Abstract
High-throughput instruments were recently developed to determine gene expression patterns on tissue sections by RNA in situ hybridization. The resulting images of gene expression patterns, chiefly of E14.5 mouse embryos, are accessible to the public at http://www.genepaint.org. This relational database is searchable for gene identifiers and RNA probe sequences. Moreover, patterns and intensity of expression in ∼100 different embryonic tissues are annotated and can be searched using a standardized catalog of anatomical structures. A virtual microscope tool, the Zoom Image Server, was implemented in GenePaint.org and permits interactive zooming and panning across ∼15 000 high-resolution images.
INTRODUCTION
The recent sequencing of the human and mouse genomes (1–3) and large-scale sequence analysis of cDNA libraries (4) provide a framework of sequences that permit construction of a sophisticated map of mammalian genomes. This achievement leads to another major task, which is determining the function of the estimated 30 000–40 000 genes that are predicted from genome maps. Biochemical function and subcellular localization of a novel protein is with a reasonable degree of certainty predictable from homologous, well-studied proteins. In contrast, understanding gene and protein function at a physiologic level depends on a spectrum of experimental approaches such as expression analysis, genetic manipulation and cell-based assays. Although expression patterns are encoded in cis-regulatory elements of genes, theoretical prediction of gene expression patterns seems currently elusive, prompting the development of suitable experimental genomic techniques for expression analysis. DNA arrays elucidate expression on a transcriptome-wide level (5) and methods to analyze gene expression in individual cells using arrays are emerging (6). In situ hybridization (ISH) is a comprehensive method of determining gene expression in cells residing in their natural environment. When automated (7–9), ISH can provide expression data on a transcriptome-wide scale, although at a slower rate than array-based methods and at higher costs. The enormous wealth of information inherent in ISH data prompted us to establish an interactive database, GenePaint.org, to make expression patterns available to the scientific community.
SYNOPSIS OF DATA GENERATION AND IMAGE ACQUISITION
Expression patterns are determined by means of non-radioactive ISH on frozen sections of mouse embryos and mouse brains (see Table 1) as described (7–9). A link (‘Manual of GenePaint’) on the home page of GenePaint.org leads to a description of the pertinent experimental techniques on which ISH data production rests. Briefly, specimens are serially sectioned and arranged in sets on standard microscope slides (Fig. 1A and B). Each set is hybridized in a flow-through chamber with a particular antisense digoxigenin-tagged RNA probe. Probes are detected by means of a two-step catalyzed reporter deposition method which increases the sensitivity at least 5- to 10-fold, perhaps more (10), and is thus comparable to radioactive protocols while preserving the distinct localization of ISH signal and offering single cell resolution. We estimate that as few as five copies of mRNA/cell can be detected. Gene expression patterns are digitally photographed at a resolution of either 1.6 µm or 3.2 µm/pixel using a customized compound microscope equipped with a motorized scanning stage and a digital camera. At 1.6 µm/pixel, a typical section of a post-coital embryonic day 14.5 (E14.5) embryo requires the collection of 195 single exposures, each of which is 500 × 668 pixels and has 24-bit color depth. The TIFF file assembled automatically from these single elements and cropped to remove blank space has a size in the range 100–200 MB.
Table 1. Data content of GenePaint.org (snapshot of August 12, 2003).
| Developmental stage | Type of specimen | Sections per set | Position of sections | Number of genes available |
|---|---|---|---|---|
| E10.5 | Whole embryo | 1–5 | Representative sections | 112 |
| E14.5 | Whole embryo | >20 | 100–175 µm constant spacing | 390 |
| E15.5 | Head | 10 | Selected standard planes | 132 |
| P7 | Brain | 11 | Selected standard planes | 142 |
| Adult (P56) | Brain | 10 | Selected standard planes | 139 |
Sections per set are the standard values that apply to the majority of sets at the respective stage. In cases where no expression is detected, only one or a few representative sections close to the midline are scanned and publicly available.
