In the last several years, the momentum towards utilizing global profiling methods has increased, and many studies have incorporated these approaches in order to gain insights into previously undetected host or microbe responses during the host-microbe encounter. Over 100 studies available on NIH Pubmed have used microarrays or similar technologies to assess the transcriptional basis for observed physiological outcomes. Further, these studies have revealed a high level of transcriptional activity that is not readily observable and, indeed, often phenotypically silent; presumably the cell’s attempt to maintain homeostasis and symbiotic harmony (12, 28). Additionally, proteomic studies which profile the entire arsenal of proteins present in a cell at a given time are now also possible. Each technique has its own advantages and limitations, and the technologies themselves are reviewed elsewhere (3, 10, 16, 19, 20, 30, 34). As also reviewed previously, the epithelium is the first line of defense to human infections initiating at mucosal membranes (4, 7), and several studies have demonstrated the specific and active nature of the epithelial response to microbial encounters. The response of the epithelium to bacterial, viral, fungal and protozoal challenge has yielded significant insights into the role of the epithelium in host defense. In addition to a passive role as a physical barrier, the epithelium actively participates in the recruitment of immune effectors, and can directly fend off microbes via defensins and other innate immune mechanisms.
Despite the useful information that can be gained from transcriptional or proteomic approaches that study the epithelium, only a limited number of studies have been performed which are specifically targeted to the oral epithelium. Even fewer are targeted specifically to probe host-pathogen interactions of the oral epithelium, as many global genomic studies focus on oral carcinogenesis. Nonetheless, profiling of the host epithelium and/or the infecting microbe has emerged as a useful reporter of specific interactions. Both in vitro and in vivo studies have been performed which have yielded potential markers for periodontal disease progression, novel targets for therapeutics, and have improved the general understanding of host-pathogen cross talk. This review will highlight some important studies which have been conducted to gain insight into cellular and bacterial transcriptional and proteomic profiles associated with the oral cavity. In general, we will consider both sides of the story; the host perspective and that of the colonizing microbe. In closing, the state of the field will be considered and some suggestions will be made to best utilize existing data and overcome existing challenges to understanding the complex interplay between host and microbe in the oral cavity.
In vitro transcriptional studies
In vitro transcriptional studies have encompassed a wide variety of experimental designs (17, 27). Microbial responses to host cell components, nutrient depletion, and antimicrobials, are some strategies designed to evaluate the microbial response to its environment. The cell culture models of epithelial cells exposed to whole bacteria, lipopolysaccharide or other microbial components—free standing or in combination with— mutant analysis, or pre-treatment of host cells with immune effectors followed by bacterial challenge, are utilized. The overall goal is to recreate the interactions in the oral cavity without introducing noise into the system that makes deciphering the transcriptome or proteome problematic. The dilemma is between recreating the complexity of the oral cavity for the sake of accuracy, and the need for simplicity in order to comprehend the profiles uncovered. This section will reflect the attempts to rectify this disconnect, as experimental designs are becoming more complex by utilizing combinations of bacteria infecting an epithelial cell line, the use of regenerated tissue systems, and the combination of epithelial cells with immune cells to tease out the cascades of signaling events that occur after the initial host-microbe encounter and the downstream ripple effect of these responses.
Transcriptional profiling of oral microbes has been performed to compare the gene expression profiles of microbes under typical in vitro culture conditions with those of microbes in contact with host cells or host cell secreted factors and cell extracts. Recently, Almeida and colleagues (2) have assessed the Candida albicans iron acquisition system by transcriptional profiling of C. albicans isogenic mutants, in comparison to wild-type, in the presence of ferritin. Initially, epithelial monolayers were manipulated to produce depleted or enhanced levels of intracellular ferritin. These monolayers were next infected with the C. albicans strains. The monolayers with low levels of intracellular ferritin were protected from invasion by C. albicans, whereas the monolayers with enhanced ferritin levels were damaged more than untreated cells, due to increased C. albicans invasion. To investigate the iron acquisition strategy of C. albicans in this model, and after determining that hyphae were responsible for ferritin binding by C. albicans, transcriptional profiling was performed on three strains of C. albicans infecting oral epithelial monolayers. The first strain was a wild-type strain, with normal ferritin binding activity. The second strain was mutant with an intermediate phenotype in regards to hyphal formation (Δhgc1), and was thus attenuated in its ability to bind ferritin. The third strain was fully deficient in hypha formation (Δras1). The rationale was that candidate genes encoding putative ferritin receptors should be up-regulated in wild-type and Δhgc1 cells, but should be unaltered or down-regulated in the Δras1 mutant. A total of 22 genes met these criteria, and three of these were known surface localized hyphal proteins and thus chosen for further study. The result of this study is a novel proposed mechanism for acidification of epithelial cells and subsequent utilization of intracellular ferritin by C. albicans, and a new avenue of study for C. albicans pathogenesis of oral epithelial cells.
