the reconstructed image. The SR image is reconstructed by adding bivariate 2-D Gaussian shapes to the SR image at the location of the position estimates. The covariance of the bivariate Gaussian is constructed using the appropriate elements of F(θ)−1 and indicate the asymmetry of the position uncertainties that arise from the multi-emitter localization process.
4. Computational and experimental methods
4.1. Hardware and software implementation of analysis routines
Numerical analyses are performed using MATLAB (The Mathworks, USA), the imaging processing toolbox, DipImage [28] and c-language codes that are compiled to MATLAB mex files and initiated from within the MATLAB environment. GPU code (Nvidia CUDA [29]) are managed through c-language codes that are also compiled to MATLAB mex files and runs within the MATLAB environment. All CPU based code runs on a single thread.
Image pre-processing and segmentation are implemented in c-code. The array of isolated sub-regions are passed into the GPU global device memory for the MFA. The MFA for each sub-region is independently carried out by a single thread on the GPU similar to that is described in previous publication [25] using 50 iteration steps in NR iteration process. The model selection is performed in the same thread as part of the MFA. The fitted parameters for successful models are passed back to the CPU. The generation of F(θ) and its inversion, by LU decomposition with back substitute method [30], are implemented on the GPU executing with one thread per sub-region. The resulting uncertainties for each parameter are passed back to the CPU. The filtering of position estimates by sub-region position and their uncertainties is performed on the CPU. Reconstruction of the SR image is performed in a manner inverse to the sub-region selection. First, in the GPU, an up-sampled sub-region is generated that corresponds to each position estimate and its uncertainties. The bivariate Gaussian shapes for the position estimates are added to the sub-region. All generated up-sampled sub-regions are passed back to the CPU and assembled into a single up-sampled SR image.
4.2. Estimator precision and algorithm performance testing
In order to demonstrate the performance of the modified Fisher information matrix in calculating the estimator precision, two types of data sets were generated and analyzed. First, a series of simulated images of two emitters that had increasing separations between their centers were generated. For each separation, 1000 identical two-emitter images were generated, corrupted by Poisson noise and fitted by MFA. Second, images of 1000 configurations of random placements of 1,2,3,4 and 5 emitters were replicated 1000 times, corrupted by Poisson noise and fitted by MFA. The Performance of the modified Fisher information matrix in providing the correct precision estimates was demonstrated by comparing the observed standard deviation of estimates and the precision of the estimator calculated using the modified Fisher information matrix. Estimator accuracies for each emitter distribution were calculated by taking the ratio between the mean of the uncertainty estimates and the observed uncertainty.
Algorithm performance was also tested on simulation data where 2D Gaussian shapes were randomly placed with uniform distribution through the image with their actual position registered for later calculation. The total expected photon count per emitter was selected from a normal distribution with μ = 800, σ = 100. A background count rate of 5 count/pixel was added to the image, and then the image was corrupted with Poisson noise. After fitting these images using MFA with a target resolution of 20 nm or 50 nm, the localization fraction was calculated by taking the ratio between the number of correctly localized emitters which is defined as having a registered emitter position near the localized emitter within the target resolution and the total number of emitters in simulation. The error rate of the algorithm was obtained by calculating the ratio between the number of mis-localized emitters which is defined as having no actual emitter position near the localized emitter within the target resolution and the total number of emitters obtained from fitting.
4.3. Synthetic data generation
Synthetic image series in a Siemens star pattern with 50 non-empty slice regions were generated such that the maximum width of each slice (on the outer diameter) equals 213 nm. A fixed labeling density ρ0=5000 μm−2 and off rate koff = 0.8 frame−1 were used, with varied kon to generate variations in active densities (ρactive) according to:
| (12) |
A blinking trace was generated for each emitter using the transition rates kon and koff for dark to active, and active to dark transitions respectively and were designed to emulate realistic photophysical properties. As in all of our simulations, the active emitters were represented as a 2D Gaussian shapes, with σ = σPSF = 1.2 pixels (127 nm). To represent the experimentally observed variation in emitter brightness, for each emitter, the total expected photon count per frame was selected from a normal distribution with μ = 800 σ = 100. Shown in Fig. 3 is the single frame intensity distribution of Alexa Fluor 647. A background count rate of 5 count/pixel was added to the image, and then the image was corrupted with Poisson noise. Calculation of the density of active emitters assumes a pixel size of 106 nm, which is the back-projected pixel size in the experimental system.
