Abstract
The biochemistry of eukaryotic homologous recombination caught fire with the discovery that Rad51 is the eukaryotic homolog of the bacterial RecA and T4 UvsX proteins; and this field is still hot. The core reaction of homologous recombination, homology search and DNA strand invasion, along with the proteins catalyzing it, are conserved throughout evolution in principle. However, the increased complexity of eukaryotic genomes and the diversity of eukaryotic cell biology pose additional challenges to the recombination machinery. It is not surprising that this increase in complexity coincided with the evolution of new recombination proteins and novel support pathways, as well as changes in the properties of those eukaryotic recombination proteins that are evidently conserved in evolution. In humans, defects in homologous recombination lead to increased cancer predisposition, underlining the importance of this pathway for genomic stability and tumor suppression. This review will focus on the mechanisms of homologous recombination in eukaryotes as elucidated by the biochemical analysis of yeast and human proteins.
1 Introduction
Homologous recombination (HR) is a ubiquitous cellular pathway that performs template-dependent, high-fidelity repair of complex DNA damage caused by endogenous and exogenous sources including DNA double-stranded breaks (DSBs), DNA gaps, and inter-strand crosslinks. Historically, the interest in HR has been sparked by its essential role during meiosis where recombination is initiated by a specific DSB delivered by the Spo11 transesterase. Moreover, HR is essential in preserving genome stability through its role in the recovery of stalled or collapsed replication forks, as well as its function in telomere maintenance. This review will discuss the mechanisms of HR and its potential regulation as elucidated by the biochemical analysis of recombination proteins from the yeast Saccharomyces cerevisiae and humans. The discussion will concentrate on the core proteins (and their homologs) defined by the RAD52 epistasis group and on a number of context-specific factors that are involved in HR in some sub-pathways. The reader is referred to excellent earlier reviews of recombination models and proteins in eukaryotes (Paques and Haber 1999; Symington 2002; Sung et al. 2003; West 2003; Krogh and Symington 2004; Wyman et al. 2004), as well as to reviews of the paradigmatic Escherichia coli and phage T4 recombination proteins (Kowalczykowski et al. 1994; Beernink and Morrical 1999), in addition to the archaeal recombination proteins (Seitz et al. 2001). The discussion will be limited to recombination in somatic (vegetative) cells as this volume provides a contribution dedicated to meiotic recombination (Hunter). Particular emphasis is given to the assembly and function of the central recombination catalyst, the Rad51-ssDNA presynaptic filament. The article devoted entirely to the late stages of HR (Whitby) will elaborate this topic in much greater detail than attempted here.
2 Homologous recombination in different contexts
HR functions in different contexts with some variations on the types of substrates and intermediates encountered by the recombination machinery. The primary realm of discussing HR has been in the repair of a frank (i.e. two-ended) DSB induced either by ionizing radiation (IR) or an endonuclease (Fig. 1A). Three sub-pathways were proposed to lead to repair of a frank DSB: double-strand break repair (DSBR), synthesis-dependent strand annealing (SDSA), and single-strand annealing (SSA). The enzymatic and mechanistic requirements for these pathways differ. DSBR and SDSA require DNA strand invasion mediated by the Rad51 filament (and the corresponding co-factors needed for filament assembly and function). On the contrary, SSA is Rad51-independent and does not involve strand invasion but rather reannealing of RPA-coated ssDNA. This rather specialized pathway requires direct repeat sequences flanking the breaks and leads to deletion of one repeat and of the intervening DNA. SSA becomes quite relevant with model substrates containing repeated DNA sequences (for review see Paques and Haber 1999; Krogh and Symington 2004), but is not discussed further here. The DSBR and SDSA pathways diverge after D-loop formation, requiring different sets of activities in postsynapsis that function on different types of junctions (Fig. 1A). The repair of a one-sided DSB, formed during break-induced replication (BIR), telomere maintenance or recovery of a broken replication fork brings variations on this theme (Fig. 1B). This pathway likely involves a single Holliday junction instead of the double Holliday junction during DSBR. Gap repair after replication fork blockage poses yet a different substrate for Rad51 filament assembly and likely involves different junction types (Fig. 1C). The different pathways leading to D-loop formation and the processing of the D-loop formed by Rad51 and its cofactors are likely reflected in the differential requirement for specific protein factors, termed here context-specific factors.
Fig. 1.
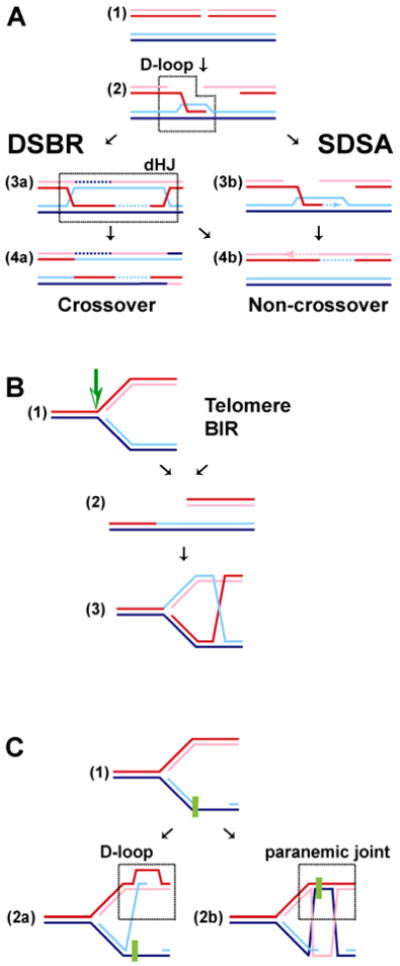
(overleaf). Pathways of homologous recombination. A. Repair of a frank DSB by double-strand break repair (DSBR) and synthesis-dependent strand annealing (SDSA). After Rad51-mediated D-loop formation, the DSBR and SDSA pathways split. In DSBR, a double Holliday junction (dHJ) is generated that can be resolved by a HJ-specific endonuclease into crossover and non-crossover products or dissolved by the action of BLM-TopoIIIα leading to non-crossover products only. In SDSA, the invading strand reanneals after DNA repair synthesis with the second DSB end without generating a dHJ intermediate, leading to non-crossover products only. B. Repair of a one-sided DSB. Cleavage of a stalled replication fork yields a one-sided DSB, a situation that is similar to break-induced replication (BIR) and recombination at chromosome ends (telomeres). Rad51-mediated DNA strand invasion (D-loop) can establish a replication fork with a single Holliday junction. C. Bypass of DNA damage blocking the lagging strand of a replication fork. Rad51-mediated DNA strand invasion using the blocked 3′ end leads to D-loop formation and may proceed either by a DSBR-type pathway (involving a dHJ) or an SDSA-type pathway (without dHJ). This pathway requires a 5′-3′ DNA helicase that peels the blocked strand off the template. Alternatively, Rad51 assembles a filament on the template strand to form a paranemic joint (no free end available), allowing the blocked end to use the new sister strand as a template, giving rise to a nicked dHJ. After DNA synthesis either a DSBR-type pathway (involving a dHJ) or an SDSA-type pathway (without dHJ) may ensue. Paranemic joint formation likely requires an additional factor(s) to stabilize the joint.
3 Biochemistry of recombination proteins
The seminal discovery that Rad51 represents the eukaryotic homolog of the bacterial homologous pairing and DNA strand exchange protein RecA represented a breakthrough in the biochemistry of eukaryotic recombination (Aboussekhra et al. 1992; Shinohara et al. 1992). The demonstration that Rad51 forms a filament as well as functions in ways that are highly similar to RecA allowed for application of the paradigms established with bacterial RecA and T4 UvsX to eukaryotes (Ogawa et al. 1993; Sung 1994). Table 1 lists HR proteins in the budding yeast S. cerevisiae and humans. The bacterial proteins are listed for comparison and their biochemistry is elaborated in another dedicated contribution in this volume (Cox). Figure 2 illustrates a number of commonly used in vitro recombination assays and a brief discussion of their properties.
Table 1. Homologous recombination proteins in E. coli, S. cerevisiae and humans.
| E. coli | S. cerevisiae | Human | |
|---|---|---|---|
| Initiation | RecBCD | - | - |
| SbcCD | Mre11-Rad50-Xrs2 | Mre11-Rad50-Nbs1 | |
| RecQ | Sgs1 | RecQL, RecQ4, RecQ5, BLM, WRN | |
| RecJ | ExoI | ExoI | |
| UvrD | Srs2 | Fbh1? | |
| Homologous pairing and DNA strand exchange | RecA | Rad51, Dmc1 | Rad51, Dmc1 |
| SSB | RPA | RPA | |
| RecF(R) | Rad55-Rad57 | Xrcc3-Rad51C | |
| Shu1-Psy3-Shu2-Csm2 | Xrcc2-Rad51D-Sws1 | ||
| Rad51B | |||
| RecO(R) | Rad52 | Rad52 | |
| - | Rad59 | - | |
| - | - | Brca2-Dss1 | |
| - | Rad54, Rdh54/Tid1 | Rad54 | |
| Rad54B | |||
| DNA Heteroduplex Extension | RuvAB | Rad54 | Rad54 |
| RecG | - | - | |
| RecQ | Sgs1 | RecQL, RecQ4, RecQ5, BLM, WRN | |
| Resolution/Dissolution | RuvC | - | Resolvase A |
| RecQ-TopoIII | Sgs1-TopIII-Rmi1 | BLM-TopoIII α-BLAP75 | |
| - | Mus81-Mms4 | Mus81-Mms4/Eme1 |
Fig. 2.

Model reactions to study homologous recombination in vitro. A. DNA annealing. Annealing of protein-free DNA is a relatively non-specific reaction that can occur when proteins aggregate DNA non-specifically. Annealing of RPA-coated ssDNA instead is a highly specific reaction, catalyzed by the UvsY, RecO, and Rad52 proteins. B. D-loop reaction with either linear ssDNA (shown) or linear tailed DNA (not shown) and a supercoiled dsDNA substrate. Note that the RPA requirement for this reaction depends on the length and secondary-structure potential of the ssDNA. Low level, non-specific apparent D-loop formation can occur in particular when the supercoiled dsDNA substrate has been prepared by procedures involving DNA denaturation. C. Three strand DNA strand exchange between circular ssDNA and linear dsDNA. A well-known potential artifact with this assay is exonucleoytic resection followed by DNA strand annealing that will lead to the formation of intermediates and products that resemble DNA strand exchange intermediates. D-loop and DNA strand exchange reactions typically follow strict order of addition protocols: ssDNA + Rad51 → +RPA → +dsDNA. Inhibition of in vitro recombination by early RPA addition is overcome by the mediator proteins. Inhibition by early addition of dsDNA can be overcome by the Rad54 motor protein. Release of the product DNA is achieved in all reactions by treatment with detergent and proteinase, thus side-stepping a requirement for turnover by the proteins.
HR can be conceptually divided into three stages: presynapsis, synapsis, and postsynapsis (see Fig. 3). In presynapsis, the DNA lesion is processed, if necessary, to form a Rad51-ATP-ssDNA filament, which is also known as the presynaptic filament. Synapsis is defined by homology search and DNA strand exchange, leading to the D-loop intermediate by DNA strand invasion, which is the hallmark of a Rad51-dependent recombination reaction. All ensuing steps constitute postsynapsis including the release of Rad51 from the heteroduplex product DNA, mismatch repair (MMR), DNA synthesis, and processing of the various junction intermediates. Our understanding of events during postsynapsis is poor, and the potential pathways display significant variety after the initial D-loop is formed (Fig. 1). We would like to use the term context-specific factors for those HR proteins that lead to and from the core reaction of Rad51 filament formation and function starting with various substrates (frank or one-sided DSBs, gaps) and process the D-loop intermediate into different pathways of resolution/dissolution or annealing. The requirement for such factors will depend on the context in which the recombination core reaction occurs, and will vary with the specific biochemical or genetic assays utilized. The discussion will focus on the proteins from S. cerevisiae but will introduce human proteins where they are unique, or when their function diverged or differs from that of their yeast counterparts.
Fig. 3.

Mechanistic stages of homologous recombination. HR can be conceptually divided into three stages. First, during presynapsis the ends are processed and the Rad51 filament is assembled. The potential functions of cofactors in Rad51 filament assembly are indicated as anchoring the non-growing end, binding to the growing end or binding the filament laterally. In addition, cofactors may control the Rad51 protomer pool. Second, in synapsis the Rad51 filament undergoes homology search and DNA strand invasion, likely in conjunction with Rad54 protein. Third, postsynapsis comprises all ensuing steps including branch migration, Rad51 turnover, and DNA synthesis, common to the DSBR and SDSA pathways, as well as the subpathway-specific functions of dHJ resolution by a putative resolvase (Resolvase A) and dHJ dissolution by BLM-TOPOIIIα. The SDSA sub-pathway requires a protein, likely a DNA helicase, to dissolve the D-loop and likely employs Rad52 in reannealing the broken ends. The representation of the proteins is for illustration purposes only and does not imply specific stoichiometries or a specific oligomeric assembly status.