A recent extensive review by Handfield et al. (12) analyzed the response of another oral epithelial cell line; human immortalized gingival keratinocytes (HIGKs) to four distinct oral microbes: Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Fusobacterium nucleatum, and Streptococcus gordonii. HIGKs were produced from primary cultures of gingival epithelial cells and express differentiation markers of junctional epithelium (31). The review built upon previous work in which Affymetrix HGU133A and U133 Plus 2.0 Genechips were used to probe the transcriptional response of the HIGK cells to co-culture with oral microbes after 2 hours (11, 13, 26). The resulting profiles were further analyzed using gene ontology tools to identify biological pathways significantly regulated. Additionally, the phenotypic outcomes were predicted and validated in several instances. The major finding was that the transcriptional profiles resulting from the infection of HIGK cells displayed very little consistency among the four species. Overall, F. nucleatum and S. gordonii perturbed the gingival epithelial cell transcriptome much less significantly than P. gingivalis and A. actinomycetemcomitans, correlating with the two latter microbes’ recognized roles as more overt pathogens. Consideration of the degree of perturbation in pathways comprising apoptosis and MAPK signaling, for example, suggests that there is a great degree of host adaptation by the less pathogenic species which are not inducing proinflammatory responses. In contrast, the recognized pathogens A. actinomycetemcomitans and P. gingivalis both perturbed the host transcriptome to greater extents than S. gordonii or F. nucleatum (12). Pathways that were impacted by A. actinomycetemcomitans and P. gingivalis include Regulation of cell cycle, Cytokine activity, and Programmed cell death. Fig. 1 illustrates the impact upon several key regulators of the host cell cycle by A. actinomycetemcomitans challenge. Further studies dissecting the contribution of specific bacterial components, such as the cytolethal distending toxin or leukotoxin, to host transcriptional regulation of the cell cycle can now be performed using mutant strains. Another finding that arose from these studies was the differential impact of each pathogen upon the p53 branch of the apoptotic pathway. A. actinomycetemcomitans was discovered to induce the pro-apoptotic molecules BBC3, GADD45A, E2F1 and ATM while repressing cMYC expression. The net effect of this transcriptional profile is a pro-apoptotic phenotype (11). Conversely, P. gingivalis induced a response in HIGK cells that was predicted to be anti-apoptotic, or pro-survival, as cMAC, SGK, Bcl2 and Bfl1 were activated. These effectors combine to inhibit apoptosis through repression of p53 as well as inhibition of cytochrome c release from the mitochondria. Importantly, these predicted phenotypes—pro-apoptotic for A. actinomycetemcomitans infection and anti-apoptotic for P. gingivalis infection—were corroborated by a DNA fragmentation assay to measure apoptotic activity in HIGK cells (11). Strikingly, P. gingivalis was also able to attenuate camptothecin-induced apoptosis in HIGK cells. Similar instances of unique and tailored responses to each organism exist, and provide new insight into the degree of perturbation in the host cells. Collectively, these findings suggest that traditional designations of “commensal” or “pathogenic” are insufficient to reflect the unique interplay that occurs between each organism and the host. Thus, the host transcriptional profiling can be a useful tool to probe the degree of microbial damage inflicted in the absence of preconceived ideas of what constitutes a pathogenic or commensal organism.
Fig. 1. HIGK Cell Cycle Transcriptional Response to Aggregatibacter actinomycetemcomitans.
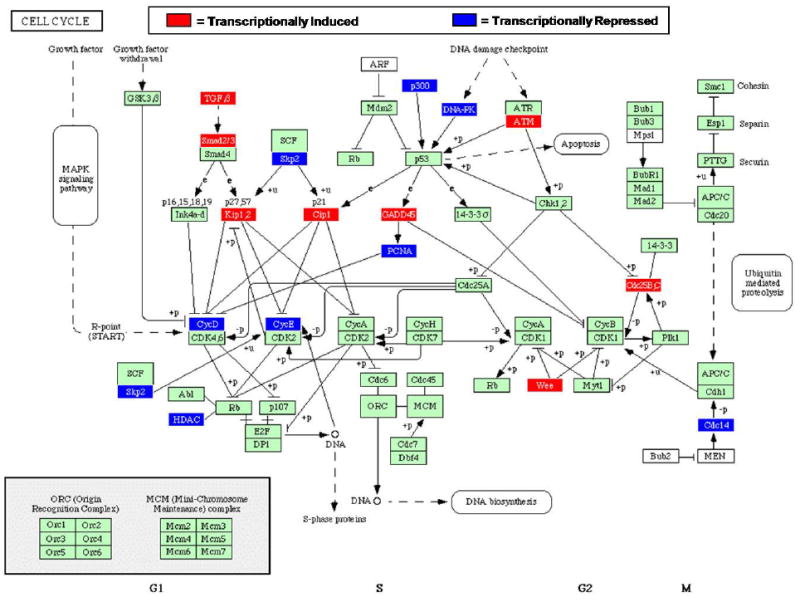
BRB Array Tools was used to generate probesets differentially regulated at the P<0.05 level of significance. The geometric mean signal intensity level for probesets passing this threshold were analyzed with Pathway Express software to populate known KEGG pathways according to transcriptional profiles obtained from GeneChip experiments. Nodes shown in red are transcriptionally induced in HIGK cells encountering wild-type A. actinomycetemcomitans compared to uninfected cells. Nodes shown in blue are transcriptionally repressed. Nodes in green are unchanged at the P<0.05 significance level.
Mutant analysis approaches have been successfully applied to transcriptional profiling experiments to probe the contribution of specific bacterial components to the host-pathogen interaction in A. actinomycetemcomitans and P. gingivalis. The aforementioned study (11) also assessed the contributions of the major fimbria of P. gingivalis, and a conserved gene with unknown function, ORF859, in A. actinomycetemcomitans. Isogenic mutant strains and wild type parental strains of these organisms were allowed to interact with oral epithelial cells, and the host transcriptomes were compared. Novel insights were gained into the pathogenic properties of these organisms, such as fimbrial-triggered regulation of genes related to the cytoskeleton and to membrane and receptor activity by P. gingivalis. The regulation of genes in these pathways corroborated earlier observations and indeed provided the transcriptional “reason” that fimbriae-deficient mutants were significantly impaired in their invasion and cytoskeletal remodeling activities in host cells (41, 42). The transcriptome of HIGK cells in contact with the ORF859 mutant in A. actinomycetemcomitans suggested the involvement of intermediate metabolism functions, signal transduction and cytokine activity by this bacterial component. This study thus provided the first insight into the function of an uncharacterized bacterial gene, although further work is necessary to corroborate the role of this bacterial component.