Fig. 3.
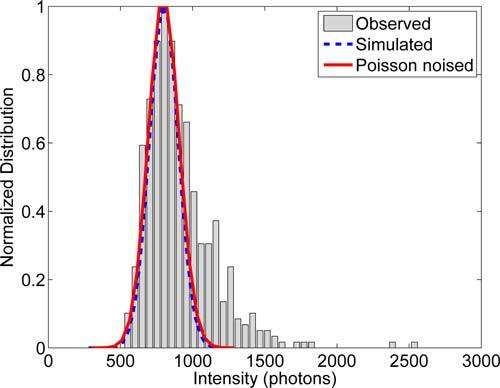
Single fluorophore intensity distribution of the organic fluorophore Alexa Fluor 647 obtained from the data set described in section 4.4.1 taken in TIRF condition. The distribution is modeled as a normal distribution with μ = 800, σ = 100.
4.4. SM-SR imaging
4.4.1. Cell culture
Human epithelial cervical cancer (HeLa) cells were cultured in Minimum Essential Media (Gibco) supplemented with Fetal Bovine Serum (HyClone), L-Glutamine and Penn-Strep. Five hours after plating at low confluency onto 8-well Labtek chambers (Nunc), cells were serum starved approximately 10 hours and fixed using 4% paraformaldehyde in phosphate buffered saline (PBS). After three washes in PBS, cells were permeabilized (0.5%v/v Triton X-100) at room temperature for 15 minutes in the presence of 3% BSA to reduce non-specific binding. Cells were again washed three times in PBS before addition of Alexa Fluor 647 Phalloidin (Invitrogen). Phalloidin was added at four times the recommended concentration (approximately 660 nM) to ensure dense labeling. Cells were washed five times and imaged in the presence of an oxygen scavenging system including 50 mM MEA [31] as a reducing agent.
4.4.2. Microscopy and data acquisition
Single molecule imaging experiments were performed in an epi-fluorescence microscope setup consisting of an inverted microscope (IX71, Olympus America Inc.), 1.45 NA TIRF objective (U-APO 150x NA 1.45, Olympus America Inc.), 635 nm diode laser (Radius 635, Coherent Inc.), and an electron multiplying CCD camera Ixon (897, Andor Technologies PLC.) with EM gain set to ≈ 200. The epi-fluorescence filter setup consisted of a dichroic mirror (650 nm, Semrock) and an emission filter (692/40, Semrock). The sample chamber was mounted in a 3D piezostage (Nano-LPS, Mad City Labs). 5000 images were taken in a TIRF configuration at 20 frames/second. Drift correction was not implemented, but from independent measurements we estimate a drift of less than 25 nm over the acquisition time. Frames were 256 × 256 pixels with a pixel size of 0.106 μm.
5. Results and discussion
5.1. Optimal sub-region size and Nmax
Various sub-region sizes ranging from 4σPSF to 8σPSF were evaluated in the aspects of both localization fraction and error rate that are defined in section 4.2. Small sub-regions tend to isolate individual emitters from one another better compared to larger sub-regions and thus results in sub-regions containing fewer emitters. However, the smaller area decreased the amount of information that could be used in fitting and thus the error rate increases compared to larger sub-regions. Large sub-regions provide more accurate estimates compared to a smaller subregion but the probability of introducing more emitters within or near the sub-region increases quadratically with the width of the square sub-region. We have tested our algorithm performance under different sub-region sizes, such as 4σPSF, 5σPSF, 6σPSF, 7σPSF, 8σPSF, various active emitter densities from 0.1 μm−2 to 10 μm−2, various emitter intensities from 200 to 5000 and various intensity variance. After comparing these plots (data not shown), we found that sub-region size of 6σPSF shows the best compromise of error rates and localization fraction. Nmax values ranging from 1 to 8 were tested. Large Nmax tend to generate a more complex likelihood surface and thus the possibility for the estimator being stuck at a local minimum increases with Nmax. The complexity introduced by multi-emitter model results in higher error rates and thus Nmax was restricted to 5 in our analysis.