3.1 Structure of the presynaptic Rad51 filament
S. cerevisiae Rad51 is a ∼43 kDa (400 amino acids) protein and shares a core ATPase domain with its homologs, RecA, UvsX, and the archaeal RadA. This domain includes the Walker A and B boxes with structural similarity to the ATPase domains of DNA motor proteins and that of the F1-ATPase. The ATP-bound form of Rad51 undergoes a conformational change necessary for DNA binding. The binding cooperativity leads to filament formation with a stoichiometry of one Rad51 protomer per three-four nucleotides. Rad51 forms a right-handed filament with a helical pitch of 130Å, as determined by crystal structure analysis of a Rad51 fragment lacking the N-terminal 79 amino acids (Conway et al. 2004). The lacking N-terminal amino acids almost exactly correspond to the budding yeast-specific N-terminal extension of Rad51 (Shinohara et al. 1993). Although the crystals were grown in the presence of DNA, the DNA was not visible in the crystal, and the ATP was replaced, probably by a sulfate. Yet, the filament likely represents the DNA-bound form of Rad51. This is consistent with electron microscopic (EM) studies of Rad51/RecA filaments, suggesting flexible filaments with a pitch that varies with the particular protein and the bound nucleotide cofactor (Ogawa et al. 1993; Yu et al. 2001). The crystal structure of the Rad51 filament likely represents the extended, active form, whereas the crystal structure of the RecA filament has a much shorter pitch (83Å) (Story et al. 1992), which is comparable to the pitch determined for inactive filaments by EM (Yu et al. 2001). The Rad51 crystal structure revealed that the nucleotide-binding pocket of one protomer is in direct contact with the ATPase domain of the neighboring subunit, providing a structural basis for coordinated ATPase activity in the Rad51 filament. The conserved N-terminal domain of Rad51, which contains a DNA binding site, was also found in contact with the ATPase domain of the next protomer. This arrangement possibly provides a basis for cooperative DNA binding and for linking the ATPase cycle to DNA binding (Galkin et al. 2006). The crystallographic analysis revealed an asymmetry in the filament due to alternating conformations of the nucleotide-binding pocket. Conway et al. (2004) noted the similarity of this arrangement to hexameric helicases where only a subset of the ATPase sites is active at any given time (Singleton et al. 2000). The crystal structure of the Rad51-ssDNA filament represents a significant advance and provides a sound basis for understanding the interaction of the accessory proteins as well as their roles in the assembly, function, and turnover of the Rad51 filament.
3.1.1 Rad51 versus RecA - cousins not brothers
While RecA and Rad51 are homologous proteins that form relatively equivalent structures, the biochemical and structural analysis has revealed interesting differences between the two proteins. These differences are the result of different evolutionary constraints and selection. Understanding the basis for these differences will enable us to elucidate the functional environment of the Rad51 filament. The ATPase core is structurally conserved between all RecA homologs with recognizable sequence similarity (Conway et al. 2004); however, the N- and C-terminal extensions vary extensively between these proteins (Shinohara et al. 1993). The equivalent of the C-terminal DNA binding site of RecA resides at the N-terminus in Rad51 (Aihara et al. 1999), but the significance of this is not understood.
In RecA, the ATPase cycle is tightly coupled to DNA binding, such that DNA binding requires ATP binding and ATP-hydrolysis triggers the release of DNA (see Cox, this volume; Bianco et al. 1998). RecA has significant preference in binding ssDNA, due to a kinetic barrier that restricts binding to dsDNA. The high cooperativity leads to filament formation in the 5′-3′ direction with the addition of ATP-bound subunits on the growing end and the much slower loss of protomers after ATP hydrolysis at the initiating end. On ssDNA, RecA hydrolyzes about 30 ATP min-1, leading to a dynamic situation of filament assembly and disassembly. After DNA strand exchange, RecA is bound to the product heteroduplex DNA and ATP hydrolysis (20 ATP min-1) results in dissociation of RecA from the dsDNA. This frees up the 3′-OH end of the invading strand, allowing access by DNA polymerase to extend the D-loop formed by RecA (Xu and Marians 2002).
Rad51's biochemical properties differ from those observed in RecA in several ways, but the significance of these differences is poorly understood. First, unlike RecA, Rad51 has only a slight preference for binding ssDNA and can readily bind dsDNA (Zaitseva et al. 1999). It is likely that co-factors, probably one or several of the mediator proteins discussed below, target Rad51 to form filaments on ssDNA. Second, Rad51 binds DNA with significantly less cooperativity than RecA, forming only short filaments at limiting protein concentrations (Kiianitsa et al. 2002). Third, RecA has a clear polarity in filament formation as well as in subsequent DNA strand exchange. RecA polymerizes in a 5′ to 3′ direction, ensuring coverage of the 3′ end to make it more invasive. The issue of polarity with Rad51 appears somewhat unresolved. Some results argue in favor of a DNA strand exchange polarity (and hence filament formation polarity) of yeast and human Rad51 opposite to that of RecA (Sung and Robberson 1995; Baumann and West 1997; Solinger and Heyer 2001). Another study using an N-terminal truncation of yeast Rad51 was shown to lack polarity (Namsaraev and Berg 2000). Fourth, yeast Rad51 exhibits significantly lower ATPase activity than RecA (0.7 ATP min-1 on ssDNA, 40 × less than RecA; 0.1 ATP min-1 on dsDNA, 200 × less than RecA) (Sung 1994), and similar values were obtained with the human protein (Tombline and Fishel 2002). The lower ATPase activity of Rad51 will significantly curtail the dynamics of the filament. This may lead to less biased polar growth and possibly allow growth in both directions depending on the substrates and factors that nucleate the filament (see below). The lower cooperativity of the Rad51-ssDNA binding may create the problem of multiple nucleation events on a given ssDNA tail. Frequent independent nucleations resulting initially in short filaments will unlikely lead to long contiguous filaments, because with a binding site size of three nucleotides there is a two in three probability to be out of register. Dynamic rearrangement of the mini-filaments would be required to form a single Rad51-ssDNA filament. Compared to RecA filaments, Rad51 filaments on duplex DNA are significantly more stable. Rad51 remains bound to DNA even in the ADP-bound form posing a problem for Rad51 turnover after DNA strand exchange (Zaitseva et al. 1999; Tombline et al. 2002; Bugreev and Mazin 2004). These biochemical properties of Rad51 have to be viewed in the context of the eukaryotic cofactors of Rad51. The specific eukaryotic cofactors may function in addition to nucleating the filament in modulating cooperativity of Rad51 DNA binding, Rad51 filament dynamics on ssDNA to form functional presynaptic filaments and dissociation of Rad51 from dsDNA after DNA strand exchange.
3.1.2 Human Rad51 versus yeast Rad51 - brothers not twins
Besides the fundamental homology between RecA and Rad51, the above discussion focused on the differences that selection imposed on the two proteins. The yeast and human Rad51 proteins exhibit significantly more sequence homology with each other (57% or 66% identity, depending on direction of comparison) than with RecA (26% and 29% identity, respectively) (Shinohara et al. 1992, 1993). However, yeast and human Rad51 proteins also exhibit significant differences. The yeast Rad51 protein is 61 amino acids longer (400 versus 339) than its human counterpart. This difference accounts for the budding yeast specific N-terminal extension of about 75 amino acids, which is the site of species-specific interaction of yeast Rad51 with the Rad52 protein (Donovan et al. 1994). Unfortunately, this segment is lacking in the Rad51 crystal structure (Conway et al. 2004). A major difference between the yeast and human Rad51 proteins was noted immediately in the much lower efficiency of the human protein in DNA strand exchange reactions (Baumann et al. 1996). The efficiency of the in vitro recombination activities of the human Rad51 protein can be significantly enhanced by curtailing its binding to dsDNA by addition of 100 mM ammonium sulfate (Sigurdsson et al. 2001a) and inhibiting its ATPase through the addition of calcium ions (Bugreev and Mazin 2004). These reaction conditions produce filaments of more regular structure with decreased dynamics as visualized by atomic force microscopy (Ristic et al. 2005). Interestingly, both protocols were reported not to stimulate the yeast Rad51 protein. Moreover, the strict nucleotide cofactor requirement for DNA binding by yeast Rad51 at neutral pH is relaxed with human Rad51 (Bianco et al. 1998; Zaitseva et al. 1999; Chi et al. 2006). These structural and biochemical changes suggest that the yeast and human Rad51 proteins will have somewhat different dynamic properties when bound to DNA and understanding these differences may provide a key to understanding the differences in the complexity and function of the cofactors for both proteins (Table 1). While the dynamics of Rad51 filament assembly and filament function is amenable to biochemical analysis, in vivo it is difficult to distinguish between functional and non-functional Rad51 assemblies by cytology or chromatin immunoprecipitation (ChIP).
3.2 Presynapsis: different pathways leading to Rad51 filament formation and the function of distinct mediator proteins
3.2.1 Different pathways to generate ssDNA for Rad51 filament formation
Context-specific factors during presynapsis are likely to be required in the processing of various types of DNA damage to single-stranded DNA amenable for Rad51 filament formation. In the context of a frank or one-sided DSB, nucleases or helicase/nuclease combinations are needed to generate a single-stranded tail. In the case of IR-induced breaks, the Mre11-Rad50-Xrs2 complex is needed to process ends with non-standard chemistry. However, with clean nuclease-induced breaks (HO, EndoSceI), the requirement for this complex appears diminished and other nucleases, including Exo1, can function. Furthermore, in E. coli RecQ/J function in end-processing; a similar role of the eukaryotic homologs Sgs1/Exo1 has not been clearly demonstrated.
Gap repair on the lagging strand of stalled replication forks (Fig. 1C) may not require further resection, but poses specific topological challenges. In the case of Rad51 filament formation on the uninterrupted ssDNA molecule, DNA strand invasion would lead to a paranemic joint that may require specific stabilization. It is also possible that gap repair involves a helicase that dissociates the stalled 3′-end from the template to create a substrate for Rad51 filament formation on the interrupted strand. This scenario requires a 5′-3′ DNA helicase translocating on the uninterrupted strand that is not stalled by the DNA damage that led to the initial stall of the replicative polymerase. The 5′-3′ requirement rules out the RecQ family and the Srs2 helicase, which display the opposite polarity (3′ to 5′).
3.2.2 The problem of forming a filament - learning from actin and tubulin
The RecA/Rad51/RadA proteins are among the few proteins in nature that acquired the propensity to form helical filaments, and their structural similarity to actin filaments has been pointed out before (Egelman 2003). The dynamic instability of the Rad51 filament, the role of the nucleotide cofactor cycle, and the roles of cofactors can be productively compared to other filament forming proteins, e.g. actin and tubulin, where these processes are understood in significant detail (reviewed in Moritz and Agard 2001; Pollard and Borisy 2003; Zigmond 2004). This comparison reveals a number of potentially general characteristics that may help in understanding the complexity and function of the cofactors required for the Rad51 filament (Table 1, see Fig. 3).
Nucleation (the binding of the first subunit) is the rate-limiting step in forming a filament. The filaments are polar with two distinctive ends, displaying dynamic instability correlated to the nucleotide cofactor cycle. The faster growing end adds triphosphate-bound protomers and the slower growing end accumulates the diphosphate-bound protomer. While the filament can grow in both directions, the difference in growth speed leads to treadmilling. Proteins can cap either end of the filament, leading to strongly biased polar filament growth. At high concentration, the protomers can nucleate filament formation independently, but in vivo and under limiting in vitro conditions, nucleation requires specific mechanisms, providing the basis for regulated filament assembly.
In actin and tubulin filaments, central nucleation factors, the Arp2/3 complex and the γ-tubulin complexes respectively, contain paralogs of the filament protomer and additional subunits to initiate filament formation. These paralogs are incapable of forming an extended polar filament. The Arp2/3 complex, as well as the γ–tubulin complexes, anchor the minus end of the protomer preventing minus end growth and depolymerization, thus leading to plus end-directed filament growth. Arp2/3 is a major regulatory target for actin filament formation. A similar role can be envisioned for the Rad51 paralogs and Rad51 mimics (Brca2), as discussed below. An alternative mode of nucleating the actin filament is catalyzed by formins, a family of proteins with no recognizable homology to actin. These proteins bind to the plus-end of the actin filament and remain processively associated with the growing end. This possibility has not yet been considered for Rad51 co-factors. The continued association of cofactors with the filament could lead to a function also downstream of filament nucleation (Rad51C-Xrcc3, see Section 3.4). Proteins binding monomeric protomers can bias filament growth and dynamics. Profilin catalyzes nucleotide exchange from ADP-G-actin to the ATP-G-actin form. In addition, by binding to the plus end of the protomer, profilin targets the minus end of the protomer to the plus end of the growing filament. The importance of regulating the nucleotide cycle of Rad51 has recently been appreciated (roles of Rad51D-Xrcc2 and Ca++, see below), but it is not known if any cofactor delivers Rad51 protomers to the growing end of the filament. Filament stability is also regulated by lateral binding of proteins that stabilize or destabilize actin filaments. Tropomyosin is one such factor that stabilizes actin filaments by lateral binding, and a conceptually similar role has been identified for Rad54 in presynapsis (see below). Actin filament dynamics are tightly linked to the nucleotide cycle. ATP hydrolysis is required for dissociation of the protomer from the filament, but the cofactor cofilin is important for efficient dissociation of ADP-actin from the filament. An active role in Rad51 filament turnover has been proposed for Rad54 protein (see Section 3.4).
Detailed understanding of the actin and tubulin systems provides guidance for identifying the exact mechanisms of Rad51 co-factors. In difference to actin and tubulin, Rad51 (and RecA) form a filament on an underlying lattice, DNA, with defined physical properties, which dictates the register of the filament and confines the filament structure to some degree. Moreover, the DNA provides a lattice for motor proteins (dsDNA: Rad54, Tid1/Rdh54 discussed in Section 3.4; ssDNA: Srs2 discussed in Chapter 4; see also Table 1) that affect Rad51 filament stability.