The design of the above studies utilized single microbes in combination with host cells, however this is not an accurate reflection of the conditions in the oral cavity. Over 700 species are capable of inhabiting the oral cavity, with around 200 colonizing any given individual in unique combinations (1). Thus, to expand the power of transcriptional profiling to unravel host-microbe interactions, we chose to compare transcriptomes of oral epithelial cells (HIGK) encountering P. gingivalis or S. gordonii mono-cultures to the response of a mixed culture of these organisms. Hence microbe-microbe interactions also entered into the equation, and the balance of these interactions affected the host cell transcriptional response in unforeseen ways. For example, the overall transcriptional profile was shifted towards that of S. gordonii infection in mixed cultures of P. gingivalis and S. gordonii. This phenotype was not attributed to S. gordonii preventing P. gingivalis attachment to host cells, indicating that S. gordonii can insulate host cells’ regulatory pathways from P. gingivalis’ attempts at reprogramming.
A similar experiment performed for F. nucleatum in combination with S. gordonii showed mixed-infection dependent novel effects upon the host transcriptional response. The transcriptional profile of HIGK cells in contact with F. nucleatum or S. gordonii alone demonstrated downregulation of actin polymerization as compared to uninfected HIGK cells. However, when both bacteria were co-cultured simultaneously with HIGK cells, actin polymerization becomes upregulated. This approach thus has the potential to yield a novel understanding of the interactions occurring in the oral cavity between epithelial cells and communities of oral microbes. Along the same lines, other groups have begun to increase the model complexity on the host side though utilization of epithelial model systems consisting of multiple layers of epithelial cells, or layers consisting of different cell types (5) to more accurately reflect gingival tissue. This strategy is likely to be pressed to the limit, in an attempt to attain the anatomical and physiological accuracy of in vivo human tissue with the controlled environment characteristic of in vitro models.
In vivo transcriptional studies
While in vivo studies have obvious advantages, as Handfield and colleagues (12) noted, clinical studies involving patient volunteers present the greatest potential for uncontrolled experimental variables. Such variables include, but are not limited to: age of subject, diet, diurnal variations in gene expression, type of anesthesia used, length of ischiemia prior to tissue removal, time from tissue removal to RNA stabilization, and method of RNA isolation. Further, the heterogeneity of cell types comprising gingival tissues which are isolated for transcriptional study complicates the deciphering of transcriptional data. Even when distinct profiles emerge in healthy tissues compared to diseased tissue, it is difficult to assign a pathway level host response to a specific cell type.
Despite the challenges, Demmer and colleagues (9) recently reported a vast and pioneering study in which 90 patients contributed 2 diseased papillae and one healthy papilla (if available) to transcriptional profiling. A total of 247 arrays were run, which identified 12,744 differentially expressed genes. This ambitious study highlights some challenges to transcriptional profiling, and also yields insight into the pathways and underlying gene regulation of these pathways that distinguish healthy from diseased periodontal tissue. As the authors reported, the tissue was heterogeneous and thus consisted of epithelial tissue, connective tissue, and inflammatory infiltrates. The outcome nonetheless was identification of novel targets for further investigation for their potential role in periodontitis, in particular CXCL6 (granulocyte chemoattractant protein 2), and confirmation of overexpression of multiple MMPs (matrix metalloproteinases) in diseased tissue. Importantly, the dataset is available for other groups to mine for meaningful expression patterns, as gene expression omnibus GSE 10334. As the vast amount of information from these experiments can be overwhelming and the potential areas of investigation often are beyond a single laboratory’s scope and area of expertise, this sharing of raw data is a critically important component for new transcriptional findings; and revisiting old datasets by outside groups is likely to generate important new information. There is a wealth of information available without the prohibitive expense of performing the actual array experiments.
In 2006, Kim and colleagues (18) sought to elucidate host factors which may explain the occurrence of refractory periodontitis in certain individuals. A total of 14 patients were involved in a study to examine the transcriptional profiles from sub-epithelial connective tissues in healthy or diseased individuals. Seven patients undergoing a crown extension procedure donated tissue as healthy controls; and another seven refractory periodontitis patients donated tissue from active periodontitis sites. The study uncovered the differential expression of 74 genes using two-fold expression changes as the cutoff, 68 of which were upregulated. As Table 1 illustrates, twelve genes were particularly interesting to the authors because the fold change ratios were pronounced as compared to healthy controls. Seven genes were confirmed by quantitative RT-PCR. The confounding variables were seemingly reduced in this tightly controlled case-matched study. Furthermore, the tissue type profiled was more stringently controlled based on histological examination. However, the small number of participants and low number of genes identified demonstrates the inherent challenges arising during in vivo studies. However, availability of this dataset for analysis by other methods may yield more genes which are differentially expressed at a level attaining statistical significance without meeting the cutoff threshold of 2-change.
Table 1.
