The full-length RNA-chaperone Hfq from Escherichia coli (102 amino acids) has been crystallized. The crystal belongs to the P1 space group. The structure was solved to 2.85 Å resolution, revealing that the disordered C-terminal resides on the proximal face of the hexamer.
Keywords: RNA chaperones, Hfq, riboregulation, crystal packing
Abstract
The structure of full-length host factor Qβ (Hfq) from Escherichia coli obtained from a crystal belonging to space group P1, with unit-cell parameters a = 61.91, b = 62.15, c = 81.26 Å, α = 78.6, β = 86.2, γ = 59.9°, was solved by molecular replacement to a resolution of 2.85 Å and refined to R work and R free values of 20.7% and 25.0%, respectively. Hfq from E. coli has previously been crystallized and the structure has been solved for the N-terminal 72 amino acids, which cover ∼65% of the full-length sequence. Here, the purification, crystallization and structural data of the full 102-amino-acid protein are presented. These data revealed that the presence of the C-terminus changes the crystal packing of E. coli Hfq. The crystal structure is discussed in the context of the recently published solution structure of Hfq from E. coli.
1. Introduction
Host factor Qβ (Hfq) is a hexameric phylogenetically conserved small bacterial RNA-binding protein that belongs to the Sm-protein superfamily (Møller et al., 2002 ▶). Hfq was first discovered in Escherichia coli as a prerequisite for replication of the RNA phage Qβ (Franze de Fernandez et al., 1972 ▶). More recently, Hfq proteins from different bacterial species have been characterized (Chao & Vogel, 2010 ▶). Hfq has been shown to be important for virulence in several pathogens, including Pseudomonas aeruginosa (Sonnleitner et al., 2003 ▶) and Vibrio cholerae (Ding et al., 2004 ▶). It is generally believed that Hfq is involved in post-transcriptional regulation. For instance, deep-sequencing analysis revealed that Hfq affects the expression of 20% of the genes of Salmonella typhimurium (Sittka et al., 2008 ▶). Based on its capacity to alter the RNA secondary structure and to facilitate the annealing of two RNA ligands (Lease & Belfort, 2000 ▶; Sledjeski et al., 2001 ▶; Zhang et al., 2002 ▶; Lease & Woodson, 2004 ▶), Hfq has been assigned to a class of proteins termed RNA chaperones (Moll et al., 2003 ▶; Rolle et al., 2006 ▶; Arluison et al., 2007 ▶). The pleiotropic effects observed in bacterial hfq deletion strains are therefore most likely to be rooted in the involvement of the protein in riboregulation of mRNAs by small regulatory RNAs.
In contrast to those from Gram-positive bacteria, Hfqs from Gram-negative bacteria belonging to the β- and γ-proteobacteria have an extended C-terminal domain. In E. coli the C-terminal extension is dispensable for binding of small RNAs, but has been implicated in the binding of longer mRNAs (Vecerek et al., 2008 ▶). Bioinformatic and biophysical studies have suggested that the C-terminus is intrinsically disordered, which may facilitate the interaction with different RNA substrates (Beich-Frandsen et al., 2011 ▶).
The three-dimensional structure of Hfq was first solved for the Staphylococcus aureus Hfq protein, establishing its close resemblance to eukaryotic Sm proteins (Schumacher et al., 2002 ▶). Since then, 13 unique structures of Hfq proteins from seven different organisms have been deposited in the Protein Data Bank: S. aureus (HfqSa; PDB entries 1kq1 and 1kq2; Schumacher et al., 2002 ▶), E. coli [HfqEc; PDB entries 1hk9 (Sauter et al., 2003 ▶) and 3gib (Link et al., 2009 ▶)], P. aeruginosa [HfqPae; PDB entries 1u1t, 1u1s (Nikulin et al., 2005 ▶), 3m4g and 3inz (Moskaleva et al., 2010 ▶)], Methanococcus jannaschii (HfqMj; PDB entry 2qtx; Nielsen et al., 2007 ▶), Synechocystis sp. (HfqCSyn; PDB entry 3hfo; Bøggild et al., 2009 ▶), Anabaena sp. (HfqCAna; PDB entry 3hfn; Bøggild et al., 2009 ▶) and Bacillus subtilis [HfqBs; PDB entries 3hsb (Baba et al., 2010 ▶) and 3hsa (Joint Center for Structural Genomics, unpublished work)]. Common to all crystal structures of Hfq proteins is the absence of structural information on the C- and N-termini of the six monomers of the functional hexamer, indicating the inherent flexibility in these regions.
Here, we present crystallographic data for full-length E. coli Hfq; the unstructured C-terminal extension is present on the proximal face of the hexameric core, but is mostly disordered in the crystal.