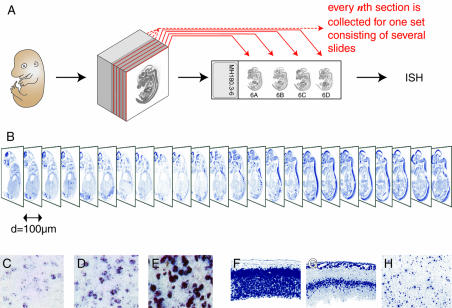
Figure 1.
GenePaint image data. (A) A day 14.5 mouse embryo, embedded in a block of cryomedium, is cryosectioned at 20 µm thickness. The first, sixth, eleventh, etc., section is collected onto standard size microscopy slides constituting one slide set. Starting with the second, third, fourth and fifth section, four more slide sets are generated in an analogous manner. Hence a single embryo gives rise to a total of five sets of slides. (B) Each slide set is hybridized with a particular riboprobe of interest and digital images are created. Typically, an E14.5 embryo is accommodated on six slides with four sections each, resulting in a 24 image stack that covers all major organ systems. (C–E) illustrate the strength of expression. Weak (C), medium (D) and strong (E) are characterized by different quantities of color precipitate within cells expressing the gene in question. (F–H) illustrate expression patterns that can either be ubiquitous, regional or scattered. Foxg1 (F) is ubiquitous in the E14.5 cerebral cortex as it is expressed by all cells (GenePaint set MH224). Expression of Cxcl12 (G) encoding a chemokine ligand is regional since its transcripts are restricted to the intermediate layer of the developing cortex (GenePaint set MH428). Neuropeptide Y (H) is expressed by a subset of cells in all layers of the adult cortex (GenePaint set HB244); expression is thus scattered.
THE DATA
GenePaint.org contains images and associated meta-data (specimen identifier, sequence of the template used for in vitro transcription of the RNA probe, hybridization conditions, etc.). In the case of E14.5 mouse embryos, all sections of a set are uploaded onto GenePaint.org while in the case of E15.5 heads, postnatal brains at postnatal day 7 (P7) and adult brains at P56, 10 (E15.5, adult) or 11 (P7) planes representing certain standard sections are available. The main features of these standard planes are described on the ‘Brain Maps’ pages accessible at GenePaint.org. ‘Brain Maps’ are annotated Nissl stains of sagittal sections of E15.5 mouse head, and postnatal P7 and P56 brains. In cases in which no expression is detected, GenePaint.org contains a single section located near the midline of the specimen. At all stages of data production, quality control steps are implemented. This includes sequence validation of templates, tissue quality assessment and inclusion of positive and negative controls for each ISH experiment. If the data fail at any of these steps, they are re-collected. After final evaluation of the quality of the digital image, data are made public. Expert annotations of embryonic expression patterns (see below) are made public as soon as available.
QUERYING GenePaint.org
GenePaint.org is a relational DB2 database and is located at the Gesellschaft für wissenschaftliche Datenverarbeitung (GWDG) in Göttingen, Germany. GenePaint.org has two components, one is public and is the theme of this article, the other is a laboratory management system employed to document all experimental steps and parameters required for the production of the data that eventually appear in the public component. During the stage of data quality control preceding publication, data are accessible by password only. GenePaint.org uses the nomenclature for genes defined in LocusLink (11) and the Mouse Genome Database (12), which follow the rules set forth by the International Committee on Standardized Genetic Nomenclature for Mice (13).
Gene expression patterns can be retrieved by several strategies.
(i) Retrieval through the ‘Gene Directory’ located on the home page. This directory is a complete and printable spreadsheet of all genes present in GenePaint.org. It provides (from left to right) (a) links leading to LocusLink and GenBank, (b) the GenBank accession number of the sequence that is used for designing the RNA probe, (c) the LocusLink ID, (d) the number of data sets present on GenePaint.org for the gene in question, (e) the status of annotation (a star symbol indicates whether sites of expression are annotated), (f) the gene symbol as defined in Mouse Genome Informatics (MGI), (g) the gene name and (h) aliases and alternative gene names and additional gene description. Note that no systematic efforts are made in GenePaint.org to record accession numbers of related sequences.