Selected Genes Significantly Regulated in Refractory Periodontitis Compared to Healthy Controls*
| Upregulated | |
|---|---|
| Lactotransferrin | LTF |
| Matrix metallinoproteinase-1 | MMP-1 |
| Matrix metallinoproteinase-3 | MMP-3 |
| Interferon induced-15 | IFI-15 |
| Hypothetical protein MGC5566 | MGC5566 |
| V-FOS FBJ murine osteosarcoma viral oncogene homolog B | FOSB |
| Caspase-10 | CASP10 |
| Interleukin-24 | IL24 |
| Rho GTPASE activating protein 8 | ARHGAP8 |
| Downregulated | |
| Keratin 2A | KRT2A |
| Desmocollin-1 | DSC-1 |
| Transmembrane 7 superfamily member 3 | TM7SF3 |
Adapted from (25)
An important technological advance that addresses the problem of tissue heterogeneity is the use of laser microdissection, which allows the isolation of a homogeneous population of cells prior to RNA extraction and processing for array hybridization and analysis. In addition to reducing the background in transcriptional profiling experiments, this approach can aid in the interpretation of the data after significance analysis. For example, in an approach that identifies multiple pathways with a diversity of functions and sharing one or more genes, judgments can be made regarding the relevance of these pathways for a given cell type. If the physiology of the isolated cell types is known, then pathways that are biologically irrelevant can be eliminated even if the statistical analysis identifies that pathway as a high probability target. One drawback to this approach however, is the low yield of RNA often limits or precludes effective processing for microarray experiments. As Clement-Ziza et al. (6) recently reported, several commercially available kits are designed to amplify the small amount of RNA uncovered from techniques such as laser microdissection prior to cDNA synthesis with good results. It is not clear however, if the sampling error introduced during the RNA amplification step is greater or equal to that expected when large amounts of RNA are harvested from heterogeneous tissue sources. One would expect the result to be better for homogeneous cell types with low RNA yield, but this has not been conclusively demonstrated. Ma and colleagues (25) used this strategy to identify 2351 differentially expressed genes in 8 tumor samples compared to 8 matched/adjacent normal tissue samples. The numbers of identified genes in this study are more comparable to that found from in vitro studies using quadruplicate biological samples for each condition, although less than the 12,000 genes identified in vivo by Demmer and colleagues (9). However, from a cost perspective, it is clear that laser microdissection in conjunction with microarray analysis can reduce the numbers of samples required to obtain a large pool of candidate genes while running significantly fewer arrays. As more advancements occur, laser microdissection and microarray analysis will become a more affordable option that would likely prove useful in studies of the oral cavity.
Simultaneous host and pathogen profiling
Few studies have been conducted with the intention of identifying crosstalk between host and microbe, although this scenario is arguably the most interesting or informative scenario in experimental design. To date this approach has not been utilized to study host-microbe interactions of the oral cavity. However, the two studies summarized below exemplify the type of information that can be gained from this strategy should it be applied to the study of periodontal health and disease. Some of the technical issues associated with this approach have been addressed by the following studies and they thus serve as a model for similar experiments to study periodontal diseases.
Motley and colleagues (29) utilized a murine granulomatous pouch model to study the host response to bacterial infection. Although this model is not anatomically accurate for several diseases, the physiological response is representative of the host response to a local infection. A number of microbial species can potentially be tested with this method, to assess the resulting immune response. Briefly, the pouch environment was created by injecting air into the mouse intracutaneously, and subsequently injecting a croton oil-olive oil mixture. This created an in vivo testing chamber into which bacteria were injected and the infection was allowed to follow a time course. Fluids from the pouch were taken at several time points, and following RNA extraction and cDNA preparation, mouse and bacterial microarrays were probed with the sample target. Probes which cross hybridized to both arrays were removed from consideration. The authors uncovered a novel bacterial stress response, as well as interesting profiles on the host side specifically related to cytokine and chemokine gene expression.
Also in a mouse model, Lovegrove and colleagues (24) utilized a model of cerebral malaria (CM) caused by Plasmodium berghei ANKA (PbA), which approximates to human CM. The authors designed “combination” microarrays to simultaneously profile PbA and mouse responses to the experimental CM infection. Expression data from brain, lung, liver, and spleen of PbA infected mice were compared between CM-susceptible (C57BL/6) and CM-resistant (BALB/c) mice, in addition to profiling of the parasite using a single microarray. Specific parasite transcriptional signatures in each tissue were uncovered, and lung tissue was identified as a potential large reservoir for metabolically active parasites. On the host side, distinct, organ-specific transcriptional profiles were also observed.
These experiments demonstrate the potential for expanding transcriptional profiling in the oral cavity to study host and microbe simultaneously. While not necessarily a component of simultaneous host and pathogen profiling, both experiments described here also attempted another dimension of infection: changes over time. Novel algorithms and experiments have been performed to identify the dynamic and temporal nature of host and pathogen interactions at the transcriptional level. New statistical analysis approaches are currently being developed with the goal of finding a ‘cause and effect’ relationship between host and pathogen gene regulation events (22, 35, 36). The experiments themselves are easy to conduct from a technical standpoint, but advances in the bioinformatics field are necessary before progress can be made in this regard. This approach has not been undertaken to study the oral cavity, however the methods designed will be applicable to such studies.