5.2. Uncertainty estimator performance
Using simulations, estimator precision calculations for various emitter configurations were calculated from our modified Fisher information matrix F(θ) of Eq. (11) and compared with observed standard deviations. Singularity of the Fisher Information matrix for the multi-component Gaussian model when 2 (or more) emitter centers that have a separation less than 100 nm results in a failure of the CRLB to correctly estimate the precision of the position estimation. This effect is demonstrated in Fig. 4(a). Figure 4(a) also shows that calculations based on F(θ) gave a correct estimator precision (compared to the observed standard deviation of the estimates) in the regions both near and far from the point of singularity of the two emitter model, with only a small but conservative deviation below 50 nm. We also show the performance of F(θ) based precision calculations for random configurations of multiple emitters by looking at the estimator accuracy, defined as the ratio of the precision calculated using F(θ) to the observed standard deviation of the estimates. The cumulative distribution (the normalized integral of the histogram) of the estimator accuracy is shown in Fig. 4(b) and demonstrates that the estimator accuracy distribution (corresponding to the derivative the CDF) of is narrowly peaked around 1 for the 1–3 emitter models (ideal) and is conservative (reported precision is larger than observed standard deviation) on the 4 and 5 emitter models where the estimator accuracy distribution is peaked around 1.1 and 1.2 respectively.
Fig. 4.
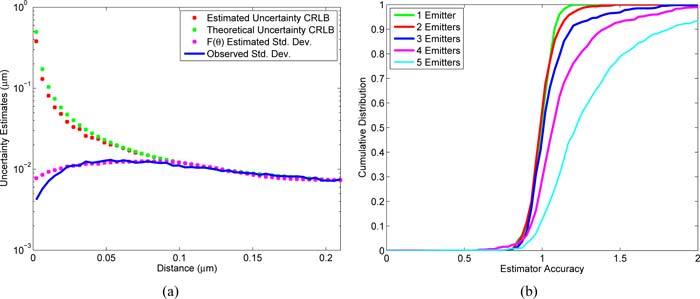
Performance of the precision estimate. (a) A comparison between the precision predicted from the CRLB and from the modified Fisher information matrix. A series of simulated images of two emitters at varoius separations between their centers were generated. MFA was performed on these images and the precision estimates calculated by the modified Fisher information matrices (F(θ) Estimated Std. Dev.) were compared with that obtained from the CRLB (Estimated Uncertainty CRLB), precisions obtained from the CRLB generated by emitter’s true position (Theoretical Uncertainty CRLB), and the observed standard deviation of the estimates (Observed Std. Dev.). (b) The CDF (integral of histogram) of the uncertainty estimator accuracy obtained using the modified Fisher information matrices for random placements of multiple emitters.
5.3. Algorithm performance versus density and intensity distribution
We have tested our algorithm on simulated data sets where emitters were randomly placed with uniform distribution in a 64 × 64 image representing an area of 46 μm2 in our microscope camera setup. By increasing the number of active emitters within the image, density increased from 0.01 μm−2 to 10 μm−2. Both single (Nmax = 1) to multi (Nmax = 5) emitter fitting algorithm were performed on these data sets and localization fraction (defined in 4.2) were calculated.
Figure 5 shows the performance of the MFA analysis for various densities and intensity distributions. The simulations show that the localization fraction improvement from Nmax = 1 to Nmax = 5 is most significant at a densities higher than 1 μm−2. We note that at high intensities with narrow intensity distribution (Figs. 5(e), 5(f)) the localization error improves, but the localization fraction does not. This is attributed to high sensitivity to model mismatches created by the fixed intensity assumption and emitters outside the fitting window.
Fig. 5.