(ii) Retrieval through entering into the query field of the home page either a gene name, gene symbol, LocusLink ID, GenBank accession number or GenePaint set ID. The latter is a unique number defined in GenePaint.org that identifies a set of sections hybridized with a particular probe and typically consists of letters followed by a number (e.g. MH99).
(iii) Entry of queries listed under (ii) above into the top query field of the ‘Advanced Search’ page (Fig. 2A, top field in gray query section).
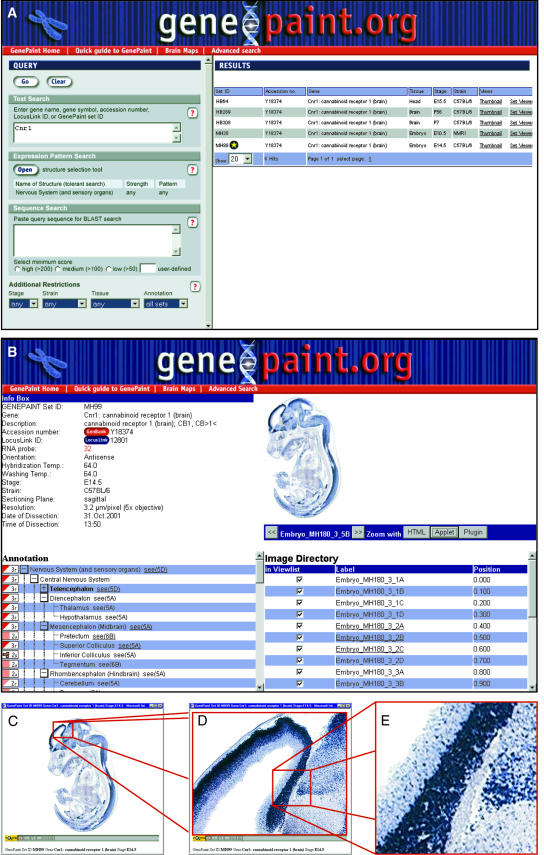
Figure 2.
Steps of a query for expression data based on a gene name. (A) In order to localize expression data for cannabinoid receptor 1, its LocusLink gene symbol ‘Cnr1’ is typed into the query field. Several data sets at different developmental stages are displayed as results. (B) Data set MH99 is selected and opened in the set viewer. Information related to this gene and data set is displayed in the left part of the page, along with the annotation of the expression pattern. Browsing through the list of images in the right part of the page, an image of interest is selected and opened in the Java applet viewer (C). (D) Any region of the image can be magnified either by stepwise zooming in or by directly selecting a rectangle of interest. (E) At the maximum zoom level, single cells are distinguishable [magnified view of the original screenshot in (D) without further resolution enhancement].
(iv) Entry of a DNA sequence into a dedicated field of the query section of the ‘Advanced Search’ page (Fig. 2A). A BLAST search (14) implemented in GenePaint.org compares query sequences with the coding strand sequences of the templates used for RNA probe synthesis. Since optimal RNA probes are complementary to non-conserved regions of the expressed sequence, BLAST searches using conserved motifs are less likely to yield a result. The results of the BLAST search are graphically displayed upon clicking the score in the ‘Score’ column of the ‘Results’ page.
(v) Search for expression in anatomical structures in E14.5 mouse embryos. In an effort to make expression patterns systematically searchable, a growing number of genes at E14.5 are annotated according to a predefined, hierarchically organized list of anatomical structures. For the most part, terms used are those suggested by the Mouse Atlas Project (15). Both the expression level and the local distribution pattern (see Fig. 1C–H) of a gene are recorded for each of the anatomical structures. In order to search for genes that are expressed in a particular region, the structure selection tool in the query section of the advanced search page is opened (Fig. 2A), producing a selection window. Defining the desired expression pattern in the selection window involves three consecutive steps. First, the extendable tree of structures (right part of the selection window) is used to select all structures of interest, which are subsequently tabulated on the left of the page. Second, for each structure the desired expression level (none, weak, medium, strong) and local distribution pattern (any, ubiquitous within the structure, regionalized to part of the structure, scattered) is specified using a selection box. Third, the search mode is selected. Hits may fulfill ‘all’ or ‘any’ of the defined criteria, corresponding to Boolean operators AND or OR. The available search strategies are ‘stringent’ (the selected expression type has to match the exact structure defined by the user) or ‘tolerant’ (the selected expression type must be present either in the structure defined by the user or in any substructure thereof).