Proteomics of in vitro Bacteria-Host Cell Interactions
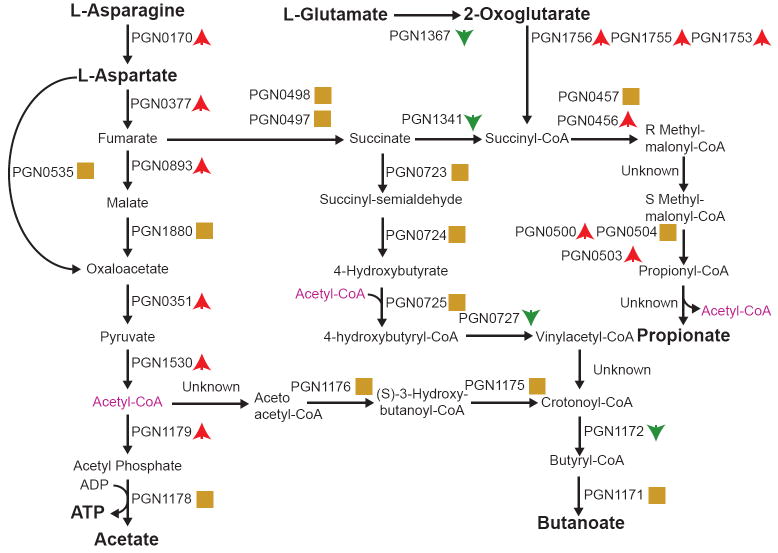
A small number of proteomics studies have been conducted to determine the response of bacteria to host epithelial cells or conditions believed to be important in the infection process. One such condition is iron availability. Indeed, access to iron is a major virulence determinant, and pathogens are unable to grow without a method of deriving iron from their host. P. gingivalis does not make use of the traditional iron binding siderophores, but rather acquires heme groups, which can be generated from hemoglobin and other host heme-containing proteins using cell surface proteinase-adhesion complexes (8). Dashper and colleagues examined the response of P. gingivalis to heme limitation using ICAT post-harvest stable isotopic labeling and mass spectrometry (8). Proteins from heme limited cells were labeled with ICAT reagent, separated on SDS gels, and subjected to in-gel tryptic digestion, affinity purification, and mass spectrometry. This study identified 70 proteins with a 2-fold or greater difference between cells grown under the two conditions. Most of these proteins, 53, showed increased levels under heme limitation. A significant change was seen in proteins related to energy metabolism. Normally, P. gingivalis favors aspartate and glutamate as metabolic substrates. Some of the metabolized aspartate passes through pyruvate to produce acetate as a byproduct, while some passes from fumarate to succinate and then on to produce propionate or butyrate as byproducts. Figure 2 shows these metabolic pathways (although not the data from this study). Fumarate reductase, which catalyzes the conversion of fumarate to succinate, is believed to require heme for activity and showed significantly decreased levels under heme limation. Consequently, aspartate would primarily feed down the pathway to acetate. Consistent with this, increased levels of enzymes converting pyruvate to acetate were observed, and acetate production doubled under heme limitation. In addition, increased levels of proteins required for conversion of glutamate to propionate and butyrate were observed, possibly compensating for the loss of succinate. Unsurprisingly, proteins of predicted heme-uptake systems, Hmu and Htr, showed increased expression. The largest increase under heme limitation was the alkyl hydroperoxide reductase protein, a peroxide-scavenging enzyme that plays a role in peroxide resistance. This may be compensating for the loss of heme that P. gingivalis uses for protection from reactive oxidative species. An increase in a number of cell envelope proteins was also observed. Demonstrating a correlation between heme and pathogenicity, a mutant in one of these surface proteins, an internalin-related protein, resulted in decreased internalization of P. gingivalis. Oxidative stress is an important component of the mammalian immune response, and the ability of pathogens to withstand or modulate oxidative stress is a key factor in determining their long-term survival and pathogenic potential. The importance of heme for oxidative stress resistance in P. gingivalis was examined by Okano and colleagues (33). P. gingivalis was grown under hemin limitation to avoid the oxygen protective effects of hemin. The cells where then stressed with oxygen and the proteome analyzed using 2-dimentional gel electrophoresis and mass spectrometry. Nineteen proteins with altered abundance were identified. Interestingly, alkyl hyperoxide reductase C, but not super oxide dismutase, showed an expression increase, although both proteins are believed to be important for oxidative stress protection. Most of the proteins with increased abundance were heat shock proteins such as DnaK, GroEL, and trigger factor. The consequence of such regulation would be an increased ability to recover or dispose of misfolded proteins, such as might be generated by oxygen exposure. The results indicate that despite being an obligate anaerobe, P. gingivalis maintains a number of different systems for dealing with oxidative stress.
Fig. 2. Metabolic Map of Energy and Cytotoxin Production.
Proteins catalyzing each step are shown by their Porphyromonas. gingivalis PGN (strain 33277) designation. Red up arrows indicate increased levels upon internalization, green down arrows decreased levels, and yellow squares no statistical change. Acetyl-CoA appears as a substrate and product at multiple points and is shown in purple. Metabolites and metabolic precursors discussed in the text are shown in bold. Reprinted from BMC Microbiology, 2009, in press.
Just as bacteria respond to environmental conditions like oxidative stress, they are known to respond to components released into the extracellular milieu by host cells. In order to examine the initial stages of host cell interaction, our laboratory examined the proteome of P. gingivalis cells exposed to tissue culture media that had been used to culture gingival epithelial cells (GECs) (43). P. gingivalis cells exposed to unconditioned culture media served as the control. 60 proteins showed differential abundance between conditioned and unconditioned media; 35 increased and 25 decreased. Several components involved in hemin uptake showed reduced expression levels. In addition, similar to P. gingivalis under oxidative stress, there were increases in the levels of Clp stress response system (ClpC, CplP, and ClpX) as well as several heat shock proteins. The general implication was that the extracellular epithelial cell environment is hemin sufficient but places more stress upon P. gingivalis.