3.2.3 The problem of forming a filament on DNA - learning from Escherichia coli
Bacterial RecA protein forms a highly cooperative filament and faces the same nucleation problem as Rad51. The two major HR pathways in E. coli employ two different strategies to nucleate the RecA filament. In the RecBCD pathway, filament nucleation is coupled to DSB processing and RecA is loaded by the resecting RecBCD helicase/nuclease (Spies and Kowalczykowski 2006). The C-terminal domain of the RecB subunit interacts with a conserved element of the RecA fold, mimicking the initiating protomer. In the RecF pathway, the RecFOR complex targets filament nucleation to the dsDNA-ssDNA transition, forcing filament growth toward the 3′ end of the ssDNA tail and limiting growth towards the duplex DNA (Morimatsu and Kowalczykowski 2003). The RecO protein facilitates SSB displacement, and RecO is able to reanneal ssDNA coated with SSB, the prokaryotic ssDNA binding protein (Fig. 2) (Kantake et al. 2002; Morimatsu and Kowalczykowski 2003). While the RecOR complex alone can nucleate RecA filament formation on SSB-coated ssDNA, RecF protein targets the nucleation to a dsDNA-ssDNA junction. RecF lacks obvious sequence homology with RecA and no structural information is available to suggest whether it is structurally related to RecA. In the RecF pathway, nucleation competes with ongoing resection by RecJ (or other nucleases), and it is unclear how this competition is balanced. The absence of a RecBCD homolog in eukaryotes and the similarity between the RecO and Rad52 proteins (Table 1) suggests that the RecF pathway may provide a paradigm for Rad51 filament nucleation anchoring the minus end of the filament to a dsDNA-ssDNA transition.
3.2.4 The problem of forming a Rad51 filament on RPA-coated single-stranded DNA
Forming a Rad51 filament on ssDNA faces two distinct but related mechanistic challenges. First, like all filament forming proteins, Rad51 has to overcome the rate-limiting step of nucleation (binding of the first protomer), which determines the time and place of filament formation. Second, Rad51 faces a more specific challenge of having to replace RPA, the eukaryotic ssDNA binding protein, from ssDNA. RPA binds to ssDNA with higher affinity and specificity than Rad51. These challenges are similar to the bacterial RecA protein, but the biochemical properties of Rad51 discussed above (little preference for ssDNA binding, low filament dynamics, lower cooperativity) create additional problems.
3.2.5 Proteins functioning in Rad51-ssDNA filament assembly and stability
RPA
RPA is a hetero-trimeric complex with subunits of about 70, 30, and 14 kDa, containing six ssDNA binding sites or OB-folds, of which four actively bind to ssDNA in a sequential fashion (for review see Wold 1997; Bochkarev and Bochkareva 2004). RPA is an abundant cellular protein with the highest known affinity to ssDNA of any cellular protein, so that any ssDNA generated during replication, repair, or recombination can be expected to be an ssDNA-RPA complex in vivo. RPA engages in a host of protein interactions to function in these contexts. In recombination, RPA serves to counteract secondary structure in ssDNA. This is particularly important for Rad51-mediated reactions, because Rad51 readily binds to dsDNA that forms as secondary structure in ssDNA. This would interfere with forming a functional presynaptic filament on ssDNA. In vitro yeast and human Rad51 are strongly stimulated by RPA in reactions employing long ssDNA substrates that have potential to form secondary structures, whereas reactions using ssDNA substrates devoid of secondary structure can largely dispense with RPA (Sung 1994; Sugiyama et al. 1997; Sigurdsson et al. 2001a). Another feature of RPA function in the DNA strand exchange reaction is that it may bind to the displaced strand, preventing the reverse reaction during DNA strand exchange (Fig. 2). It is unclear whether this property is relevant in vivo or whether RPA would have access to the displaced strand in a D-loop (Fig. 2). Lastly, reannealing of RPA coated ssDNA by Rad52 protein is a highly specific in vitro reaction that distinguishes Rad52 from many other proteins that non-specifically reanneal protein-free DNA (Fig. 2) (Sugiyama et al. 1998). This reaction lies at the heart of the SSA pathway that is not discussed here, but is reviewed elsewhere (Krogh and Symington 2004). Moreover, this mechanism is likely to be relevant for the capture of the second end in the formation of the double Holliday junction and in the annealing step of SDSA (Fig. 1).
While RPA is required for efficient DNA strand exchange by Rad51, the stimulation is only observed using a strict order of addition protocol, in which RPA is added to the ssDNA after Rad51 (Sung 1994; Sugiyama et al. 1997). This allows Rad51 to nucleate on ssDNA, and the binding cooperativity of Rad51 displaces RPA. Simultaneous addition of both proteins to ssDNA or pre-incubation of ssDNA with RPA strongly inhibits Rad51-mediated DNA strand exchange. Since both proteins are expected to be present simultaneously in vivo, this poses a problem and calls for catalysts of this reaction to displace RPA from ssDNA by Rad51. These catalysts are called recombination mediators (Beernink and Morrical 1999; Sung et al. 2003) (Table 1).
The inhibition of Rad51-mediated DNA strand exchange reactions by the early addition of RPA provided an assay to search for mediator proteins. Two types of mediators, Rad52 (Sung 1997a; New et al. 1998; Shinohara and Ogawa 1998) and the Rad55-Rad57 heterodimer (Sung 1997b), were identified by biochemical analysis in budding yeast that appear to address the specific problems of Rad51 nucleation on RPA-coated ssDNA.
Rad52
S. cerevisiae Rad52 protein features an N-terminal DNA binding domain and a C-terminal Rad51 interaction domain. The 52.4 kDa budding yeast protein forms a multimeric ring-shaped structure (Shinohara et al. 1998). Three-dimensional image reconstruction of the human Rad52 ring structure revealed a heptameric ring around a central channel that can bind ssDNA on the outside face of the ring (Stasiak et al. 2000; Kagawa et al. 2002; Singleton et al. 2002). S. cerevisiae Rad52 specifically interacts with Rad51 and RPA (Shinohara et al. 1992; Hays et al. 1998), and is critical for the ejection of RPA from ssDNA by Rad51 (Sugiyama and Kowalczykowski 2002). The interaction of Rad52 with Rad51 was demonstrated to be critical for its mediator function using a small internal Rad52 deletion mutant specifically affecting this interaction (Krejci et al. 2002). The detailed model for the mediator function of the T4 UvsY protein, a Rad52 homolog in T4 UvsX-mediated recombination, suggests that binding of RPA covered ssDNA to the outside of the Rad52 ring kinks DNA sufficiently to favor binding of the DNA strand exchange protein (Beernink and Morrical 1999; Liu et al. 2006). The genetics and cytology are consistent with Rad52 functioning as a mediator in Rad51 filament assembly (Symington 2002; Krogh and Symington 2004). Rad51 foci, which likely represent Rad51 filaments or later pairing intermediates, do not form or very poorly form in the absence of Rad52 (Gasior et al. 1998, 2001; Lisby et al. 2004). The recombination defect in rad52 mutants is the most extreme in budding yeast and significantly stronger than in rad51 mutants (Symington 2002; Krogh and Symington 2004). This reflects the dual function of Rad52 as a mediator for Rad51 and in Rad51-independent SSA (see above).
Rad55-Rad57
Rad55 and Rad57 are two Rad51 paralogs in S. cerevisiae with 46.3 and 52.2 kDa, respectively. Both proteins share the RecA core with Rad51 but maintain different N- and C-terminal extensions (Symington 2002; Krogh and Symington 2004). Rad55 and Rad57 form a tight heterodimer, and the available biochemical and genetic evidence suggests that both proteins exclusively function as a complex (Sung 1997b). The heterodimer does not catalyze DNA strand exchange itself, but was shown to function as a mediator, like Rad52 protein, allowing DNA strand exchange when RPA was added to ssDNA at the same time as Rad51 instead of after Rad51 as in standard reactions (Sung 1997b). Unlike Rad52, Rad55-Rad57 are not known to interact with RPA physically, but Rad55 interacts directly with Rad51 (Hays et al. 1995; Johnson and Symington 1995; Sung 1997b). Based on the conceptual similarity with microtubules and actin filaments, one might speculate that the specialized paralogs Rad55-Rad57 nucleate the filament of the Rad51 protomer like γ–tubulin and the Arp2/3 complex. However, at present there is no direct mechanistic biochemical evidence to support this model. It has also been speculated that like RecFOR, Rad55-Rad57 targets nucleation to the dsDNA-ssDNA junction to force mono-directional Rad51 filament formation on ssDNA (Morimatsu and Kowalczykowski 2003). However, such substrates have not been directly tested yet with Rad55-Rad57. The use of a heterodimer (Rad55-Rad57) for nucleation may reflect that the functional unit of Rad51 in the filament appears to be a dimer as proposed from the crystal structure (Conway et al. 2004). The genetics and cytology are consistent with a role of Rad55-Rad57 in Rad51 filament formation (Symington 2002; Krogh and Symington 2004), although the requirement in the formation of DNA damage-induced Rad51 foci is less strict than for meiotic Rad51 foci (Gasior et al. 1998, 2001; Lisby et al. 2004). A particularly elegant demonstration of the presynaptic Rad55-Rad57 function was the isolation of a partially Rad55-independent Rad51 mutant, Rad51-I345T, that increased its intrinsic capacity for displacing RPA from ssDNA (Fortin and Symington 2002). In S. cerevisiae the general requirement of filament forming proteins for a nucleation factor and the specific requirement of Rad51 to displace RPA appear to be fulfilled by the Rad55-Rad57 and Rad52 mediators. However, it is unclear how these two mediators cooperate, and no reactions have been reconstituted containing both mediators.
Shu1-Psy1-Shu2-Csm2
Another complex containing two highly divergent Rad51 paralogs, Rdl1 and Rlp1, has been identified in the fission yeast Schizosaccharomyces pombe together with a third subunit Sws1 (Martin et al. 2006). The genetic and cytological characterization of the genes establish an early function in HR. Rdl1 and Rlp1 appear to be reduced versions of the human Rad51 paralogs Rad51D and Xrcc2 proteins, respectively (Table 1). Identification of this homology was complicated by the loss of the Walker A box in Rdl1 and the loss of the Walker B box in Rlp1. The authors also noted significant homology to components of the Shu1-Psy3-Shu2-Csm2 complex of budding yeast, proposing that Shu1 represents the S. cerevisiae Xrcc2 (Rlp1) homolog, Psy3 the Rad51D (Rdl1) homolog, and Shu2 the budding yeast Sws1 (Table1) (Martin et al. 2006). The Shu1-Psy3-Shu2-Csm2 complex was identified in S. cerevisiae as suppressors of the slow growth phenotype of top3 cells and demonstrated to be factors in the RAD52 epistasis group required for efficient HR (Shor et al. 2005). While their contribution does not appear to match the effect of the other Rad51 paralogs, Rad55 and Rad57, deletion of SHU1 (and presumably of the other subunits as well) prolongs the half-life of MMS-induced Rad52 foci (Shor et al. 2005), suggesting this complex may be required for normal kinetics in Rad51 filament assembly. Linking the Shu1-Psy3-Shu2-Csm2 complex to Rad51 paralogs (Martin et al. 2006) provides a great impetus to initiate biochemical studies with these proteins.
In humans, the presynaptic situation is somewhat different and surprisingly more complex. In the discussion of the yeast and human Rad51 proteins it is argued that this is in part a reflection of the biochemical differences between both eukaryotic Rad51 proteins and in part a reflection of the greater complexity of human cells vis-à-vis pathway regulation and tissue differentiation. A major difference lies with the Rad52 protein. Rad52 is critically required for Rad51 filament formation and SSA in yeast, causing the most extreme recombination defects as a single mutant, whereas the mouse mutant causes only a very mild recombination defect (Rijkers et al. 1998). Has the role of Rad52 been usurped by other factors (Brca2, Rad51 paralogs)? Which protein is mediating reannealing of RPA-coated ssDNA during mammalian SSA?
Human Rad52
Human Rad52 was found to stimulate human Rad51-mediated DNA strand exchange. This effect was observed in the absence of RPA (Benson et al. 1998) and may involve mechanisms other than the mediator function demonstrated for yeast Rad52. In support of a mediator function speaks the genetic observation that double mutants of XRCC3 and RAD52 are lethal in chicken DT40 cells (Fujimori et al. 2001), suggesting a shift in balance between the individual mediators in different eukaryotes.
Human Rad51 paralogs
The number of Rad51 paralogs increased to five in vertebrates (Rad51B/Rad51L1, Rad51C/Rad51L2, Rad51D/Rad51L3, Xrcc2, Xrcc3; Table 1) (Thacker 2005). Each of the five paralogs is required for Rad51 focus formation in vivo in chicken DT40 cells and in mammalian cells. The individual single mutants display near identical phenotypes in chicken DT40 cells, where they were studied in parallel (Takata et al. 2000, 2001). Also the mammalian Rad51 paralog mutants display highly similar, though possibly not identical, phenotypes (Thacker 2005). The presynaptic function of the Rad51 paralogs is underscored by the significant rescue of the individual paralog mutants by Rad51 overexpression (Takata et al. 2000, 2001), recapitulating the same finding with the yeast paralogs Rad55-Rad57 (Hays et al. 1995; Johnson and Symington 1995). The initial model proposed a function of all Rad51 paralogs acting in a single complex to support Rad51 filament assembly. It is now clear that the situation is more complex. The five paralogs form different sub-assemblies with the main complexes being Rad51B-Rad51C-Rad51D-Xrcc2 and Rad51C-Xrcc3 (Masson et al. 2001b). Genetic analysis of rad51B, rad51D, and xrcc3 single and double mutants in DT40 cells is consistent with both complexes having distinct functions (Yonetani et al. 2005). The biochemistry of the paralogs is hampered by their poor solubility, and the existence of further subassemblies (Rad51B-Rad51C, Rad51D-Xrcc2) complicates the interpretation. Moreover, Rad51D-Xrcc2 associates with an additional protein, Sws1. RNAi knockdown of Sws1 reduced but not eliminated spontaneous and IR-induced Rad51 focus, suggesting that Sws1 is another protein with a function in Rad51 filament formation/stabilization (Martin et al. 2006). The presence of novel subunits in Rad51 paralog complexes could explain the difficulties in expressing soluble forms of such complexes in the absence of these subunits. The purified Rad51B-Rad51C-Rad51D-Xrcc2 and Rad51C-Xrcc3 complexes preferentially bind ssDNA with no preference for tailed DNA and little to no reported dsDNA binding activity (Masson et al. 2001a, 2001b). [The potential function of the Rad51C-Xrcc3 complex in Holliday junction resolution is discussed below and more extensively in the contribution by Whitby.] It is generally believed that the Rad51 paralogs neither form filaments nor catalyze DNA strand invasion reactions, although this view has been challenged by the visualization of Rad51D-Xrcc2 and Rad51C-Xrcc3 filaments on ssDNA (Kurumizaka et al. 2001, 2002). The significance of the observed structures remains unclear. The low activity in D-loop assays (Fig. 2) identified for both complexes in these studies may be a reflection of a strand annealing activity rather than true DNA strand invasion. Such an activity may result from destabilizing duplex DNA, followed by random renaturation of single-strands, as demonstrated for the Rad51C protein (Lio et al. 2003). The Rad51B-Rad51C subassembly, but not the tetrameric complex also containing Rad51D and Xrcc3 (Masson et al. 2001b), were found to preferentially bind 3′-end tailed DNA (Lio et al. 2003). The Rad51B-Rad51C complex also partially overcame the inhibition imposed by early addition of RPA in the DNA strand exchange reaction (Fig. 2A) displaying mediator function similar to the Rad55-Rad57 complex (Sigurdsson et al. 2001b). These studies did not yet test whether Rad51B-Rad51C target filament formation to the dsDNA-ssDNA junction, as suggested by its DNA binding specificity.