DISPLAY OF SEARCH RESULTS
Execution of any of the search strategies outlined above produces a ‘Results’ table with as many lines as data sets exist in GenePaint.org that meet the search criteria. The ‘Results’ table contains the following data (from left to right, Fig. 2A): (a) BLAST score if a BLAST search was conducted, (b) GenePaint set ID, (c) accession number of gene sequence used for template production, (d) gene symbol and name (e) type of tissue examined, (f) stage of specimen, (g) mouse strain used. The two rightmost fields are links to either a thumbnail representation of images contained in the data set or to the ‘Set Viewer’, which is described in the next section.
VIEWING DATA SETS AND IMAGE DATA
The ‘Set Viewer’ page is the most important information display page of the GenePaint database and offers four types of data.
(i) An info box with metadata such as GenePaint set ID, gene name, link to template sequence, specimen data (Fig. 2B, top left).
(ii) A directory of images for the selected GenePaint set (Fig. 2B, bottom right). Each directory line represents one image whose name is defined in the left column. The right-hand column specifies the position in millimeters of a particular section within the image stack (Fig. 1B) with the lateralmost section being defined as zero.
(iii) A low-resolution image of individual sections in the stack through which the user can browse using the arrow buttons underneath the thumbnail (Fig. 2B, top right). The image changes as the user browses through the image stack. Instead of using forward and backward arrows, thumbnail images can be accessed directly by activating an image label in the image directory.
(iv) An ‘Annotation’ table (Fig. 2B, bottom left), which lists each of the anatomical structures annotated. Icons situated on the left-hand side of this table provide for quick viewing of intensity and pattern of expression. Structures are organized in the same hierarchical fashion as used for the anatomy search [see point (v) in the preceding section] and the resulting tree of structures can be either expanded or collapsed. Sections that typically illustrate the expression pattern are accessed by a link provided on the right-hand side of the structure in question.
The primary TIFF image can exceed 100 MB for one single section, since it is assembled as a mosaic from multiple single exposures. In order to view such images through the internet at reasonable speed, images are stored on the GenePaint server in a multi-resolution file format. The GenePaint.org server uses the Zoom Image Server (www.iseemedia.com), an internet imaging viewer that allows dynamic reloading of user-defined regions of interest (Fig. 2C–E). Thus, it is possible to zoom into any region of an image up to its original resolution. Currently, three different options for this ‘virtual microscope’ function are available. A plain HTML viewer is compatible with all platforms, while the Java applet or the plug-in viewer (link for download is found on the home page of GenePaint.org) provide additional convenient features. Various self-explanatory buttons at the bottom of each viewer allow for zooming in and out of images, panning and activation of a magnifying lens (applet only). Images can be downloaded as large JPEGs (5–10 MB) as described in the Quick Guide manual.
CITING GENEPAINT ENTRIES
For general references to GenePaint.org, please either provide the full address (http://www.GenePaint.org) or, if references to URLs are not permitted, please cite this article. For referring to specific data sets contained in GenePaint.org, please provide the GenePaint set ID (MH99). The correct GenePaint set ID is the first line of the info box of the ‘Set Viewer’ and should not be confused with the label of single sections, which may have a similar format (e.g. Embryo_MH180_3_3C).
PERSPECTIVES
Currently, expression data for several hundred genes are available at GenePaint.org (Table 1) and additional image data for E14.5 embryos and the corresponding annotations of sites, strength and pattern of expression are continuously added to GenePaint.org. This database already contains ∼15 000 sections, and this repertoire is expanded by more than 1000 sections each month, eventually providing a transcriptome-wide gene expression atlas of the E14.5 mouse embryo. As GenePaint.org becomes more comprehensive, interoperability with other major biology databases is anticipated.