The proteome of internalized P. gingivalis has also been examined (14, 40). Proteins were extracted from P. gingivalis cells 18 hours after internalization into GECs and analyzed by MudPIT. MudPIT, or multidimensional protein identification technology (14, 38), is the current state of the art for proteomics techniques because it combines two phase separation steps with mass spectrometric methods on an automated platform (reviewed in (15, 21, 23, 39)). This stringent separation prior to mass spectrometry is necessary for analysis of complex mixtures of proteins, such as those isolated from host-microbe interaction experiments. Similar to cells exposed to GEC conditioned media, the internalized P. gingivalis showed decreases in hemin uptake proteins as well as increases in stress-related proteins including heat shock proteins, alkyl hydroperoxide reductases, and two Clp proteases, ClpB and ClpC. Internalized cells also showed a significant reduction in the major fimbriae FimA, presumably as fimbriae are superfluous after internalization. Proteases were also downregulated in internalized P. gingivalis, indicating that the organism may control expression of compounds that are toxic to the host cells in order to prolong survival. There were also extensive changes in metabolism (Fig. 2) (14). The main energy producing pathway from asparagine/ aspartate to acetate and ATP had increased levels of protein components. Increased abundance was also seen for proteins in the pathway for production of the cytotoxin propionate, while the butyrate pathway had reduced or unchanged protein levels. The overall trend is to an increase in energy production in internalized cells and a shift in the production of cytotoxic metabolic byproducts from butyrate to propionate. Given that butyrate is a more potent apoptosis inducing agent than propionate, this too is indicative of a shift to a physiology that supports long-term survivability of host cells. Internalized P. gingivalis showed a general increase in the translational machinery. Most translation initiation, elongation and termination proteins as well as tRNA synthetases and ribosomal proteins had increased abundance in internalized P. gingivalis. The transcriptional machinery also showed increased abundance. Combined with the increase in energy metabolism, these results indicate that the interior of GECs is an energy rich environment for P. gingivalis.
The future
The inescapable conclusion from systems based methodologies is that genes and proteins operate within extensively interconnected networks. Studies of single genes/proteins in isolation is rapidly being rendered obsolete, and global expression profiling provides a more natural platform whereby bacterial components are studied in the context of their interconnectivity such as occurs in vivo. As mentioned earlier, an abundance of transcriptional and proteomic data is currently available to the public through various repositories. Table 2 identifies several genomic and proteomic databases with potential application to the study of host-microbe interactions of the oral cavity. A broader listing of data repositories is available at OmicsWorld (www.omicsworld.com). The nature of these global studies results in more data than could ever be reasonably examined by any single group, especially when the research laboratory is focused on specific fields. For this reason, it is necessary that transcriptional and proteomic data be revisited periodically, given advances in data analysis which regularly occur. Furthermore, the costs typically associated with conducting genome wide profiling experiments can be avoided by revisiting deposited datasets, and meta-analysis of several experiments done by different researchers can yield new findings, or evaluate the consistency of core profiles. For example, in the case of two deposited oral carcinoma datasets, GDS1584 and GDS1062 in the GEO (32, 37), it is possible to reanalyze both datasets and identify predictive markers. This highlights the need for researchers to responsibly deposit their datasets in a repository according to MIAME standards in a timely fashion. Seemingly unrelated datasets might be joined in the future, such as host pathogen interaction dataset and a carcinoma dataset, much like H. pylori and HPV are known to contribute to carcinogenesis.
Table 2.
Public Repositories for Genomic or Proteomic Data with Potential Relevance to Host-Pathogen Interactions of the Oral Cavity*
| Proteomics Databases | Website Address |
|---|---|
| Open Proteomics Database (University of Texas) | http://bioinformatics.icmb.utexas.edu/OPD/ |
| Proteome Database of Lactococcus lactis (Technische Universität München) | http://www.wzw.tum.de/proteomik/lactis/ |
| University of Tennessee Proteome and Transcriptome Database | http://www.utmem.edu/proteomics/ |
| PHCI-2DPAGE (University of Aarhus) | http://www.gram.au.dk/ |
| Expert Protein Analysis System (ExPASy) - Swiss Institute of Bioinformatics | http://us.expasy.org/ |
| Proteome Database System for Microbial Research (Max Planck Institute) | http://www.mpiib-berlin.mpg.de/2D-PAGE/ |
| OGP-WWW Database (Oxford GlycoProteomics, UK) | http://proteomewww.glycob.ox.ac.uk/2d/2d.html |
| Phosphosite Plus (Cell Signaling Technology, USA) | http://www.phosphosite.org/homeAction.do |
| Toothprint Dental Tissue Database (University of Otago, New Zealand) | http://toothprint.mdhs.unimelb.edu.au/fmi/xsl/toothprint/home.xsl |