Brca2
Brca2, a human breast cancer tumor suppressor protein, is also required for DNA damage-induced Rad51 focus formation in vivo and was identified as another mediator for Rad51 filament formation (Tarsounas et al. 2003; Pellegrini and Venkitaraman 2004). The protein contains eight BRC repeats that bind Rad51 with varying degree of affinity. In addition, Brca2 contains a unique Rad51 binding domain in the extreme C-terminus. Crystal structure analysis identified three OB-folds in the C-terminal portion of the protein, a typical ssDNA-binding motif also found in RPA, and a helix-turn-helix domain, a dsDNA-binding motif found in many transcription factors, suggesting that Brca2 might bind at an ssDNA-dsDNA transition (Yang et al. 2002). Brca2 is associated with the small 70-residue protein, Dss1, whose function remains largely mysterious. The enormous size of Brca2 with 3,418 amino acids has precluded analysis of the full-length human protein, but the analysis of Brca2 fragments and of Brca2 homologs with a smaller size than the human protein has afforded significant insights into Brca2 function. The Ustilago maydis Brca2 homolog Brh2 represents a diet version of human Brca2 with 1,075 amino acids containing a single BRC repeat plus the C-terminal Rad51 binding domain. Elegant biochemical experiments demonstrated that Brh2 targets Rad51 filament formation to a dsDNA-ssDNA transition and overcomes inhibition of early RPA addition (Yang et al. 2005). This is fully consistent with the analyses of human Brca2 fragments, which also support a mediator function of Brca2 for Rad51 nucleation (Yang et al. 2002; San Filippo et al. 2006). Of particular interest for the mechanism of Brca2 function is the structure of the BRC repeats that appear to mimic the Rad51 subunit interface in the filament (Yang et al. 2002), although no apparent sequence homology between the BRC repeat and the structurally corresponding Rad51 sequence can be detected. This suggests a potential mechanism for Rad51 filament nucleation in that Brca2 provides a polymerization interface for the first Rad51 protomer, similar to the Arp2/3 and γ-tubulin complexes with actin filaments and microtubules.
Why do mammalian cells need so many mediators: five Rad51 paralogs, Brca2, Sws1, and to some degree Rad52? A simple model would be to suggest that they serve as mediators on different substrates. However, the observation that all proteins are required (with the exception of Rad52 as a single mutant) for IR-induced Rad51 focus formation suggests otherwise. If Brca2 nucleates the Rad51 filament at a dsDNA-ssDNA transition and anchors the filament at the non-growing end, what do the Rad51 paralogs do? Why does human Brca2 have eight BRC repeats, and what is the function of the C-terminal Rad51 binding site? In the actin filament, two cofactors impinge on the nucleotide cycle of actin. ATP hydrolysis by actin occurs relatively fast, but dissociation of the phosphate and release of actin-ADP is slow and aided by cofilin. Instead, profilin catalyzes nucleotide exchange from ADP-G-actin to the ATP-G-actin form. Similar proteins could be expected to function in the dynamics of the Rad51 filament and its nucleotide cycle. First evidence for such a function comes from the analysis of the effect of the Rad51D-Xrcc2 complex on the ATPase cycle of Rad51 (Shim et al. 2004). Xrcc2 enhanced the Rad51 ATPase activity and the function of Rad51 filaments as assayed by DNA unwinding and DNA strand exchange, whereas the Rad51D-Xrcc2 complex exhibited less stimulation of the Rad51 ATPase activity. Such an activity would enhance the dynamic turnover of the Rad51 filament and might be critical to maintain a functional and growing Rad51 filament. This effect of Xrcc2 appears opposite to the observed stimulation of Rad51 DNA pairing activities by Ca++, which was accompanied by an inhibition of the Rad51 ATPase activity that keeps the filament protomers in the active, ATP-bound state (Bugreev and Mazin 2004). It is unclear whether this is a physiological role of Ca++ or whether Ca++ replaces a protein cofactor(s). These biochemical studies are still in their infancy, but clearly suggest that protein cofactors, possibly the Rad51 paralogs, affect the nucleotide cycle of Rad51 and hence the dynamics of the Rad51 filament. Thus, it is possible that these proteins might also associate with the growing end of the filament, as found for profilin in the actin filament. This role may result in a continuous requirement for such a protein in filament formation, in addition to or instead of a requirement solely in the nucleation of the Rad51 filament. Cytologically both defects would be indistinguishable leading to the absence of detectable Rad51 foci. Physical monitoring of DNA intermediates during DSB repair may have identified a signature of such a late role. S. cerevisiae rad57 mutants exhibit a delay in the occurrence of DNA synthesis products from the invading strand of the D-loop, consistent with a defect in DNA strand invasion (Aylon et al. 2003). The accumulation of this intermediate (D-loop + DNA synthesis; Fig. 3, Step 6) was interpreted as evidence for a late, second role of Rad57 protein in processing the extended D-loop to conversion products by the SDSA pathway (Fig. 1, Fig. 3), but this phenotype could also be a late consequence of a Rad51 filament that is less dynamic, interfering with the strand annealing step.
Rad54
Rad54 is another protein with a function in presynapsis, which appears quite different from the mediator proteins introduced before. Due to the structure of this review, the presynaptic function of Rad54 is discussed here, whereas the analysis of the roles of Rad54 protein in synapsis (see Section 3.3) and postsynapsis (see Section 3.4) is deferred to later. The extensive biochemical analyses of this protein has been more fully evaluated in other reviews (Tan et al. 2003; Heyer et al. 2006). Rad54 is a dsDNA motor protein (see below), whose ATPase activity is essential for its in vivo function (Clever et al. 1999). However, budding yeast Rad54 was identified to also exhibit an ATPase-independent function in presynap-sis (Mazin et al. 2003; Wolner and Peterson 2005). Effects on Rad51 filament formation are difficult to distinguish from effects on filament stability using in vivo methods (cytology, ChIP), but biochemical experiments demonstrated that Rad54 stabilizes Rad51 filaments rather than helping in their assembly (Solinger et al. 2002; Mazin et al. 2003). The stabilization likely occurs through inhibition of Rad51 protomer dissociation, but it is unclear whether Rad54 binds to the end of the Rad51-ssDNA filament or binds laterally. The association with the presynaptic filament targets Rad54 protein to the pairing site (Mazin et al. 2000a), which is likely critical in the positioning of the motor on dsDNA for its function in the later stages of HR.
In summary, the surprising complexity of pre-synapsis and Rad51 filament formation, particularly in mammalian cells, still needs to be fully explored biochemically. Formation of a functional filament is not only mechanistically complex, but its nucleation is also a likely regulatory target. Nucleation defines the transition from RPA-covered ssDNA to a Rad51 filament. While RPA-coated ssDNA may have multiple fates (NHEJ vs. SSA vs. HR for a DSB; HR vs. translesion synthesis [TLS] vs. fork regression for a gap), a Rad51 filament is committed to HR. The genetic and biochemical differences between the yeast and the human proteins suggest that different variations on the same underlying fundamental theme are at work.
3.3 Synapsis: homology search and DNA strand invasion
The ternary complex of Rad51-ATP-ssDNA, otherwise known as the presynaptic filament, has a secondary binding site for the duplex DNA used during the homology search. The DNA in the presynaptic filament is stretched to an extended state with 5.1Å per base equaling 18 nt/bp per helical repeat (Yu et al. 2001), which is believed to facilitate the homology search by a base flipping mechanism, allowing the single-strand to sample homology on the duplex DNA (Gupta et al. 1999). However, the exact mechanism of the homology search still remains to be determined. Rad51 filament formation, homology search and DNA strand exchange do not require ATP hydrolysis (Sung and Stratton 1996). In RecA protein, ATP hydrolysis serves to release the heteroduplex DNA product of the DNA strand exchange reaction and turnover of RecA, which is critical for filament dynamics (Bianco et al. 1998). Unlike RecA, Rad51 cannot catalyze a four-stranded reaction or by-pass heterology. This inability is likely a consequence of the impaired dynamics of Rad51 filaments compared to RecA, suggesting that specific cofactors (maybe a function of the mediator proteins) will be required to reconstitute such reactions in vitro.
Rad54
While Rad51 can perform synapsis on its own, synapsis is greatly stimulated by the dsDNA motor protein Rad54 (for review see Tan et al. 2003; Heyer et al. 2006). In budding yeast, Rad54 is an 898 amino acid (human Rad54: 748 amino acids) dsDNA motor protein/translocase and exhibits significant dsDNA-specific ATPase activity of about 1,000/min per Rad54 molecule (Petukhova et al. 1998; Swagemakers et al. 1998; Ristic et al. 2001). Rad54 translocates on dsDNA at an astounding pace of 300 bp/sec with significant processivity (Amitani et al. 2006). Although not all experiments have been performed with both the yeast and human Rad54 proteins, there is currently no reason to suggest a difference in function between both proteins. The specific physical interaction between the Rad51 and Rad54 proteins has significant functional consequences for the biochemical activities of either protein (Jiang et al. 1996; Clever et al. 1997; Golub et al. 1997). Rad51 in its DNA-bound form stimulates the ATPase and motor activity of Rad54, and vice versa Rad54 stimulates the pairing activities of Rad51 (Petukhova et al. 1999; Mazin et al. 2000b; Van Komen et al. 2000; Solinger et al. 2001; Kiianitsa et al. 2002; Sigurdsson et al. 2002; Mazin and Mazin 2004). In particular the D-loop reaction (Fig. 2) essentially requires the presence of Rad54 to be catalyzed by Rad51. The mechanism by which Rad54 stimulates Rad51-mediated pairing remains to be determined. It may involve sliding of the target duplex DNA during homology search, topological opening of the target duplex DNA, or the clearing of Rad51 protein bound to the target DNA. Rad54 is a member of the Snf2 family of DNA translocases, which contains paradigmatic chromatin remodeling factors. Another function of Rad54 may be its ability to remodel chromatin structure. This activity was identified in biochemical assays with nucleosomal substrates (Alexeev et al. 2003; Jaskelioff et al. 2003) and may help to overcome the inherent inhibition imposed by nucleosomes (Alexiadis and Kadonaga 2003). Rdh54/Tid1, another Snf2-like motor protein primarily involved in meiotic recombination, also stimulates Rad51-mediated DNA strand exchange in a manner that appears similar to Rad54 (Petukhova et al. 2000).
3.4 Postsynapsis: many subpathways call for context-specific factors
A multitude of processes occur after DNA strand invasion. Distinct subpathways of HR (Fig. 1, Fig. 3) require common factors for Rad51 dissociation, DNA synthesis, and branch migration from the invading 3′-end, but also distinct, context-specific factors that act on the specific intermediates generated by the discrete pathways (DSBR: second end capture, dHJ resolution/dissolution; SDSA: D-loop dissolution, reannealing; BIR: single HJ resolution; gap repair: paranemic joint processing). Little is known about these processes in eukaryotes and a dedicated chapter in this volume (Whitby) focuses on this theme; two reasons to keep the discussion brief here.
3.4.1 Rad51 turnover by Rad54
After DNA strand invasion, RecA requires ATP hydrolysis to release the resulting heteroduplex DNA to allow DNA polymerase access to the invading 3′-OH end, as shown in a reconstituted reaction (Xu and Marians 2002). Similarly, Rad51 requires turnover and its DNA binding properties suggest that its intrinsic ATPase activity is insufficient for efficient product release. The Rad54 motor protein dissociates Rad51-dsDNA filaments in a reaction that requires specific protein interaction and the dsDNA-specific ATPase activity of Rad54 (Solinger et al. 2002). The Rad54 ATPase is significantly (six-fold) stimulated by Rad51 filaments partially occupying dsDNA (Kiianitsa et al. 2002; Solinger et al. 2002), suggesting that Rad54 associates with the terminus and catalyzes processive dissociation of terminal protomers. This terminal interaction between the motor and the Rad51 filament has been visualized by EM (Kiianitsa et al. 2006), and this mechanism is consistent with the ability of Rad54 to translocate on dsDNA (Amitani et al. 2006). In addition to its ATPase-independent function in presynapsis and ATP-dependent functions during synapsis and chromatin remodeling, ATP-dependent Rad51 turnover after DNA strand invasion is the fourth role assigned to Rad54 by biochemical experiments. Data from in vivo assays (ChIP, cytology, genetics) are consistent with a function of Rad54 in synapsis and postsynapsis but presently these assays cannot distinguish between both possibilities. An evaluation of this evidence would exceed the frame of this review and the interested reader is referred to dedicated reviews on this subject (Tan et al. 2003; Heyer et al. 2006). It is interesting to note that E. coli does not have a Rad54 homolog (Table 1), possibly because RecA is self-sufficient in turnover. A role of Rad54 in Rad51 turnover helps explain some biochemical differences between RecA and Rad51 with regards to dsDNA binding and ATPase activity.