Also note that the home page of GenePaint.org provides a link (‘Request’) to a form and instructions to submit requests to have one or more ISH analyses done at no cost. Such analyses will initially be limited to E14.5 mouse embryo sections. The benefit of an exhaustive analysis of this particular stage is that at E14.5 most of the major organ systems characteristic of an adult mammal are already present. It has long been thought that the genetic program of pathologic states such as dysplasia and cancers in the adult organism resembles that ongoing in developing tissues. Thus gene expression in the embryo may be a rich resource for understanding the pathology of the adult organ. GenePaint.org is therefore not only aimed at developmental biologists but will also contribute to cancer biology and tissue regeneration research.
Acknowledgments
ACKNOWLEDGEMENTS
Funding for GenePaint is provided by Burroughs Wellcome Fund, the European Union, the German Ministry of Research, the Max Planck Society, the Merck Genome Research Institute and the National Institute of Neurological Disorders and Stroke (USA). A.V. is supported by a Boehringer Ingelheim Fonds scholarship.
REFERENCES
- 1.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 2.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 3.Waterston R.H., Lindblad-Toh,K., Birney,E., Rogers,J., Abril,J.F., Agarwal,P., Agarwala,R., Ainscough,R., Alexandersson,M., An,P. et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature, 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 4.Okazaki Y., Furuno,M., Kasukawa,T., Adachi,J., Bono,H., Kondo,S., Nikaido,I., Osato,N., Saito,R., Suzuki,H. et al. (2002) Analysis of the mouse transcriptome based on functional annotation of 60 770 full-length cDNAs. Nature, 420, 563–573. [DOI] [PubMed] [Google Scholar]
- 5.Barlow C. and Lockhart,D.J. (2002) DNA arrays and neurobiology—what’s new and what’s next? Curr. Opin. Neurobiol., 12, 554–561. [DOI] [PubMed] [Google Scholar]
- 6.Tietjen I., Rihel,J.M., Cao,Y., Koentges,G., Zakhary,L. and Dulac,C. (2003) Single-cell transcriptional analysis of neuronal progenitors. Neuron, 38, 161–175. [DOI] [PubMed] [Google Scholar]
- 7.Carson J.P., Thaller,C. and Eichele,G. (2002) A transcriptome atlas of the mouse brain at cellular resolution. Curr. Opin. Neurobiol., 12, 562–565. [DOI] [PubMed] [Google Scholar]
- 8.Herzig U., Cadenas,C., Sieckmann,F., Sierralta,W., Thaller,C., Visel,A. and Eichele,G. (2001) Development of high-throughput tools to unravel the complexity of gene expression patterns in the mammalian brain. Novartis Found. Symp., 239, 129–146. [DOI] [PubMed] [Google Scholar]
- 9.Visel A., Ahdidan,J. and Eichele,G. (2003) A gene expression map of the mouse brain. In Koetter,R. (ed.), Neuroscience Databases. Kluwer Academic Publishers, Boston, MA, USA, pp. 19–36. [Google Scholar]
- 10.Speel E.J. (1999) Detection and amplification systems for sensitive, multiple-target DNA and RNA in situ hybridization: looking inside cells with a spectrum of colors. Histochem. Cell Biol., 112, 89–113. [DOI] [PubMed] [Google Scholar]
- 11.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blake J.A., Richardson,J.E., Bult,C.J., Kadin,J.A. and Eppig,J.T. (2003) MGD: the Mouse Genome Database. Nucleic Acids Res., 31, 193–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maltais L.J., Blake,J.A., Chu,T., Lutz,C.M., Eppig,J.T. and Jackson,I. (2002) Rules and guidelines for mouse gene, allele and mutation nomenclature: a condensed version. Genomics, 79, 471–474. [DOI] [PubMed] [Google Scholar]
- 14.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bard J.L., Kaufman,M.H., Dubreuil,C., Brune,R.M., Burger,A., Baldock,R.A. and Davidson,D.R. (1998) An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature. Mech. Dev., 74, 111–120. [DOI] [PubMed] [Google Scholar]