| Genomics Databases | Website Address |
| Array Express (European Bioinformatics Institute) | http://www.ebi.ac.uk/microarray-as/ae/ |
| ExpressDB (Harvard) | http://arep.med.harvard.edu/ExpressDB/ |
| Gene Expression Omnibus (GEO) (National Center for Biotechnology Information) | http://www.ncbi.nlm.nih.gov/geo/ |
| MUSC DNA Microarray Database (Medical University of South Carolina) | http://proteogenomics.musc.edu/ma/musc_madb.php?page=home&act=manage |
| Stanford Microarray Database (SMD) | http://smd.stanford.edu/ |
| BioGPS (Genomics Institute of the Novartis Research Foundation) | http://biogps.gnf.org/#goto=welcome |
| Yale Microarray Database | http://info.med.yale.edu/microarray/yale_links.htm |
Adapted from Omicsworld (http://www.omicsworld.com/)
References
- 1.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almeida RS, Brunke S, Albrecht A, Thewes S, Laue M, Edwards JE, Filler SG, Hube B. the hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog. 2008;4:e1000217. doi: 10.1371/journal.ppat.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auer H, Newsom DL, Kornacker K. Expression Profiling Using Affymetrix GeneChip Microarrays. Methods Mol Biol. 2009;509:35–46. doi: 10.1007/978-1-59745-372-1_3. [DOI] [PubMed] [Google Scholar]
- 4.Bosshardt DD, Lang NP. The junctional epithelium: from health to disease. J Dent Res. 2005;84:9–20. doi: 10.1177/154405910508400102. [DOI] [PubMed] [Google Scholar]
- 5.Carlson MW, Alt-Holland A, Egles C, Garlick JA. Three-dimensional tissue models of normal and diseased skin. Curr Protoc Cell Biol. 2008;Chapter 19(Unit 19):19. doi: 10.1002/0471143030.cb1909s41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clement-Ziza M, Gentien D, Lyonnet S, Thiery JP, Besmond C, Decraene C. Evaluation of methods for amplification of picogram amounts of total RNA for whole genome expression profiling. BMC Genomics. 2009;10:246. doi: 10.1186/1471-2164-10-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dale BA. Periodontal epithelium: a newly recognized role in health and disease. Periodontol 2000. 2002;30:70–78. doi: 10.1034/j.1600-0757.2002.03007.x. [DOI] [PubMed] [Google Scholar]
- 8.Dashper SG, Ang CS, Veith PD, Mitchell HL, Lo AW, Seers CA, Walsh KA, Slakeski N, Chen D, Lissel JP, Butler CA, O’Brien-Simpson NM, Barr IG, Reynolds EC. Response of Porphyromonas gingivalis to heme limitation in continuous culture. J Bacteriol. 2009;191:1044–1055. doi: 10.1128/JB.01270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demmer RT, Behle JH, Wolf DL, Handfield M, Kebschull M, Celenti R, Pavlidis P, Papapanou PN. Transcriptomes in healthy and diseased gingival tissues. J Periodontol. 2008;79:2112–2124. doi: 10.1902/jop.2008.080139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dufva M. Introduction to microarray technology. Methods Mol Biol. 2009;529:1–22. doi: 10.1007/978-1-59745-538-1_1. [DOI] [PubMed] [Google Scholar]
- 11.Handfield M, Mans JJ, Zheng G, Lopez MC, Mao S, Progulske-Fox A, Narasimhan G, Baker HV, Lamont RJ. Distinct transcriptional profiles characterize oral epithelium-microbiota interactions. Cell Microbiol. 2005;7:811–823. doi: 10.1111/j.1462-5822.2005.00513.x. [DOI] [PubMed] [Google Scholar]
- 12.Handfield M, Baker HV, Lamont RJ. Beyond good and evil in the oral cavity: insights into host-microbe relationships derived from transcriptional profiling of gingival cells. J Dent Res. 2008;87:203–223. doi: 10.1177/154405910808700302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasegawa Y, Mans JJ, Mao S, Lopez MC, Baker HV, Handfield M, Lamont RJ. Gingival epithelial cell transcriptional responses to commensal and opportunistic oral microbial species. Infect Immun. 2007;75:2540–2547. doi: 10.1128/IAI.01957-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendrickson E, Xia Q, Wang T, Lamont R, Hackett M. Pathway analysis for intracellular Porphyromonas gingivalis using a strain ATCC 33277 specific database. BMC Microbiology. 2009 doi: 10.1186/1471-2180-9-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendrickson EL, Lamont RJ, Hackett M. Tools for interpreting large-scale protein profiling in microbiology. J Dent Res. 2008;87:1004–1015. doi: 10.1177/154405910808701113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang SH, Triche T, Jong AY. Infectomics: genomics and proteomics of microbial infections. Funct Integr Genomics. 2002;1:331–344. doi: 10.1007/s10142-002-0048-4. [DOI] [PubMed] [Google Scholar]
- 17.Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- 18.Kim DM, Ramoni MF, Nevins M, Fiorellini JP. The gene expression profile in refractory periodontitis patients. J Periodontol. 2006;77:1043–1050. doi: 10.1902/jop.2006.050254. [DOI] [PubMed] [Google Scholar]
- 19.Kozarova A, Petrinac S, Ali A, Hudson JW. Array of informatics: Applications in modern research. J Proteome Res. 2006;5:1051–1059. doi: 10.1021/pr050432e. [DOI] [PubMed] [Google Scholar]
- 20.Kuo WP. Overview of bioinformatics and its application to oral genomics. Adv Dent Res. 2003;17:89–94. doi: 10.1177/154407370301700121. [DOI] [PubMed] [Google Scholar]
- 21.Lamont RJ, Meila M, Xia Q, Hackett M. Mass spectrometry-based proteomics and its application to studies of Porphyromonas gingivalis invasion and pathogenicity. Infect Disord Drug Targets. 2006;6:311–325. doi: 10.2174/187152606778249935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leek JT, Monsen E, Dabney AR, Storey JD. EDGE: extraction and analysis of differential gene expression. Bioinformatics. 2006;22:507–508. doi: 10.1093/bioinformatics/btk005. [DOI] [PubMed] [Google Scholar]