3.4.2 DNA synthesis and DNA polymerase η
DNA synthesis after DNA strand invasion is critical in restoring the genetic information compromised by a DSB and the continuity of strands in gaps. In E. coli, a coupled reaction between D-loop formation and extension has been reconstituted using Pol III holoenzyme, the highly processive bacterial replicative DNA polymerase (Xu and Marians 2002). Which polymerase accesses the invading 3′-end in vivo remains unclear. In eukaryotes this question encounters significant complexity with the identification of an entire new suite of translesion synthesis DNA polymerases (Rattray and Strathern 2003). Fractionation of human cell extracts identified that human DNA polymerase η (eta), known for its role in bypass of UV photoproducts and the UV syndrome Xeroderma pigmentosum, extends the invading strand in a D-loop (Fig. 2) in a reaction that could not be supported by other DNA polymerases (McIlwraith et al. 2005). This reaction may involve an interaction between Polη and Rad51 (McIlwraith et al. 2005) and its in vivo significance is underlined by the finding in chicken DT40 cells that Polη is required for efficient gene conversion between immunoglobulin-variable genes (Kawamoto et al. 2005). The involvement of a low fidelity polymerase in the high fidelity HR process is somewhat surprising and suggests a hand-off to another more processive, high-fidelity polymerase might occur. Genetic studies in yeast had suggested earlier the potential involvement of TLS polymerases in recombination-mediated DSB repair (Holbeck and Strathern 1997).
3.4.3 Branch migration: Rad54 and BLM
In bacteria, RuvA protein targets the RuvB motor protein to Holliday junctions. RuvB positions as hexameric rings to two opposite arms of the Holliday junction where they act as coordinated DNA pumps providing the motor force for branch migration (West 2003). Considering the comparably low ATPase and DNA strand activity of Rad51, eukaryotes would appear to require a DNA motor protein for branch migration. However, it is unclear from genetic analyses whether branch migration occurs in eukaryotes or whether the extent of heteroduplex DNA is entirely determined by the extent of the DSB resection. The Rad54 motor protein was shown to enhance branch migration in the three-stranded DNA strand exchange reaction (Fig. 2) by up to six-fold. This activity of Rad54 depended on its ATPase activity and a specific interaction with the Rad51 proteins, as branch migration in reactions catalyzed by RecA or human Rad51 was not stimulated (Solinger and Heyer 2001). The human Rad54 protein displays both specificity in DNA binding and ATPase activity for several types of junction substrates, including X and partial X-junctions, as well as forked DNA (Bugreev et al. 2006). Both yeast and human Rad54 proteins could branch migrate protein-free three-stranded and four-stranded branches in a bi-directional manner, but not branched substrates still associated with Rad51 strand exchange protein (Bugreev et al. 2006). It is unclear whether Rad54 is responsible for the ATP-dependent branch migration activity observed in human extract and partially purified fractions of resolvase A (see below) (Constantinou et al. 2002). A potential role of Rad54 in branch migration is inconsistent with in vivo data measuring conversion track length under conditions where Rad54 or an ATPase-defective Rad54 mutant protein are overexpressed (Kim et al. 2002). Overexpression of wild type Rad54 protein led to a reduction in conversion tract length, not an increase as expected if Rad54 drives branch migration. Whereas overexpression of Rad54-K391R, an ATPase-deficient mutant, led to an increase in conversion tract length, inconsistent with a model that the Rad54 motor drives branch migration in vivo. Branch migration of Holliday junctions formed by RecA-mediated four-stranded DNA strand exchange was demonstrated for the BLM helicase, an activity that is underpinned by the binding preference of this DNA helicase to model Holliday junctions (Karow et al. 2000). During HR, BLM is possibly targeted to junctions by its interactions with Rad51 and the Rad51 paralog, Rad51D (Wu et al. 2001; Braybrooke et al. 2003). BLM is a 3′-5′ DNA helicase of the RecQ family (Table 1), and it will be interesting to learn whether the other RecQ homologs in humans display similar biochemical activities.
3.4.4 Second end capture/DNA annealing by Rad52 and Rad59
The capture of the second end during DSBR could conceivably be accomplished by a second Rad51-mediated DNA strand invasion event or by reannealing the second end to the displaced strand from the initial DNA strand invasion (Fig. 1, Fig. 3). DNA strand annealing in DSBR and SDSA (Figs. 1, 2) likely involves RPA-coated ssDNA, which can be reannealed by Rad52 protein in a highly specific reaction (Sugiyama et al. 1998). It is unclear how such an asymmetry between an invading end (Rad51 filament assembly) and non-invading end (strand annealing) can be mechanistically accomplished. Rad52-mediated reannealing in S. cerevisiae likely involves Rad59 (Table 1), a budding yeast specific Rad52 paralog, that enhances the Rad52 annealing reaction (Bai and Symington 1996; Davis and Symington 2001; Wu et al. 2006b).
3.4.5 Junction resolution/dissolution: the roles of BLM-TOPOIIIα, WRN, and Mus81-Mms4/Eme1
The SDSA subpathway requires dissociation of the D-loop, and this activity has been identified with two RecQ-like DNA helicases, BLM and WRN (van Brabant et al. 2000; Orren et al. 2002). BLM favored a D-loop structure with 5′ invading ssDNA, a non-productive recombination intermediate, but also dissociated D-loops with 3′ invading end (van Brabant et al. 2000). The possible functions of the other three human RecQ-like enzymes in such assays remain to be tested. In vivo analysis in yeast suggested that Srs2, another 3′-5′ DNA helicase (Table 1), plays a crucial role enabling the SDSA pathway (Ira et al. 2003). However, the biochemical analysis of Srs2 does not presently support a function in dissociating D-loops (Krejci et al. 2003; Veaute et al. 2003). See Chapter 4 for a more extended discussion of Srs2.
Processing of double Holliday junctions (dHJs) lies at the heart of crossover formation in present recombination models (Fig. 1, Fig. 3). While single Holliday junctions require endonucleolytic processing, dHJs may be processed by an endonuclease, following the RuvC paradigm of bacteria (see chapter by Cox), or by a combination of a helicase and type I topoisomerase. Elegant biochemical work using an oligonucleotide-based dHJ substrate demonstrated that BLM helicase in conjunction with TOPOIIIα can process dHJs by collapsing the junctions to a hemi-catenane (Fig. 3) that is resolved by the topoisomerase activity (Wu and Hickson 2003). This activity has been termed dHJ dissolution, as opposed to resolution by an endonuclease, and leads to non-crossovers exclusively. A third subunit of the BLM-TOPOIIIα complex, BLAP75/RMI1, appears to recruit TOPOIIIα to dHJs (Raynard et al. 2006; Wu et al. 2006a). The elevated level of sister chromatid exchanges found in BLM-deficient cells can be nicely explained as a direct consequence of a failure to process dHJs to non-crossovers.
Resolvase A
RuvC provides a paradigm for the resolution of HJs and the formation of crossovers (see contribution by Cox). Eukaryotes do not contain an obvious RuvC homolog (Table 1) and the quest to identify the eukaryotic Holliday junction resolvase has been long and arduous (for reviews see Heyer et al. 2003; West 2003). Besides a mitochondrial activity, termed CCE1 (Kleff et al. 1992), the identity of the nuclear resolvase is still elusive. Biochemical fractionation of human cell extracts identified a HJ-specific endonuclease activity, termed Resolvase A, which displays striking similarity to RuvC (Liu et al. 2004). Two of the Rad51 paralogs, Rad51C and Xrcc3, are required for Resolvase A activity, but the complex in its purified form does not exhibit resolvase activity (Liu et al. 2004) suggesting these factors might be required to target the nuclease to the junction but do not constitute the nuclease function themselves. Such a late function of the Rad51 paralogs was surprising, given their suggested role in Rad51 filament formation (see above). However, these Rad51 filament co-factors might stay associated with the growing and/or nucleating end of the Rad51 filament (Fig. 3), which may position them to direct a nuclease to a junction. Identifying the nuclease of Resolvase A will be a major breakthrough, and it will be interesting to test this activity on dHJs, the likely intermediate in the DSBR pathway (Fig. 1, Fig. 3).
Mus81-Mms4/Eme1
Mus81 contains the nuclease function of the heterodimeric, DNA structure-selective endonuclease Mus81-Mms4/Eme1 (Table 1). Mus81 was first identified by its interaction with the Rad54 motor protein in S. cerevisiae (Interthal and Heyer 2000) and as potential substrate of the Cds1 checkpoint kinase in S. pombe (Boddy et al. 2000). Genetic analysis puts this enzyme squarely in the RAD52 epistasis group, but the mutant is not sensitive to DSBs or IR damage, two classic substrates for the HR pathway. This rather confusing genetic behavior suggests that Mus81-Mms4/Eme1 is a context-specific factor required only in certain sub-pathways of HR. The chapter by Whitby will provide an in-depth analysis of this topic, and previous reviews have exhaustively discussed this subject (Haber and Heyer 2001; Heyer et al. 2003; Hollingsworth and Brill 2004). The biochemical activity of Mus81-Mms4/Eme1 (cleavage of Holliday junction versus other junction substrates including D-loops, replication forks, and 3′-flaps) appeared rather unsettled depending on the source and purity of the enzymatic preparation. A consensus has emerged to suggest that nicked HJs are the preferred in vitro substrate, whereas cleavage of intact HJs by the purified enzyme is rather poor (Gaillard et al. 2003; Osman et al. 2003; Fricke et al. 2005). The cleavage of junctions by Mus81-Mms4/Eme1 does not conform with the RuvC paradigm and the question remains: Can this enzyme resolve HJs? Is there a missing co-factor or post-translational modification? In what context does the nuclease function with the HR machinery and does Rad54 position the nuclease at the relevant joints? Regardless of the mechanism, Mus81-Eme1 controls the vast majority of meiotic crossovers in S. pombe (Smith et al. 2003), whereas it makes a more subtle contribution to crossover formation in S. cerevisiae (de los Santos et al. 2003).
3.4.6 Postsynaptic processing of terminal heterologies
During homologous recombination, a 3′ end invades a homologous donor sequence and initiates new DNA synthesis. Using the invading DNA strand as a primer for DNA synthesis requires that non-homologous bases at the 3′ end be removed. Removal of such non-homologous nucleotides can be critical when the strand interruption occurred in an area of non-homology utilizing internal homology for DNA strand invasion, as well as in strand annealing and second end capture, or in gene targeting experiments with DNA fragments that contain terminal heterology. Two pathways have been identified in Saccharomyces cerevisiae that process terminal heterologies on the invading strand. For heterologies greater than 30 nucleotides, the XPF family structure-selective endonuclease Rad1-Rad10 functions as 3′ flap endonuclease in conjunction with the MMR proteins Msh2 and Msh3 and the Srs2 helicase (Ivanov and Haber 1995; Paques and Haber 1997). It is unclear whether the related Mus81-Mms4 endonuclease that can cleaves such substrates well in vitro, functions also in this capacity in vivo. Smaller heterologies are processed by the 3′-to-5′ proofreading activity of DNA polymerase δ (Paques and Haber 1997) and possibly other DNA polymerases. The exact biochemical mechanisms of these pathways have not been analyzed yet.
4 Regulation of recombination
All fundamental DNA metabolic processes, including DNA replication and transcription, are strictly regulated, and HR is no exception. Mechanistically, HR is the pathway in which RPA-coated ssDNA is targeted for assembly of the Rad51 filament. Hence, any reaction that involves RPA needs to shield against Rad51 filament assembly, specifically DNA replication that involves large stretches of RPA-coated ssDNA on the lagging strand. In addition, chromatin modification and remodeling are required to overcome the inherently repressing character of chromatin on DNA transactions. These mechanisms will not only impact the efficiency of HR in vivo but may also be involved in regulating the hierarchy between the individual DSB response pathways (HR, NHEJ, translesion synthesis, fork regression, apoptosis). The mechanisms regulating HR are beginning to emerge and likely involve actively inhibiting pathways (Srs2, MMR), as well as the post-translational modification of HR proteins. Modulation of chromatin structure may provide another regulatory dimension in HR, but a discussion here would go beyond the scope of this review.
4.1 Negative regulation of HR and the roles of the Srs2 DNA helicase and MMR
The isolation of mutations that increase HR, so called hyper-rec mutations, provide genetic evidence for negative regulation of HR. Hyper-rec mutations are often identified in proteins with a normal function in DNA replication or in the recovery of stalled forks by translesion synthesis or fork regression, whose absence leads to an increased probability of lesions initiating HR. DNA damage induced by IR, UV, oxidative stress or agents that lead to fork stalling may also provide such lesions, and these agents were also found to induce recombination (Paques and Haber 1999). In these cases, recombination appears to occur as an indirect consequence of excess RPA-coated ssDNA.