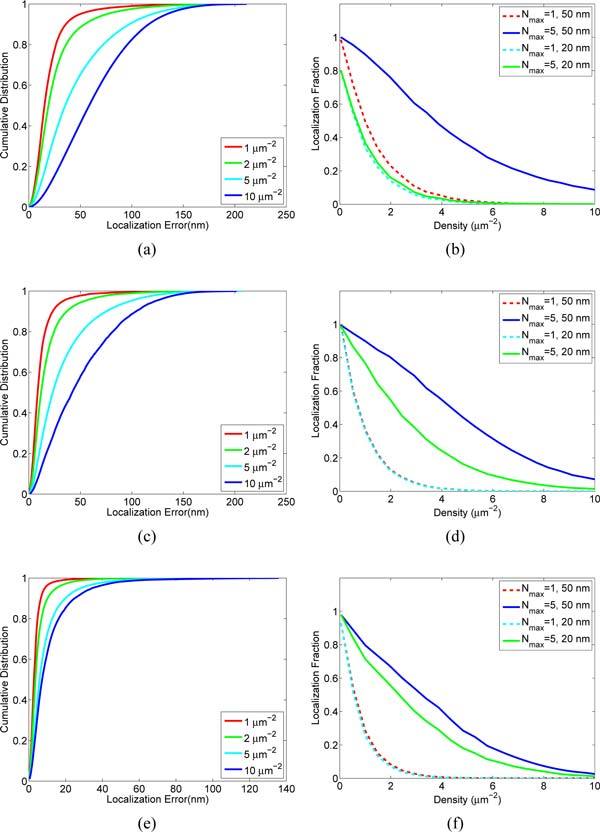
Performance versus active emitter density and intensity distribution. Shown are the results of MFA analysis of images with spatially random distributed emitters with normally distributed intensities of 300 ± 30 (a), (b), 800 ± 100 (c), (d), and 5000 ± 30 (e), (f). Localization error is calculated as the distance from the estimated position to the found position and in all cases assumes Nmax = 5. The median localization error is where the cumulative distribution reaches 0.5. Localization fraction is the fraction of emitters that are correctly localized as determined by being found within either 20 nm or 50 nm from the known position.
5.4. Pattern simulation results
Simulated Siemens star pattern images were generated such that the labeled region active emitter density is 1.0 μm−2 and 6 μm−2. These two sets of data were analyzed using Nmax = 1 and Nmax = 5. Results of the analysis are shown in Fig. 6c through Fig. 6f.
Fig. 6.
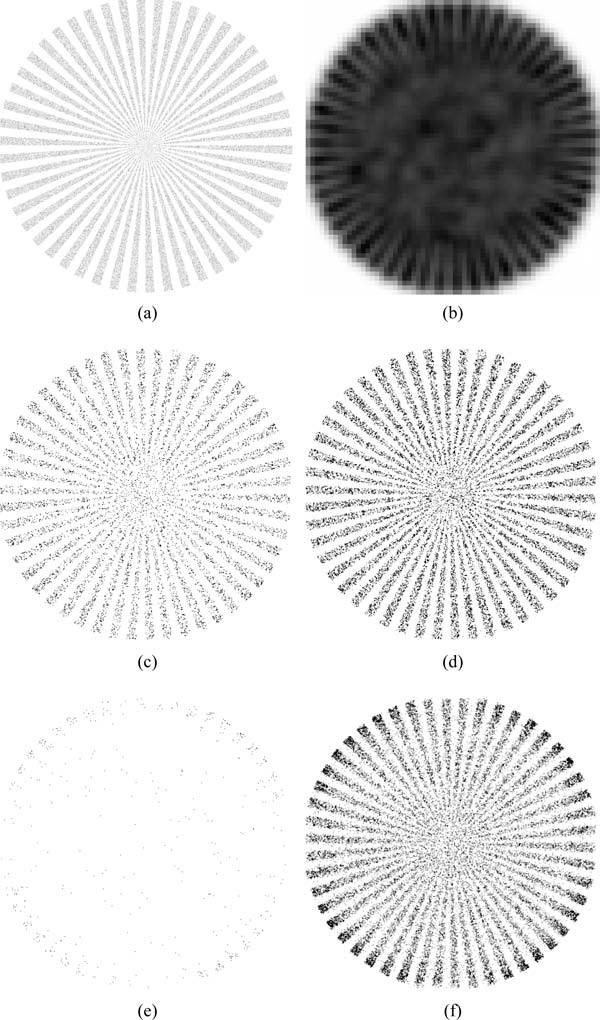
(a) The emitter position histogram used in generating synthetic data. (b) Sum projection of the generated image. (c) Single emitter fitting result at a density of 1 μm−2 with Nmax = 1. (d) Multiple emitter fitting result at a density of 1 μm−2 with Nmax = 5. (e) Single emitter fitting result at a density of 6 μm−2 with Nmax = 1. (f) Multiple emitter fitting result at a density of 6 μm−2 with Nmax = 5. At 1 μm−2 case, Nmax = 1 resulted in 12848 emitters localized while Nmax = 5 localized 30354 emitters. While in 6 μm−2 case, Nmax = 1 resulted in 519 emitters localized while Nmax = 5 localized 33580 emitters. The contrast of images (c) to (f) were globally adjusted across all images for optimal display.