2. Materials and methods
2.1. Cloning, expression and purification of Hfq
The plasmid pUH5 carrying the hfq gene of E. coli has been described previously (Vecerek et al., 2003 ▶). For production of full-length HfqEc protein, the plasmid was transformed in E. coli BL21 (DE3) (Novagen) and the cells were grown in Luria–Bertani medium supplemented with 100 µg ml−1 ampicillin to an OD of ∼0.6–0.8. Expression of the hfq gene was induced by the addition of 0.5 mM IPTG. After 4 h, the cells were harvested by centrifugation and used for protein purification. Hfq was purified as recently described by Beich-Frandsen et al. (2011 ▶). The Hfq protein is heat-stable and the purification involved an initial fractionation by heating and subsequent processing by FPLC. The purification scheme comprised four steps: (i) an initial washing step by Ni2+-affinity chromatography, (ii) filtration over an anion-exchange column to remove nucleic acids, (iii) a concentration step by Ni2+-affinity chromatography and (iv) size-exclusion chromatography. The Ni2+-affinity purification was performed with wild-type protein, which contains 2 × 2 histidine residues in the C-terminal tail, amounting to 24 histidines in the hexamer. Fractions collected from size-exclusion column were concentrated with stirring to ∼20 mg ml−1 in the same buffer using a 10 ml Amicon nitrogen pressure cell equipped with a Millipore Ultrafiltration membrane (10 kDa cutoff).
2.2. Crystallization
Initial crystallization conditions were identified by robot-assisted vapour-diffusion experiments in commercial sparse-matrix screens. Experiments were set up at 285 K and then stored at 277 K. Of 1000 conditions, only condition No. 54 of Hampton Research Index screen produced crystals at 277 K. This condition was subsequently scaled up and optimized at 277 K in 24-well Linbro plates (Walter et al., 2005 ▶). This produced larger crystals; diffraction-quality crystals were grown from 0.05 M calcium chloride dihydrate (Fluka), 0.1 M Bis-Tris pH 6.5 (Fluka), 30%(v/v) polyethylene glycol monomethyl ether 550 (Sigma–Aldrich).
Diffraction data were collected from a plate-shaped crystal with approximate dimensions of 250 × 150 × 5 µm on the ESRF microfocus beamline ID23.2 using a 10 µm beam with a wavelength of 0.873 Å. Owing to rather anisotropic diffraction, multiple data sets covering 200° were successively collected along the rotation axis, with intermittent translations of 20 µm. The data were processed using the XDS program package (Kabsch, 2010 ▶) and the five data sets with the best statistics were scaled together with XSCALE to a highest resolution of 2.85 Å.
2.3. Structure solution and refinement
The phase problem was solved by molecular replacement using the structure of hexameric E. coli Hfq (amino acids 7–65; PDB entry 1hk9; Sauter et al., 2003 ▶) as a search model employing MOLREP in the CCP4 suite (Winn et al., 2011 ▶). The initial model, which was composed of two hexamers in the unit cell, was refined in REFMAC (Murshudov et al., 2011 ▶), initially with noncrystallographic symmetry (NCS) restraints for the regions consisting of amino acids 7–45 and 53–66. On visual inspection in Coot (Emsley & Cowtan, 2004 ▶) it became evident that implying NCS affected the electron density in the terminal regions and the NCS restraints were therefore removed. Several rounds of further refinement were performed with the PHENIX package (Adams et al., 2010 ▶) with H atoms in riding positions and were alternated with iterative manual building and corrections in the program Coot. A final polishing refinement step was performed in REFMAC to adjust the weighting between X-ray and stereochemical terms in order to minimize the refinement residual, keeping the optimal stereochemistry of the model. Data-collection and refinement statistics are reported in Table 1 ▶. Stereochemistry and structure quality were checked using the program MolProbity (Chen et al., 2010 ▶). Superpositions and calculation of the r.m.s. deviations of structures and structural figures were generated using MacPyMOL (DeLano, 2007 ▶).
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Beamline | ID23-2, ESRF |
| Wavelength () | 0.873 |
| Resolution () | 80.02.85 (2.922.85) |
| Space group | P1 |
| Unit-cell parameters (, ) | a = 61.91, b = 62.15, c = 81.26, = 78.6, = 86.2, = 59.9 |
| Molecules per asymmetric unit | 12 |
| Unique reflections | 156571 (9573) |
| Completeness (%) | 92.9 (83.7) |
| R meas † | 0.138 (0.351) |
| R merge ‡ | 0.128 (0.323) |
| Multiplicity | 7.03 (6.43) |
| I/(I) | 14.22 (5.60) |
| R work §/R free ¶ | 0.207/0.250 |
| R.m.s.d. bonds () | 0.010 |
| R.m.s.d. angles () | 1.055 |
| Atoms (non-H) | 6563 |
| Solvent molecules | 71 |
| Ramachandran plot (from MolProbity) | |
| Outliers (%) | 0 |
| Favoured (%) | 92.9 |
| Rotamer outliers (%) | 0.4 |
| PDB code | 3qhs |
R
meas = 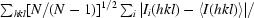
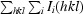 .