A more specific mechanism of negative regulation of recombination is represented by the Srs2 DNA helicase. SRS2 was identified genetically as a hyper-rec mutation and functions as an active anti-recombinator (Aguilera and Klein 1988; Aboussekhra et al. 1989). The Srs2 protein exhibits a 3′ to 5′ polarity like its homologs UvrD and Rep from bacteria (Rong and Klein 1993) and specifically dismantles the Rad51 presynaptic filament (Krejci et al. 2003; Veaute et al. 2003). When added to an ongoing DNA strand exchange reaction catalyzed by Rad51 and RPA (Fig. 2), Srs2 inhibited product formation by dissociating the Rad51 presynaptic filament, as demonstrated by EM analysis (Krejci et al. 2003; Veaute et al. 2003). However, Srs2 was unable to dissociate Rad51-made DNA joint molecules (Veaute et al. 2003). Hence, the requirement for Srs2 in the repair of a site-specific DSB by SDSA (Aylon et al. 2003; Ira et al. 2003) and the proposal that Srs2 may reverse the D-loop to allow reannealing with the second end cannot be explained by these biochemical data, suggesting that Srs2 might require other co-factors or conditions for such an activity. The biochemical data show that Rad51 presynaptic filament assembly is in a dynamic balance between nucleation/filament extension and disassembly by Srs2. Surprisingly, Srs2 lacks an obvious homolog in mammals, but an Srs2-related protein, Fbh1, is present in mammals and in S. pombe (Morishita et al. 2005; Osman et al. 2005). The fbh1 mutation does suppress the requirement for mediators of Rad51 filament assembly in fission yeast, similar to the phenotype of srs2 in S. cerevisiae. Furthermore, the fbh1 mutation confers a synthetic growth defect with srs2 or rqh1 (encodes a RecQ homolog) mutations in S. pombe, which can be suppressed by a rad51 mutation indicating the accumulation of toxic recombination intermediates in the helicase double mutants (Morishita et al. 2005; Osman et al. 2005). However, it has not yet been shown biochemically that Fbh1 can disrupt Rad51-ssDNA filaments. It is also possible that one of the mammalian RecQ-like helicases can substitute for Srs2 function, because Sgs1 overexpression can rescue an srs2 defect in budding yeast (Mankouri et al. 2002).
The anti-recombination activity of Srs2 is recruited to replication forks by the specific interaction of Srs2 with sumolyated PCNA (Papouli et al. 2005; Pfander et al. 2005). PCNA is sumoylated on lysine 164 during S-phase as well as on lysine 127 after DNA damage. The singly and doubly sumoylated PCNA species specifically interact with Srs2, whereas unmodified PCNA does not. The Srs2 recruitment to origins of replication after hydroxy urea (HU)-induced arrest was also demonstrated in vivo by ChIP, and failure to recruit Srs2 led to an increase in Rad51 protein at those forks. The biochemical and genetic data of Srs2 as well as its interaction with PCNA suggests that HR is actively antagonized at replication forks, and suggests that alternative pathways of processing stalled replication forks, like translesion synthesis and fork regression, are preferred over HR. It is unclear how the Srs2 biochemical activity of Rad51 presynaptic filament dissociation relates to its other functions in DNA damage checkpoint signaling (Liberi et al. 2000), as well as in adaptation and recovery from DNA damage (Vaze et al. 2002).
Recombination, specifically the DNA strand invasion step, is also negatively regulated by the MMR system to avoid recombination between sequences that exceed a certain heterology threshold (also known as homeologous recombination) as demonstrated for E. coli RecA and the bacterial MMR (Worth et al. 1994). Genetic evidence for a similar mechanism in yeast meiosis has been provided (Hunter et al. 1996). The RecQ-like helicase Sgs1 functions in conjunction with the MMR pathway in suppressing homeologous recombination (Myung et al. 2001; Spell and Jinks-Robertson 2004), but the biochemistry of this pathway remains to be worked out.
4.2 Post-translational modification of HR proteins
The importance of post-translational modifications in molecular regulation of HR is evident, and the sumoylation-dependent interaction of Srs2 with PCNA illustrates how a biochemical activity can be targeted to its preferred substrate by induction of a specific protein interaction through post-translational modification. A growing number of HR proteins have been identified to be post-translationally modified, mostly by phosphorylation, often carried out by cell cycle or DNA damage checkpoint kinases (Zhou and Elledge 2000). The discussion here is limited to a few proteins (RPA, Rad51, Rad55-Rad57, Brca2) due to space constraints, and it is anticipated that these examples just scratch the surface of an enormous modification iceberg, which is waiting to be uncovered.
RPA
The eukaryotic ssDNA binding protein, RPA, works at the nexus of replication, repair, and recombination, and its middle subunit (RPA32) has been found to be phosphorylated in a cell-cycle-dependent manner and in response to DNA damage (reviewed in Wold 1997). The cell-cycle-dependent phosphorylation involves cyclin-dependent kinases, but the functional consequences remain unclear. The DNA damage-induced phosphorylation of RPA32 (and likely the large subunit RPA70) is dependent on the PI3 kinase-like kinases DNA-PK, ATM, and perhaps ATR (Mec1, Tel1 in S. cerevisiae), and there was considerable uncertainty about the functional consequences of DNA damage-induced RPA phosphorylation using reconstituted in vitro assays (Wold 1997). The confusion has been clarified to some degree by showing that DNA damage-induced RPA2 phosphorylation targets RPA to repair centers and excludes the phosphorylated RPA from active replication centers (Vassin et al. 2004). Phosphorylated RPA does associate with stalled replication forks, likely involving a phosphorylation-dependent interaction with Mre11-Rad50-Nbs1 complex (Robison et al. 2004; Vassin et al. 2004), but it is unclear whether this interaction is direct or mediated by a bridging protein. Whether RPA phosphorylation only targets the protein to a specific nuclear location or also changes some intrinsic biochemical property of RPA remains unsolved.
Human Rad51
Human Rad51 was found to be phosphorylated by the c-Abl kinase on tyrosine 54 after exposure to IR in cells transiently transfected with HsRad51 and c-Abl (Yuan et al. 1998). Rad51 phosphorylation correlated with decreased DNA binding by phosphorylated Rad51. Tyrosine 54 (tyrosine 112 in yeast Rad51) stacks against another tyrosine in the protomer interface in the crystal structure of the yeast Rad51 protein (Conway et al. 2004). Surface exposure of this site would be expected to exert a negative effect on filament formation, consistent with the observed DNA binding defect. The biological significance of this phosphorylation is unclear, and the effect is somewhat unexpected, because it is predicted to inhibit HR in response to a target lesion of this pathway. c-Abl kinase activated by IR phosphorylated human Rad51 on tyrosine 315 in vitro (Chen et al. 1999). The reason for this difference in the phosphorylation patterns is unclear. Tyrosine 315 phosphorylation correlated with enhanced Rad52 interaction, but the effect on Rad51 function has not been explored (Chen et al. 1999). Tyrosine 315 phosphorylation in response to DNA damage was confirmed using a phosphotyrosine 315-specific antibody, and this residue is constitutively phosphorylated by the oncogenic BCR-ABL fusion protein found in chronic myelogenous leukemia and some acute lymphocytic leukemia patients (Slupianek et al. 2001). Biochemical analysis showed that tyrosine 315 of human Rad51 is required for DNA binding and filament formation (Takizawa et al. 2004), suggesting that phosphorylation of this residue should have a negative effect on Rad51 function. Downregulation of Rad51 in response to a target lesion of the HR pathway is unexpected, and the observed drug resistance in BCR-ABL expressing cells might be more the consequence of the increased Rad51 and Rad51 paralog expression (Slupianek et al. 2001) than an effect of Rad51 tyrosine 315 phosphorylation.
Rad55-Rad57
The Rad55-Rad57 proteins, which are critical for assembly of the Rad51 filament in yeast, are phosphorylated in response to genotoxic stress. Phosphorylation depended on the DNA damage checkpoint (Bashkirov et al. 2000). Mapping of the sites identified a complex pattern of phosphorylation sites, and analysis of three sites in Rad55 (serines 2,8,14) showed that these phosphorylation events positively regulate Rad55-Rad57 function, particularly in the recovery of stalled replication forks (Bashkirov et al. 2006). The biochemical consequences of Rad55-Rad57 phosphorylation have not been identified yet. Human Brca2 is phosphorylated on S3291 by CDK2-cyclin A during the G2/M phase of the cell cycle, which appears to negatively regulate Brca2 function in HR (Esashi et al. 2005). S3291 is part of the C-terminal Rad51 binding site of Brca2 and phosphorylation abrogates the interaction of the site with the Rad51 protein. IR counteracts S3291 phosphorylation, consistent with the expectation that Brca2 activity is required after ionizing radiation. Taking the analogy of the regulation of actin filament assembly through the Arp2/3 nucleating complex, these data suggest that also in HR the proteins controlling Rad51 filament nucleation are important regulatory targets.
These few examples should highlight that HR is not a constitutive repair pathway but is under elaborate negative and likely positive control. Cells avoid HR when unnecessary and in fact unwanted, i.e. during ongoing replication, whereas they promote HR when necessary and critical in the repair of DNA damage and in the recovery of stalled forks. These mechanisms are able to localize HR to the DNA damage site or induce general changes to the recombination machinery by a pan-nuclear response.
5 Conclusion
Homology search and DNA strand invasion are central during HR. The assembly, function, and disassembly of the Rad51 filament are critical for the central reaction. The conservation of this step in the form of the RecA/Rad51/RadA protein underlines its fundamental importance. The multitude of pathways leading to and from the central reaction in processing distinct lesions and resolving the strand invasion product in various ways are catalyzed by context-specific factors, whose requirements may vary with the lesion or postsynaptic pathway. The differences between the RecA and Rad51 cousins, as well as between the yeast and human brothers of eukaryotic Rad51 proteins, will provide a key to understanding the significant differences in the number and function of the other HR proteins that work in conjunction with the central DNA strand exchange proteins. Analogies to other filament forming proteins like actin and tubulin may elucidate the mechanistic details of these cofactors. The greater complexity of the eukaryotic genome structure and the requirement for more intricate regulation of HR in complex cells and organisms will likely be reflected in the biochemical properties of the recombination machinery and its modulation by post-translational modifications. This will provide enough fuel to keep the fire of studying recombination burning for a long while. The individual approaches to understanding homologous recombination in eukaryotes range from single-molecule biophysics to structural analysis, biochemistry, genetics, and cell biology. Their discussion is often segregated as reflected by the individual chapters in this volume. However, it is the combination of all approaches that will ultimately succeed in elucidating the mechanism and regulation of this fascinating cellular pathway.
Acknowledgments
The support of our research by the National Institutes of Health, the Susan G. Komen Breast Cancer Foundation, and University of California BIOSTAR program is gratefully acknowledged. I thank Shannon Ceballos, Tammy Doty, Kirk Ehmsen, Kristina Herzberg, Neil Hunter, Xuan Li, Jie Liu, Eugene Nadezhdin, Mike Rolfsmeier, John Roth, Erin Schwartz, William Wright, and Xiao-Ping Zhang for their critical comments on the manuscript. My sincere apologies for omitting all the references and great work that could not be discussed here due to space constraints.