At relatively low densities, results from Nmax = 1 and Nmax = 5 are similar. For Nmax = 1 shown in Fig. 6(c), 12848 emitters were localized and accepted for use in the SR reconstruction, and for Nmax = 5 shown in Fig. 6(d), 30354 emitters were accepted and used. In the high density case, shown in Fig. 6(e) and Fig. 6(f), there was nearly two orders of magnitude (519 versus 33580) more position estimations accepted and used in the reconstruction. As shown in the projections of the SR images, the Nmax = 1 fitting performs better near the edges of the structures where the local active emitter density is smaller. It is interesting to note that at the low density, Nmax = 5 fits almost 3 times more emitters than Nmax = 1 case, and thus the pattern result shows a better resolved structure near the center and provides better resolution compared to Nmax = 1 fitting result.
5.5. Algorithm speed
Algorithm speed was tested under conditions including various active densities and Nmax. Tests were performed on two set of data (data size: 128×128×1000) with densities 1 μm−2 and 5 μm−2. Algorithm execution was divided into several major sections and the run times for each section were recorded. As shown in Table 1, the operation time for MFA Nmax = 5 was 176 s for the 1 μm−2 case and 408 s for the 5 μm−2 case. When performing single emitter operation (Nmax = 1), this run time decreased dramatically to 17 s and 30 s respectively. This dramatic difference is caused by the complexity introduced by fitting multiple emitters, such as fitting to multiple models, the deflation process, NR iteration on more parameters, the Fisher information modification and also a larger Fisher information matrix. However, the fraction of localization also dramatically increased when comparing single fitting results to multi fitting results as over 100 times more emitters were localized at a density of 5 μm−2 and almost 3 times more at a density of 1 μm−2.
Table 1.
Time Consumption and Performance*
| Processing Time (s) | Performance | |||||
|---|---|---|---|---|---|---|
| Fitting Routine | Preprocessing CPU | Fitting GPU | Uncertainty GPU | Reconstruction GPU | Total Time | Localized emitters |
| Low Density: 1 μm−2, Total Simulated Emitters: 181222 | ||||||
| Nmax = 1 | 5.69 | 8.41 | 2.89 | 0.65 | 17.64 | 62267 |
| Nmax = 5 | 5.72 | 162.25 | 6.04 | 2.62 | 176.63 | 168735 |
| High Density: 5 μm−2, Total Simulated Emitters: 906127 | ||||||
| Nmax = 1 | 5.59 | 19.69 | 5.35 | 0.09 | 30.73 | 3939 |
| Nmax = 5 | 5.7 | 378.56 | 16.0 | 7.70 | 408.07 | 489137 |
Images (128×128×1000) of emitters that were randomly placed with uniform distribution are generated using two active emitter densities. Both were fitted by Nmax = 1 and Nmax = 5 and the speed for each part of the algorithm was recorded. Total numbers of fitting sub-regions are 146,597 and 336,959 in low and high density case respectively.