.
R
merge = 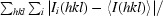
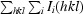 .
.
R
cryst = 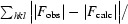
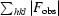 .
.
R free is the cross-validation R factor computed for a test set of reflections (5%) which were omitted in the refinement process.
Given the hexagonal packing of hexamers in layers (see below), we investigated the presence of possible higher symmetry in the crystal lattice using the program POINTLESS from the CCP4 package (Evans, 2006 ▶) and by data processing with XDS. Careful analysis did not show the presence of any higher symmetry or lattice centring.
Twinning analysis of the diffraction data with phenix.xtriage from the PHENIX package (Zwart et al., 2005 ▶) and with SFCHECK from the CCP4 package (Vaguine et al., 1999 ▶) did not indicate the presence of twinning. Visual inspection of the plate-shaped crystals did not reveal any macroscopic imperfections on the surface or zigzagged edges.
2.4. Western-blot analysis
A single crystal was harvested, washed three times in reservoir solution and subsequently dissolved in 10 µl water. Samples were retrieved from the drop and reservoir and run on 16% SDS–PAGE (1 h/250 V). The gel was blotted onto a nitrocellulose membrane [Schleicher & Schuell; 1 h/100 V in 40 mM glycine, 50 mM Tris–HCl, 0.04%(w/v) sodium dodecyl sulfate, 20%(v/v) methanol], which was blocked with 2%(w/v) bovine serum albumin (BSA) in 50 mM Tris–HCl, 150 mM NaCl, 50 mM KCl, 0.1%(v/v) Tween-20 overnight. Immunodetection was performed with rabbit primary antibody against Hfq (Vecerek et al., 2008 ▶) and a secondary antibody (goat–antirabbit) conjugated to alkaline phosphatase (Abcam). The blot was developed in 100 mM Tris–HCl pH ∼9.5, 0.5 mM MgCl2 using 0.015%(v/v) 5-bromo-4-chloro-3-indolyl-phosphate (BCIP; Sigma–Aldrich) and 0.03% nitro blue tetrazolium (NBT; Sigma–Aldrich).
3. Results and discussion
3.1. Crystallization and solution of the phase problem
Previous crystallization studies on Hfq from E. coli were hampered by protein degradation. However, optimization of the purification protocol (Beich-Frandsen et al., 2011 ▶) led to successful crystallization of the full-length Hfq protein and subsequent data collection to a resolution of 2.85 Å. The structure was solved by molecular replacement using the known hexameric structure of HfqEc as a search model and refined to R work and R free values of 20.7% and 25.0%, respectively, for diffraction data between 80.0 and 2.85 Å resolution. Data-collection, processing and refinement statistics can be found in Table 1 ▶.
3.2. Crystal structure of full-length E. coli Hfq
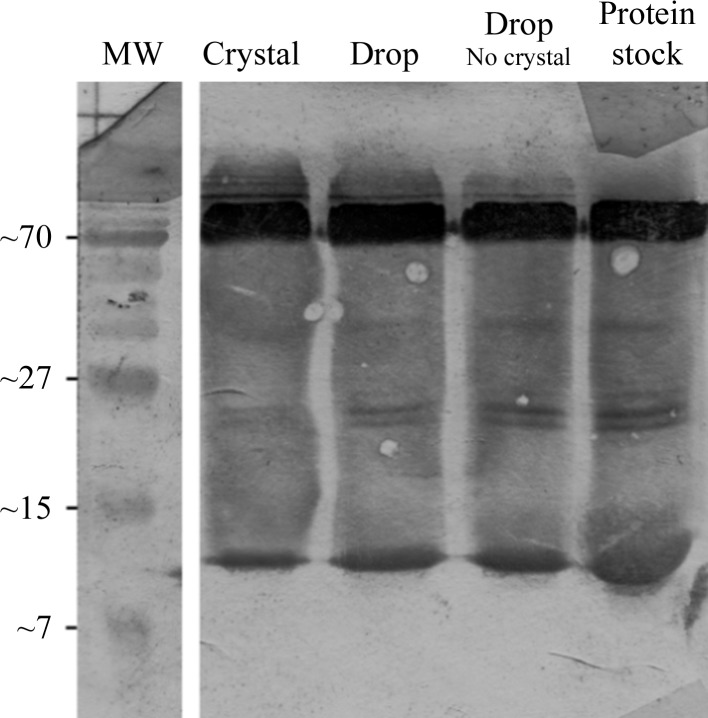
In the original study by Sauter et al. (2003 ▶), full-length E. coli Hfq was reported to crystallize in a tetragonal crystal form with a very low solvent content (18%) as a result of protein degradation. Here, we report the structure of full-length E. coli Hfq in a triclinic crystal form with two hexamers in the unit cell and a calculated solvent content of ∼40% based on the full-length HfqEc sequence. Fig. 1 ▶ displays a Western blot of the HfqEc crystal, the corresponding crystallization drop, a drop in which no crystallization occurred and the protein stock. The protein migrates predominantly in a hexameric form (∼67.2 kDa) and in a monomeric form (∼11.2 kDa). This analysis revealed no proteolytic fragmentation of the protein either in the crystallized HfqEc or in the sample remaining in the crystallization drop.