References
- Aboussekhra A, Chanet R, Adjiri A, Fabre F. Semi-dominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA protein. Mol Cell Biol. 1992;12:3224–3234. doi: 10.1128/mcb.12.7.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboussekhra A, Chanet R, Zgaga Z, Cassier Chauvat C, Heude M, Fabre F. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucl Acids Res. 1989;17:7211–7219. doi: 10.1093/nar/17.18.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A, Klein HL. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics. 1988;119:779–790. doi: 10.1093/genetics/119.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aihara H, Ito Y, Kurumizaka H, Yokoyama S, Shibata T. The N-terminal domain of the human Rad51 protein binds DNA: Structure and a DNA binding surface as revealed by NMR. J Mol Biol. 1999;290:495–504. doi: 10.1006/jmbi.1999.2904. [DOI] [PubMed] [Google Scholar]
- Alexeev A, Mazin A, Kowalczykowski SC. Rad54 protein possesses chromatin-remodeling activity stimulated by a Rad51-ssDNA nucleoprotein filament. Nature Struct Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- Alexiadis V, Kadonaga JT. Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev. 2003;16:2767–2771. doi: 10.1101/gad.1032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitani I, Baskin RJ, Kowalczykowski S. Direct visualization of a chromatin-remodeling protein, Rad54, translocating along single-molecules of double-stranded DNA. Mol Cell. 2006;23:143–148. doi: 10.1016/j.molcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Aylon Y, Liefshitz B, Bitan-Banin G, Kupiec M. Molecular dissection of mitotic recombination in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:1403–1417. doi: 10.1128/MCB.23.4.1403-1417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Symington LS. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 1996;10:2025–2037. doi: 10.1101/gad.10.16.2025. [DOI] [PubMed] [Google Scholar]
- Bashkirov VI, Herzberg K, Rolfsmeier M, Heyer WD. Rad55 phopshorylation on serines 2,8,14 is required for efficient recombinational DNA repair and recovery of stalled replication forks. Mol Cell Biol. 2006 doi: 10.1128/MCB.01317-06. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov VI, King JS, Bashkirova EV, Schmuckli-Maurer J, Heyer WD. DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol Cell Biol. 2000;20:4393–4404. doi: 10.1128/mcb.20.12.4393-4404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- Baumann P, West SC. The human Rad51 protein: polarity of strand transfer and stimulation by hRP-A. EMBO J. 1997;16:5198–5206. doi: 10.1093/emboj/16.17.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beernink HTH, Morrical SW. RMPs: Recombination/replication mediator proteins. Trends Biochem Sci. 1999;24:385–389. doi: 10.1016/s0968-0004(99)01451-6. [DOI] [PubMed] [Google Scholar]
- Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front Biosci. 1998;3:570–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- Bochkarev A, Bochkareva E. From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr Opin Struct Biol. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer WD, Russell P. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol Cell Biol. 2000;20:8758–8766. doi: 10.1128/mcb.20.23.8758-8766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braybrooke JP, Li JL, Wu L, Caple F, Benson FE, Hickson ID. Functional interaction between the Bloom's syndrome helicase and the RAD51 paralog, RAD51L3 (RAD51D) J Biol Chem. 2003;278:48357–48366. doi: 10.1074/jbc.M308838200. [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc Natl Acad Sci USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Mazina OM, Mazin A. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006;442:590–593. doi: 10.1038/nature04889. [DOI] [PubMed] [Google Scholar]
- Chen G, Yuan SSF, Liu W, Xu Y, Trujillo K, Song BW, Cong F, Goff SP, Wu Y, Arlinghaus R, Baltimore D, Gasser PJ, Park MS, Sung P, Lee E. Radiation-induced assembly of Rad51 and Rad52 recombination complex requires ATM and c-Abl. J Biol Chem. 1999;274:12748–12752. doi: 10.1074/jbc.274.18.12748. [DOI] [PubMed] [Google Scholar]
- Chi P, Van Komen S, Sehorn MG, Sigurdsson S, Sung P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amst) 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Clever B, Interthal H, Schmuckli-Maurer J, King J, Sigrist M, Heyer WD. Recombinational repair in yeast: Functional interactions between Rad51 and Rad54 proteins. EMBO J. 1997;16:2535–2544. doi: 10.1093/emboj/16.9.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clever B, Schmuckli-Maurer J, Sigrist M, Glassner B, Heyer WD. Specific negative effects resulting from elevated levels of the recombinational repair protein Rad54p in Saccharomyces cerevisiae. Yeast. 1999;15:721–740. doi: 10.1002/(SICI)1097-0061(19990630)15:9<721::AID-YEA414>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Constantinou A, Chen XB, McGowan CH, West SC. Holliday junction resolution in human cells: two junction endonucleases with distinct substrate specificities. EMBO J. 2002;21:5577–5585. doi: 10.1093/emboj/cdf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway AB, Lynch TW, Zhang Y, Fortin GS, Fung CW, Symington LS, Rice PA. Crystal structure of a Rad51 filament. Nat Rev Struc Mol Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- Davis AP, Symington LS. The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics. 2001;159:515–525. doi: 10.1093/genetics/159.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164:81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan JW, Milne GT, Weaver DT. Homotypic and heterotypic protein associations control Rad51 function in double-strand break repair. Genes Dev. 1994;8:2552–2562. doi: 10.1101/gad.8.21.2552. [DOI] [PubMed] [Google Scholar]
- Egelman EH. A tale of two polymers: New insights into helical filaments. Nat Rev Mol Cell Biol. 2003;4:621–630. doi: 10.1038/nrm1176. [DOI] [PubMed] [Google Scholar]
- Esashi F, Christ N, Gannon J, Liu YL, Hunt T, Jasin M, West SC. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- Fortin GS, Symington LS. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51-DNA complexes. EMBO J. 2002;21:3160–3170. doi: 10.1093/emboj/cdf293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke WM, Bastin-Shanower SA, Brill SJ. Substrate specificity of the Saccharomyces cerevisiae Mus81-Mms4 endonuclease. DNA Repair. 2005;4:243–251. doi: 10.1016/j.dnarep.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Fujimori A, Tachiiri S, Sonoda E, Thompson LH, Dhar PK, Hiraoka M, Takeda S, Zhang Y, Reth M, Takata M. Rad52 partially substitutes for the Rad51 paralog XRCC3 in maintaining chromosomal integrity in vertebrate cells. EMBO J. 2001;20:5513–5520. doi: 10.1093/emboj/20.19.5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard PH, Noguchi E, Shanahan P, Russell P. The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol Cell. 2003;12:747–759. doi: 10.1016/s1097-2765(03)00342-3. [DOI] [PubMed] [Google Scholar]
- Galkin VE, Wu Y, Zhang XP, Qian X, He Y, Yu X, Heyer WD, Luo Y, Egelman EH. The RAD51/RadA N-terminal domain activates nucleoprotein filaments. Structure. 2006;14:983–992. doi: 10.1016/j.str.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Gasior SL, Olivares H, Ear U, Hari DM, Weichselbaum R, Bishop DK. Assembly of RecA-like recombinases: Distinct roles for mediator proteins in mitosis and meiosis. Proc Natl Acad Sci USA. 2001;98:8411–8418. doi: 10.1073/pnas.121046198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior SL, Wong AK, Kora Y, Shinohara A, Bishop DK. Rad52 associates with RPA and functions with Rad55 and Rad57 to assemble meiotic recombination complexes. Genes Dev. 1998;12:2208–2221. doi: 10.1101/gad.12.14.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub EI, Kovalenko OV, Gupta RC, Ward DC, Radding CM. Interaction of human recombination proteins Rad51 and Rad54. Nucleic Acids Res. 1997;25:4106–4110. doi: 10.1093/nar/25.20.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RC, Folta-Stogniew E, O'Malley S, Takahashi M, Radding CM. Rapid exchange of A : T base pairs is essential for recognition of DNA homology by human Rad51 recombination protein. Mol Cell. 1999;4:705–714. doi: 10.1016/s1097-2765(00)80381-0. [DOI] [PubMed] [Google Scholar]
- Haber JE, Heyer WD. The fuss about Mus81. Cell. 2001;107:551–554. doi: 10.1016/s0092-8674(01)00593-1. [DOI] [PubMed] [Google Scholar]
- Hays SL, Firmenich AA, Berg P. Complex formation in yeast double-strand break repair: Participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc Natl Acad Sci USA. 1995;92:6925–6929. doi: 10.1073/pnas.92.15.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays SL, Firmenich AA, Massey P, Banerjee R, Berg P. Studies of the interaction between Rad52 protein and the yeast single-stranded DNA binding protein RPA. Mol Cell Biol. 1998;18:4400–4406. doi: 10.1128/mcb.18.7.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD, Ehmsen KT, Solinger JA. Holliday junctions in the eukaryotic nucleus: Resolution in sight? Trends Biochem Sci. 2003;10:548–557. doi: 10.1016/j.tibs.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Heyer WD, Li XR, Rolfsmeier M, Zhang XP. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl481. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbeck SL, Strathern JN. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics. 1997;147:1017–1024. doi: 10.1093/genetics/147.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth NM, Brill SJ. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 2004;18:117–125. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N, Chambers SR, Louis EJ, Borts RH. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 1996;15:1726–1733. [PMC free article] [PubMed] [Google Scholar]
- Interthal H, Heyer WD. MUS81 encodes a novel Helix-hairpin-Helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol Gen Genet. 2000;263:812–827. doi: 10.1007/s004380000241. [DOI] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov EL, Haber JE. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:2245–2251. doi: 10.1128/mcb.15.4.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskelioff M, Van Komen S, Krebs JE, Sung P, Peterson CL. Rad54p is a chromatin remodeling enzyme required for heteroduplex joint formation with chromatin. J Biol Chem. 2003;278:9212–9218. doi: 10.1074/jbc.M211545200. [DOI] [PubMed] [Google Scholar]
- Jiang H, Xie YQ, Houston P, Stemke-Hale K, Mortensen UH, Rothstein R, Kodadek T. Direct association between the yeast Rad51 and Rad54 recombination proteins. J Biol Chem. 1996;271:33181–33186. doi: 10.1074/jbc.271.52.33181. [DOI] [PubMed] [Google Scholar]
- Johnson RD, Symington LS. Functional differences and interactions among the putative RecA homologs RAD51, RAD55, and RAD57. Mol Cell Biol. 1995;15:4843–4850. doi: 10.1128/mcb.15.9.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa W, Kurumizaka H, Ishitani R, Fukai S, Nureki O, Shibata T, Yokoyama S. Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Mol Cell. 2002;10:359–371. doi: 10.1016/s1097-2765(02)00587-7. [DOI] [PubMed] [Google Scholar]
- Kantake N, Madiraju M, Sugiyama T, Kowalczykowski C. Escherichia coli RecO protein anneals ssDNA complexed with its cognate ssDNA-binding protein: A common step in genetic recombination. Proc Natl Acad Sci USA. 2002;99:15327–15332. doi: 10.1073/pnas.252633399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow JK, Constantinou A, Li JL, West SC, Hickson ID. The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc Natl Acad Sci USA. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T, Araki K, Sonoda E, Yamashita YM, Harada K, Kikuchi K, Masutani C, Hanaoka F, Nozaki K, Hashimoto N, Takeda S. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol Cell. 2005;20:793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Kiianitsa K, Solinger JA, Heyer WD. Rad54 protein exerts diverse modes of ATPase activity on duplex DNA partially and fully covered with Rad51 protein. J Biol Chem. 2002;277:46205–46215. doi: 10.1074/jbc.M207967200. [DOI] [PubMed] [Google Scholar]
- Kiianitsa K, Solinger JA, Heyer WD. Terminal association of the Rad54 protein with the Rad51-dsDNA filament. Proc Natl Acad Sci USA. 2006;103:9767–9772. doi: 10.1073/pnas.0604240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PM, Paffett KS, Solinger JA, Heyer WD, Nickoloff JA. Spontaneous and double-strand break-induced recombination, and gene conversion tract lengths, are differentially affected by overexpression of wild-type or ATPase-defective yeast Rad54. Nucleic Acids Res. 2002;30:2727–2735. doi: 10.1093/nar/gkf413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleff S, Kemper B, Sternglanz R. Identification and characterization of yeast mutants and the gene for a cruciform cutting endonuclease. EMBO J. 1992;11:699–704. doi: 10.1002/j.1460-2075.1992.tb05102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L, Song BW, Bussen W, Rothstein R, Mortensen UH, Sung P. Interaction with Rad51 is indispensable for recombination mediator function of Rad52. J Biol Chem. 2002;277:40132–40141. doi: 10.1074/jbc.M206511200. [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Kurumizaka H, Ikawa S, Nakada M, Eda K, Kagawa W, Takata M, Takeda S, Yokoyama S, Shibata T. Homologous-pairing activity of the human DNA-repair proteins Xrcc3.Rad51C. Proc Natl Acad Sci USA. 2001;98:5538–5543. doi: 10.1073/pnas.091603098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurumizaka H, Ikawa S, Nakada M, Enomoto R, Kagawa W, Kinebuchi T, Yamazoe M, Yokoyama S, Shibata T. Homologous pairing and ring and filament structure formation activities of the human Xrcc2*Rad51D complex. J Biol Chem. 2002;277:14315–14320. doi: 10.1074/jbc.M105719200. [DOI] [PubMed] [Google Scholar]
- Liberi G, Chiolo I, Pellicioli A, Lopes M, Plevani P, Muzi-Falconi M, Foiani M. Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and Cdk1 activity. EMBO J. 2000;19:5027–5038. doi: 10.1093/emboj/19.18.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lio YC, Mazin AV, Kowalczykowski SC, Chen DJ. Complex formation by the human Rad51B and Rad51C DNA repair proteins and their activities in vitro. J Biol Chem. 2003;278:2469–2478. doi: 10.1074/jbc.M211038200. [DOI] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Liu J, Bond JP, Morrical SW. Mechanism of presynaptic filament stabilization by the bacteriophage T4 UvsY recombination mediator protein. Biochemistry. 2006;45:5493–5502. doi: 10.1021/bi0525167. [DOI] [PubMed] [Google Scholar]
- Liu YL, Masson JY, Shah R, O'Regan P, West SC. RAD51C is required for Holliday junction processing in mammalian cells. Science. 2004;303:243–246. doi: 10.1126/science.1093037. [DOI] [PubMed] [Google Scholar]
- Mankouri HW, Craig TJ, Morgan A. SGS1 is a multicopy suppressor of srs2: functional overlap between DNA helicases. Nucleic Acids Res. 2002;30:1103–1113. doi: 10.1093/nar/30.5.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin V, Chahwan C, Gao H, Blais V, Wohlschlegel J, Yates JR, 3rd, McGowan CH, Russell P. Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. EMBO J. 2006;25:2564–2574. doi: 10.1038/sj.emboj.7601141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson JY, Stasiak AZ, Stasiak A, Benson FE, West SC. Complex formation by the human RAD51C and XRCC3 recombination repair proteins. Proc Natl Acad Sci USA. 2001a;98:8440–8446. doi: 10.1073/pnas.111005698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson JY, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, McIlwraith MJ, Benson FE, West SC. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 2001b;15:3296–3307. doi: 10.1101/gad.947001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin AV, Alexeev AA, Kowalczykowski SC. A novel function of Rad54 protein - Stabilization of the Rad51 nucleoprotein filament. J Biol Chem. 2003;278:14029–14036. doi: 10.1074/jbc.M212779200. [DOI] [PubMed] [Google Scholar]