5.6. Imaging of actin structures
Imaging the actin mesh-work within HeLa cells demonstrates the improvements in SM-SR fitting made possible by the MFA’s multi emitter analysis (Nmax = 5) compared with single emitter analysis (Nmax = 1). For samples with high labeling densities, such as those possible when using small molecule fluorescent probes such as Alexa Fluor 647 phalloidin, regions of interest that could be seen using conventional microscopy (Fig. 7(b)), may appear to be discontinuous when analyzed using Nmax = 1 that can not process high active densities (Fig. 7(d)). By analyzing these data sets using MFA (Nmax = 5), less events were discarded. The reconstructed SR image from Nmax = 5 showed more continuous structures and ultimately, enabled finer detail of the underlying protein distributions to be revealed (Fig. 7(e)). It is shown in Fig. 7(c) that although the branching structures were resolved nicely using Nmax = 1, structures toward the middle backbone can’t be resolved, because the backbone structure are composed of intercrossing actin fibers and thus possessed a higher local emitter density than isolated line structures. As shown in Fig. 7(e), MFA (Nmax = 5) achieved to resolve the backbone structure better than single emitter fitting algorithm (Nmax = 1).
Fig. 7.
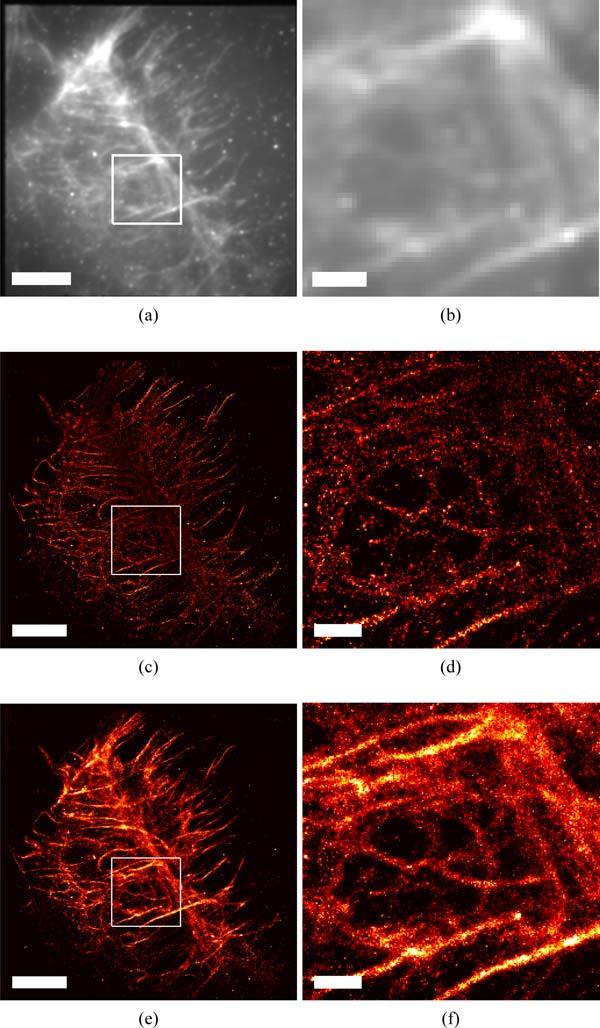
Comparison of SM-SR fitting routines for imaging the actin mesh-work within a HeLa cell labeled with Alexa 647 phalloidin. Conventional TIRF microscopy, (a) and (b), compared with SM-SR images generated using both a Nmax = 1, (c) and (d), and Nmax = 5, (e) and (f). Actin rich regions, seen in top right of (b),(d),(f) are missing using single emitter routines (Nmax = 1) (d), but successfully fit using the MFA (Nmax = 5) (f). The increase in molecular density found using the MFA (Nmax = 5) routine also reveals a more complete depiction of the underlying actin structure, outlining possible actin corrals seen in the center of (f). Scales bars represent 5 μm in (a), (c), (e) and 1 μm in (b), (d), (f).