- 23.Lohrig K, Wolters D. Multidimensional protein identification technology. Methods Mol Biol. 2009;564:143–153. doi: 10.1007/978-1-60761-157-8_8. [DOI] [PubMed] [Google Scholar]
- 24.Lovegrove FE, Pena-Castillo L, Mohammad N, Liles WC, Hughes TR, Kain KC. Simultaneous host and parasite expression profiling identifies tissue-specific transcriptional programs associated with susceptibility or resistance to experimental cerebral malaria. BMC Genomics. 2006;7:295. doi: 10.1186/1471-2164-7-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma LJ, Li W, Zhang X, Huang DH, Zhang H, Xiao JY, Tian YQ. Differential gene expression profiling of laryngeal squamous cell carcinoma by laser capture microdissection and complementary DNA microarrays. Arch Med Res. 2009;40:114–123. doi: 10.1016/j.arcmed.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Mans JJ, Baker HV, Oda D, Lamont RJ, Handfield M. Distinctive characteristics of transcriptional profiles from two epithelial cell lines upon interaction with Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 2006;21:261–267. doi: 10.1111/j.1399-302X.2006.00281.x. [DOI] [PubMed] [Google Scholar]
- 27.Mans JJ, Lamont RJ, Handfield M. Microarray analysis of human epithelial cell responses to bacterial interaction. Infect Disord Drug Targets. 2006;6:299–309. doi: 10.2174/187152606778249926. [DOI] [PubMed] [Google Scholar]
- 28.Marsh PD. Are dental diseases examples of ecological catastrophes? Microbiology. 2003;149:279–294. doi: 10.1099/mic.0.26082-0. [DOI] [PubMed] [Google Scholar]
- 29.Motley ST, Morrow BJ, Liu X, Dodge IL, Vitiello A, Ward CK, Shaw KJ. Simultaneous analysis of host and pathogen interactions during an in vivo infection reveals local induction of host acute phase response proteins, a novel bacterial stress response, and evidence of a host-imposed metal ion limited environment. Cell Microbiol. 2004;6:849–865. doi: 10.1111/j.1462-5822.2004.00407.x. [DOI] [PubMed] [Google Scholar]
- 30.Nazmul-Hossain AN, Patel KJ, Rhodus NL, Moser KL. Microarrays: applications in dental research. Oral Dis. 2008;14:25–29. doi: 10.1111/j.1601-0825.2006.01342.x. [DOI] [PubMed] [Google Scholar]
- 31.Oda D, Bigler L, Lee P, Blanton R. HPV immortalization of human oral epithelial cells: a model for carcinogenesis. Exp Cell Res. 1996;226:164–169. doi: 10.1006/excr.1996.0215. [DOI] [PubMed] [Google Scholar]
- 32.O’Donnell RK, Kupferman M, Wei SJ, Singhal S, Weber R, O’Malley B, Cheng Y, Putt M, Feldman M, Ziober B, Muschel RJ. Gene expression signature predicts lymphatic metastasis in squamous cell carcinoma of the oral cavity. Oncogene. 2005;24:1244–1251. doi: 10.1038/sj.onc.1208285. [DOI] [PubMed] [Google Scholar]
- 33.Okano S, Shibata Y, Shiroza T, Abiko Y. Proteomics-based analysis of a counter-oxidative stress system in Porphyromonas gingivalis. Proteomics. 2006;6:251–258. doi: 10.1002/pmic.200401338. [DOI] [PubMed] [Google Scholar]
- 34.Pan S, Aebersold R, Chen R, Rush J, Goodlett DR, McIntosh MW, Zhang J, Brentnall TA. Mass spectrometry based targeted protein quantification: methods and applications. J Proteome Res. 2009;8:787–797. doi: 10.1021/pr800538n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Storey JD, Xiao W, Leek JT, Tompkins RG, Davis RW. Significance analysis of time course microarray experiments. Proc Natl Acad Sci U S A. 2005;102:12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storey JD, Dai JY, Leek JT. The optimal discovery procedure for large-scale significance testing, with applications to comparative microarray experiments. Biostatistics. 2007;8:414–432. doi: 10.1093/biostatistics/kxl019. [DOI] [PubMed] [Google Scholar]
- 37.Toruner GA, Ulger C, Alkan M, Galante AT, Rinaggio J, Wilk R, Tian B, Soteropoulos P, Hameed MR, Schwalb MN, Dermody JJ. Association between gene expression profile and tumor invasion in oral squamous cell carcinoma. Cancer Genet Cytogenet. 2004;154:27–35. doi: 10.1016/j.cancergencyto.2004.01.026. [DOI] [PubMed] [Google Scholar]
- 38.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 39.Xia Q, Hendrickson EL, Wang T, Lamont RJ, Leigh JA, Hackett M. Protein abundance ratios for global studies of prokaryotes. Proteomics. 2007;7:2904–2919. doi: 10.1002/pmic.200700267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia Q, Wang T, Taub F, Park Y, Capestany CA, Lamont RJ, Hackett M. Quantitative proteomics of intracellular Porphyromonas gingivalis. Proteomics. 2007;7:4323–4337. doi: 10.1002/pmic.200700543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yilmaz O, Watanabe K, Lamont RJ. Involvement of integrins in fimbriae-mediated binding and invasion by Porphyromonas gingivalis. Cell Microbiol. 2002;4:305–314. doi: 10.1046/j.1462-5822.2002.00192.x. [DOI] [PubMed] [Google Scholar]
- 42.Yilmaz O, Young PA, Lamont RJ, Kenny GE. Gingival epithelial cell signalling and cytoskeletal responses to Porphyromonas gingivalis invasion. Microbiology. 2003;149:2417–2426. doi: 10.1099/mic.0.26483-0. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, Wang T, Chen W, Yilmaz O, Park Y, Jung IY, Hackett M, Lamont RJ. Differential protein expression by Porphyromonas gingivalis in response to secreted epithelial cell components. Proteomics. 2005;5:198–211. doi: 10.1002/pmic.200400922. [DOI] [PMC free article] [PubMed] [Google Scholar]