- Mazin AV, Bornarth CJ, Solinger JA, Heyer WD, Kowalczykowski SC. Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol Cell. 2000a;6:583–592. doi: 10.1016/s1097-2765(00)00057-5. [DOI] [PubMed] [Google Scholar]
- Mazin AV, Zaitseva E, Sung P, Kowalczykowski SC. Tailed duplex DNA is the preferred substrate for Rad51 protein-mediated homologous pairing. EMBO J. 2000b;19:1148–1156. doi: 10.1093/emboj/19.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin OM, Mazin AW. Human Rad54 protein stimulates DNA strand exchange activity of hRad51 protein in the presence of Ca2+ J Biol Chem. 2004;279:52041–52051. doi: 10.1074/jbc.M410244200. [DOI] [PubMed] [Google Scholar]
- McIlwraith MJ, Vaisman A, Liu YL, Fanning E, Woodgate R, West SC. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol Cell. 2005;20:783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Morimatsu K, Kowalczykowski SC. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: A universal step of recombinational repair. Mol Cell. 2003;11:1337–1347. doi: 10.1016/s1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- Morishita T, Furukawa F, Sakaguchi C, Toda T, Carr AM, Iwasaki H, Shinagawa H. Role of the Schizosaccharomyces pombe F-Box DNA helicase in processing recombination intermediates. Mol Cell Biol. 2005;25:8074–8083. doi: 10.1128/MCB.25.18.8074-8083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Agard DA. Gamma-tubulin complexes and microtubule nucleation. Curr Opin Struct Biol. 2001;11:174–181. doi: 10.1016/s0959-440x(00)00187-1. [DOI] [PubMed] [Google Scholar]
- Myung K, Datta A, Chen C, Kolodner RD. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat Genet. 2001;27:113–116. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- Namsaraev EA, Berg P. Rad51 uses one mechanism to drive DNA strand exchange in both directions. J Biol Chem. 2000;275:3970–3976. doi: 10.1074/jbc.275.6.3970. [DOI] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- Orren DK, Theodore S, Machwe A. The Werner syndrome helicase/exonuclease (WRN) disrupts and degrades D-loops in vitro. Biochemistry. 2002;41:13483–13488. doi: 10.1021/bi0266986. [DOI] [PubMed] [Google Scholar]
- Osman F, Dixon J, Barr AR, Whitby MC. The F-Box DNA helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Mol Cell Biol. 2005;25:8084–8096. doi: 10.1128/MCB.25.18.8084-8096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Dixon J, Doe CL, Whitby MC. Generating crossovers by resolution of nicked Holliday junctions: A role of Mus81-Eme1 in meiosis. Mol Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- Papouli E, Chen SH, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L, Venkitaraman A. Emerging functions of BRCA2 in DNA recombination. Trends Biochem Sci. 2004;29:310–316. doi: 10.1016/j.tibs.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- Petukhova G, Sung P, Klein H. Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev. 2000;14:2206–2215. doi: 10.1101/gad.826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova G, Van Komen S, Vergano S, Klein H, Sung P. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J Biol Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Rattray AJ, Strathern JN. Error-prone DNA polymerases: When making a mistake is the only way to get ahead. Annu Rev Genet. 2003;37:31–66. doi: 10.1146/annurev.genet.37.042203.132748. [DOI] [PubMed] [Google Scholar]
- Raynard S, Bussen W, Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase III alpha, and BLAP75. J Biol Chem. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- Rijkers T, VandenOuweland J, Morolli B, Rolink AG, Baarends WM, VanSloun PPH, Lohman PHM, Pastink A. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol Cell Biol. 1998;18:6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristic D, Modesti M, van der Heijden T, van Noort J, Dekker C, Kanaar R, Wyman C. Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Res. 2005;33:3292–3302. doi: 10.1093/nar/gki640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristic D, Wyman C, Paulusma C, Kanaar R. The architecture of the human Rad54-DNA complex provides evidence for protein translocation along DNA. Proc Natl Acad Sci USA. 2001;98:8454–8460. doi: 10.1073/pnas.151056798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison JG, Elliott J, Dixon K, Oakley GG. Replication protein A and the Mre11 •Rad50•Nbs1 complex co-localize and interact at sites of stalled replication forks. J Biol Chem. 2004;279:34802–34810. doi: 10.1074/jbc.M404750200. [DOI] [PubMed] [Google Scholar]
- Rong L, Klein HL. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J Biol Chem. 1993;268:1252–1259. [PubMed] [Google Scholar]
- San Filippo J, Chi P, Sehorn MG, Etchin J, Krejci L, Sung P. Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J Biol Chem. 2006;281:11649–11657. doi: 10.1074/jbc.M601249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz EM, Haseltine CA, Kowalczykowski SC. DNA recombination and repair in archaea. Adv Appl Microbiol. 2001;50:101–169. doi: 10.1016/s0065-2164(01)50005-2. [DOI] [PubMed] [Google Scholar]
- Shim KS, Schmutte C, Tombline G, Heinen CD, Fishel R. hXRCC2 enhances ADP/ATP processing and strand exchange by hRAD51. J Biol Chem. 2004;279:30385–30394. doi: 10.1074/jbc.M306066200. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikea K, Ogawa T. Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nat Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T. Rad52 forms ring structures and co-operates with RPA in single-strand annealing. Genes Cells. 1998;3:145–156. doi: 10.1046/j.1365-2443.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- Shor E, Weinstein J, Rothstein R. Genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3 and CSM2: Four genes involved in error-free DNA repair. Genetics. 2005;169:1275–1289. doi: 10.1534/genetics.104.036764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson S, Trujillo K, Song BW, Stratton S, Sung P. Basis for avid homologous DNA strand exchange by human Rad51 and RPA. J Biol Chem. 2001a;276:8798–8806. doi: 10.1074/jbc.M010011200. [DOI] [PubMed] [Google Scholar]
- Sigurdsson S, Van Komen S, Bussen W, Schild D, Albala JS, Sung P. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev. 2001b;15:3308–3318. doi: 10.1101/gad.935501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson S, Van Komen S, Petukhova G, Sung P. Homologous DNA pairing by human recombination factors Rad51 and Rad54. J Biol Chem. 2002;277:42790–42794. doi: 10.1074/jbc.M208004200. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Sawaya MR, Ellenbeger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Wentzell LM, Liu YL, West SC, Wigley DB. Structure of the single-strand annealing domain of human RAD52 protein. Proc Natl Acad Sci USA. 2002;99:13492–13497. doi: 10.1073/pnas.212449899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupianek A, Schmutte C, Tombline G, Nieborowska-Skorska M, Hoser G, Nowicki MO, Pierce AJ, Fishel R, Skorski T. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol Cell. 2001;8:795–806. doi: 10.1016/s1097-2765(01)00357-4. [DOI] [PubMed] [Google Scholar]
- Smith GR, Boddy MN, Shanahan P, Russell P. Fission yeast Mus81 •Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics. 2003;165:2289–2293. doi: 10.1093/genetics/165.4.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger JA, Heyer WD. Rad54 protein stimulates the postsynaptic phase of Rad51 protein-mediated DNA strand exchange. Proc Natl Acad Sci USA. 2001;98:8447–8453. doi: 10.1073/pnas.121009898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger JA, Kiianitsa K, Heyer WD. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol Cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- Solinger JA, Lutz G, Sugiyama T, Kowalczykowski SC, Heyer WD. Rad54 protein stimulates heteroduplex DNA formation in the synaptic phase of DNA strand exchange via specific interactions with the presynaptic Rad51 nucleoprotein filament. J Mol Biol. 2001;307:1207–1221. doi: 10.1006/jmbi.2001.4555. [DOI] [PubMed] [Google Scholar]
- Spell RM, Jinks-Robertson S. Examination of the roles of Sgs1 and Srs2 helicases in the enforcement of recombination fidelity in Saccharomyces cerevisiae. Genetics. 2004;168:1855–1865. doi: 10.1534/genetics.104.032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies M, Kowalczykowski SC. The RecA binding locus of RecBCD is a general domain for recruitment of DNA strand exchange proteins. Mol Cell. 2006;21:573–580. doi: 10.1016/j.molcel.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Stasiak AZ, Larquet E, Stasiak A, Muller S, Engel A, Van Dyck E, West SC, Egelman EH. The human Rad52 protein exists as a heptameric ring. Curr Biol. 2000;10:337–340. doi: 10.1016/s0960-9822(00)00385-7. [DOI] [PubMed] [Google Scholar]
- Story RM, Weber IT, Steitz TA. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Kowalczykowski SC. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J Biol Chem. 2002;277:31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, New JH, Kowalczykowski SC. DNA annealing by Rad52 Protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc Natl Acad Sci USA. 1998;95:6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Zaitseva EM, Kowalczykowski SC. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J Biol Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997a;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997b;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. J Biol Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- Sung P, Stratton SA. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J Biol Chem. 1996;271:27983–27986. doi: 10.1074/jbc.271.45.27983. [DOI] [PubMed] [Google Scholar]
- Swagemakers SMA, Essers J, deWit J, Hoeijmakers JHJ, Kanaar R. The human Rad54 recombinational DNA repair protein is a double-stranded DNA-dependent AT-Pase. J Biol Chem. 1998;273:28292–28297. doi: 10.1074/jbc.273.43.28292. [DOI] [PubMed] [Google Scholar]
- Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Fukushima T, Morrison C, Albala JS, Swagemakers SMA, Kanaar R, Thompson LH, Takeda S. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol Cell Biol. 2000;20:6476–6482. doi: 10.1128/mcb.20.17.6476-6482.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa Y, Kinebuchi T, Kagawa W, Yokoyama S, Shibata T, Kurumizaka H. Mutational analyses of the human Rad51-Tyr315 residue, a site for phosphorylation in leukaemia cells. Genes Cells. 2004;9:781–790. doi: 10.1111/j.1365-2443.2004.00772.x. [DOI] [PubMed] [Google Scholar]
- Tan TLR, Kanaar R, Wyman C. Rad54, a Jack of all trades in homologous recombination (vol 2, pg 787, 2003) DNA Repair (Amst) 2003;2:1293. doi: 10.1016/s1568-7864(03)00070-3. [DOI] [PubMed] [Google Scholar]
- Tarsounas M, Davies D, West SC. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 2003;22:1115–1123. doi: 10.1038/sj.onc.1206263. [DOI] [PubMed] [Google Scholar]
- Thacker J. The RAD51 gene family, genetic instability and cancer. Cancer Lett. 2005;219:125–135. doi: 10.1016/j.canlet.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Tombline G, Fishel R. Biochemical characterization of the human RAD51 protein -I. ATP hydrolysis. J Biol Chem. 2002;277:14417–14425. doi: 10.1074/jbc.M109915200. [DOI] [PubMed] [Google Scholar]
- Tombline G, Heinen CD, Shim KS, Fishel R. Biochemical characterization of the human RAD51 protein - III. - Modulation of DNA binding by adenosine nucleotides. J Biol Chem. 2002;277:14434–14442. doi: 10.1074/jbc.M109917200. [DOI] [PubMed] [Google Scholar]
- van Brabant AJ, Ye T, Sanz M, German JL, Ellis NA, Holloman WK. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- Van Komen S, Petukhova G, Sigurdsson S, Stratton S, Sung P. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol Cell. 2000;6:563–572. doi: 10.1016/s1097-2765(00)00055-1. [DOI] [PubMed] [Google Scholar]
- Vassin VM, Wold MS, Borowiec JA. Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol Cell Biol. 2004;24:1930–1943. doi: 10.1128/MCB.24.5.1930-1943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaze MB, Pellicioli A, Lee SE, Ira G, Liberi G, Arbel-Eden A, Foiani M, Haber JE. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell. 2002;10:373–385. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Wolner B, Peterson CL. ATP-dependent and ATP-independent roles for the Rad54 chromatin remodeling enzyme during recombinational repair of a DNA double strand break. J Biol Chem. 2005;280:10855–10860. doi: 10.1074/jbc.M414388200. [DOI] [PubMed] [Google Scholar]
- Worth L, Clark S, Radman M, Modrich P. Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. Proc Natl Acad Sci USA. 1994;91:3238–3241. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Bachrati CZ, Ou JW, Xu C, Yin JH, Chang M, Wang WD, Li L, Brown GW, Hickson ID. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci USA. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Davies SL, Levitt NC, Hickson ID. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J Biol Chem. 2001;276:19375–19381. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Hickson ID. The Bloom's syndrome helicase suppresses crossing-over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Wu Y, Sugiyama T, Kowalczykowski SC. DNA annealing mediated by Rad52 and Rad59 proteins. J Biol Chem. 2006;281:15441–15449. doi: 10.1074/jbc.M601827200. [DOI] [PubMed] [Google Scholar]
- Wyman C, Ristic D, Kanaar R. Homologous recombination-mediated double-strand break repair. DNA Repair (Amst) 2004;3:827–833. doi: 10.1016/j.dnarep.2004.03.037. [DOI] [PubMed] [Google Scholar]
- Xu L, Marians KJ. A dynamic RecA filament permits DNA polymerase-catalyzed extension of the invading strand in recombination intermediates. J Biol Chem. 2002;277:14321–14328. doi: 10.1074/jbc.M112418200. [DOI] [PubMed] [Google Scholar]
- Yang HJ, Jeffrey PD, Miller J, Kinnucan E, Sun YT, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- Yang HJ, Li QB, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- Yonetani Y, Hochegger H, Sonoda E, Shinya S, Yoshikawa H, Takeda S, Yamazoe M. Differential and collaborative actions of Rad51 paralog proteins in cellular response to DNA damage. Nucleic Acids Res. 2005;33:4544–4552. doi: 10.1093/nar/gki766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Jacobs SA, West SC, Ogawa T, Egelman EH. Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc Natl Acad Sci USA. 2001;98:8419–8425. doi: 10.1073/pnas.111005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan ZM, Huang YY, Ishiko T, Nakada S, Utsugisawa T, Kharbanda S, Wang R, Sung P, Shinohara A, Weichselbaum R, Kufe D. Regulation of Rad51 function by c-Abl in response to DNA damage. J Biol Chem. 1998;273:3799–3802. doi: 10.1074/jbc.273.7.3799. [DOI] [PubMed] [Google Scholar]
- Zaitseva EM, Zaitsev EN, Kowalczykowski SC. The DNA binding properties of Saccharomyces cerevisiae Rad51 protein. J Biol Chem. 1999;274:2907–2915. doi: 10.1074/jbc.274.5.2907. [DOI] [PubMed] [Google Scholar]
- Zhou BBS, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- Zigmond SH. Formin-induced nucleation of actin filaments. Curr Opin Cell Biol. 2004;16:99–105. doi: 10.1016/j.ceb.2003.10.019. [DOI] [PubMed] [Google Scholar]