6. Conclusion
The MFA method we have developed allows the analysis of images with average active emitter densities up to 10 μm−2. This capability relaxes an important constraint in SM-SR, allowing an order of magnitude improvement in the speed of acquisition and/or the maximum supported duty cycle of the emitters. Although our approach is based on a maximum likelihood estimate, robust estimation of multiple emitter positions also requires strategies such as making good initial estimates, making accurate uncertainty estimates and the model selection and rejection algorithm. Higher density imaging allows shorter acquisition times, but results in more computational complexity in analysis. By implementing key analysis steps in GPU hardware, we show the MFA method can complete the analysis of real data sets on the time scale of minutes.
Acknowledgments
We want to thank Marcel Bruchez and Yan Qi from Carnegie Mellon University for providing HeLa cell line for this work. Bernd Rieger and Robert Nieuwenhuizen are thanked for the helpful suggestions and discussions for the manuscript. We also thank W. Duncan Wadsworth for insightful discussions about test statistics. This work was supported by NIH grant #R21RR024438, NIH grant #1P50GM085273, NIH grant #2U54RR022241 and NSF grant #0954836.
References and links
- 1.Hell S. W., “Far-field optical nanoscopy,” Science 316, 1153–1158 (2007). 10.1126/science.1137395 [DOI] [PubMed] [Google Scholar]
- 2.Chi K. R., “Microscopy: ever-increasing resolution,” Nature 462, 675–678 (2009). 10.1038/462675a [DOI] [PubMed] [Google Scholar]
- 3.Huang B., Bates M., Zhuang X. W., “Super-resolution fluorescence microscopy,” Annu. Rev. Biochem. 78, 993–1016 (2009). 10.1146/annurev.biochem.77.061906.092014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patterson G., Davidson M., Manley S., Lippincott-Schwartz J., “Superresolution imaging using single-molecule localization,” Annu. Rev. Phys. Chem. 61, 345–367 (2010). 10.1146/annurev.physchem.012809.103444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schermelleh L., Heintzmann R., Leonhardt H., “A guide to super-resolution fluorescence microscopy,” J. Cell Biol. 190, 165–175 (2010). 10.1083/jcb.201002018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lidke K. A., Rieger B., Jovin T. M., Heintzmann R., “Superresolution with quantum dots: enhanced localization in fluorescence microscopy by exploitation of quantum dot blinking,” Biophys. J. 88, 346a–346a (2005). [Google Scholar]
- 7.Lagerholm B. C., Averett L., Weinreb G. E., Jacobson K., Thompson N. L., “Analysis method for measuring submicroscopic distances with blinking quantum dots,” Biophys. J. 91, 3050–3060 (2006). 10.1529/biophysj.105.079178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Betzig E., Patterson G. H., Sougrat R., Lindwasser O. W., Olenych S., Bonifacino J. S., Davidson M. W., Lippincott-Schwartz J., Hess H. F., “Imaging intracellular fluorescent proteins at nanometer resolution,” Science 313, 1642–1645 (2006). 10.1126/science.1127344 [DOI] [PubMed] [Google Scholar]
- 9.Hess S. T., Girirajan T. P. K., Mason M. D., “Ultra-high resolution imaging by fluorescence photoactivation localization microscopy,” Biophys. J. 91, 4258–4272 (2006). 10.1529/biophysj.106.091116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rust M. J., Bates M., Zhuang X. W., “Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (storm),” Nat. Methods 3, 793–795 (2006). 10.1038/nmeth929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogelsang J., Steinhauer C., Forthmann C., Stein I. H., Person-Skegro B., Cordes T., Tinnefeld P., “Make them blink: probes for super-resolution microscopy,” Chemphyschem 11, 2475–2490 (2010). 10.1002/cphc.201000189 [DOI] [PubMed] [Google Scholar]
- 12.Hell S. W., Wichmann J., “Breaking the diffraction resolution limit by stimulated-emission—stimulated-emission-depletion fluorescence microscopy,” Opt. Lett. 19, 780–782 (1994). 10.1364/OL.19.000780 [DOI] [PubMed] [Google Scholar]
- 13.Hell S., Stelzer E. H. K., “Fundamental improvement of resolution with a 4pi-confocal fluorescence microscope using 2-photon excitation,” Opt. Commun. 93, 277–282 (1992). 10.1016/0030-4018(92)90185-T [DOI] [Google Scholar]
- 14.Gustafsson M. G. L., “Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution,” Proc. Natl. Acad. Sci. U. S. A. 102, 13081–13086 (2005). 10.1073/pnas.0406877102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Small A. R., “Theoretical limits on errors and acquisition rates in localizing switchable fluorophores,” Biophys. J. 96, L16–L18 (2009). 10.1016/j.bpj.2008.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ram S., Ward E. S., Ober R. J., “Beyond rayleigh’s criterion: A resolution measure with application to single-molecule microscopy,” Proc. Natl. Acad. Sci. U.S.A. 103, 4457–4462 (2006). 10.1073/pnas.0508047103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chao J., Ram S., Ward E. S., Ober R. J., “A comparative study of high resolution microscopy imaging modalities using a three-dimensional resolution measure,” Opt. Express 17, 24377–24402 (2009). 10.1364/OE.17.024377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Högbom J. A., “Aperture synthesis with a non-regular distribution of interferometer baselines,” Astron. Astrophys. Suppl. 15, 417 (1974). [Google Scholar]
- 19.Serge A., Bertaux N., Rigneault H., Marguet D., “Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes,” Nat. Methods 5, 687–694 (2008). 10.1038/nmeth.1233 [DOI] [PubMed] [Google Scholar]
- 20.Qu X. H., Wu D., Mets L., Scherer N. F., “Nanometer-localized multiple single-molecule fluorescence microscopy,” Proc. Natl. Acad. Sci. U.S.A. 101, 11298–11303 (2004). 10.1073/pnas.0402155101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon M. P., Ha T., Selvin P. R., “Single-molecule high-resolution imaging with photobleaching,” Proc. Natl. Acad. Sci. U.S.A. 101, 6462–6465 (2004). 10.1073/pnas.0401638101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang B., Zerubia J., Olivo-Marin J. C., “Gaussian approximations of fluorescence microscope point-spread function models,” Appl. Opt. 46, 1819–1829 (2007). 10.1364/AO.46.001819 [DOI] [PubMed] [Google Scholar]
- 23.Stallinga S., Rieger B., “Accuracy of the gaussian point spread function model in 2d localization microscopy,” Opt. Express 18, 24461–24476 (2010). 10.1364/OE.18.024461 [DOI] [PubMed] [Google Scholar]
- 24.Van den Bos A., Ebooks C., Parameter Estimation for Scientists and Engineers (Wiley Online Library, 2007). [Google Scholar]
- 25.Smith C. S., Joseph N., Rieger B., Lidke K. A., “Fast, single-molecule localization that achieves theoretically minimum uncertainty,” Nat. Methods 7, 373–U52 (2010). 10.1038/nmeth.1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ober R. J., Ram S., Ward E. S., “Localization accuracy in single-molecule microscopy,” Biophys. J. 86, 1185–1200 (2004). 10.1016/S0006-3495(04)74193-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoica P., Marzetta T. L., “Parameter estimation problems with singular information matrices,” IEEE Trans. Sig. Process. 49, 87–90 (2001). 10.1109/78.890346 [DOI] [Google Scholar]
- 28.Hendriks C. L. L., van Vliet L. J., Rieger B., van Kempen G. M. P., van Ginkel M., “Dipimage: a scientific image processing toolbox for MATLAB,” Quantitative Imaging Group, Faculty of Applied Sciences, Delft University of Technology, Delft, The Netherlands (1999).
- 29.NVIDIA , “Compute unified device architecture (CUDA),” http://www.nvidia.com/object/cuda_home.html (2007).
- 30.Press W. H., Teukolsky S. L. A., Flannery B. N. P., Vetterling W. M. T., Numerical Recipes: FORTRAN (Cambridge University Press, 1990). [Google Scholar]
- 31.Heilemann M., van de Linde S., Schuttpelz M., Kasper R., Seefeldt B., Mukherjee A., Tinnefeld P., Sauer M., “Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes,” Angew. Chem. Int. Ed. 47, 6172–6176 (2008). 10.1002/anie.200802376 [DOI] [PubMed] [Google Scholar]