Figure 1.
Western blot of Hfq crystal, growth conditions and protein stock. On a 16% SDS–PAGE gel (60 min, 200 mV) the protein migrates predominantly as a hexamer (upper band; 67.2 kDa), with the monomer band visible at the bottom of each lane (11.2 kDa). No significant protein degradation was observed.
Clear electron density was observed in all protein chains for residues Glu6–Asn71 and in isolated cases for residues Gly4–Asn74. Presumably owing to high intrinsic disorder in the N- and C-terminal regions as indicated by bioinformatics analysis and by experimental studies (Vecerek et al., 2008 ▶; Beich-Frandsen et al., 2011 ▶), amino-acid residues 75–102 are not visible in the electron-density maps.
3.3. Conservation of the Sm core and structural comparisons with other Hfq proteins
In a previous study (Arluison et al., 2004 ▶), full-length and C-terminally truncated HfqEc (lacking the last 19 amino acids) were compared using equilibrium unfolding, electron-microscopy (EM) analysis and attenuated total reflectance Fourier transform infrared spectroscopy. Structural rearrangements between the subunits were observed upon truncation, coupled with a reduction in β-strand content, as well as lower thermal stability of truncated constructs. Furthermore, EM analysis showed the C-terminal segment to reside on the proximal face on top of the interface between monomers.
The comparison of our crystal structure with other crystal structures of HfqEc encompassing amino-acid residues 1–72 (PDB entry 1hk9) and 2–69 (PDB entry 3gib) and with corresponding subunits of other known Hfq protein structures (HfqSa, PDB entries 1kq1 and 1kq2; HfqPs, PDB entries 1u1t and 1u1s; HfqMj, PDB entry 2qtx; HfqCSyn, PDB entry 3hfo; HfqCana, PDN entry 3hfn) gave an overall r.m.s.d. of 0.85 Å for 354 pairs of structurally equivalent Cα atoms. The low r.m.s.d. values suggest that there are no distinct structural rearrangements in the Sm core in the presence of the C-terminus or of the hexameric architecture and indicate high structural conservation throughout distantly related phyla, therefore contrasting with the previously mentioned study.
3.4. Crystal packing
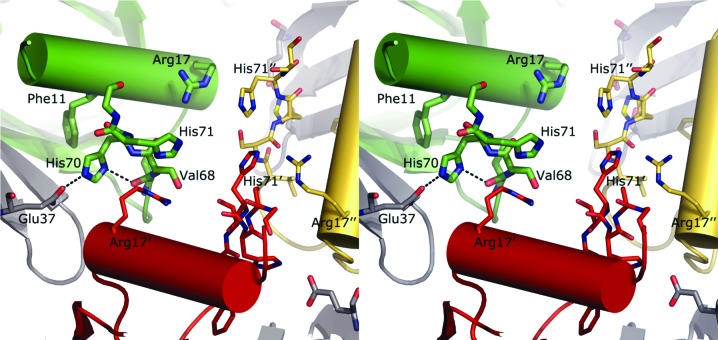
In the original crystal structure of E. coli Hfq (amino acids 1–72; PDB entry 1hk9; Sauter et al., 2003 ▶), the C-termini, which are visible in electron density up to amino-acid residue 69 or 70, are tethered in an extended conformation against the core of the HfqEc hexamer. The C-termini reside in a groove on the proximal side of the Sm core formed by the N-terminal α-helix and β-sheets of the adjacent subunit (amino acids 34–55). In the crystal structure reported here, we observe the C-terminal amino-acid residues up to position 74 in an extended conformation in which His70 and His71 act as mediators of inter-protomer crystal contacts between symmetry-related hexamers and intra-protomer contacts within the hexamer. The His70 side chain is stabilized in its orientation by hydrogen-bonding interactions with the main-chain carbonyl O atom of Val68 of the same subunit and with the side chain of Glu37 of the neighbouring subunit of the same hexamer. His70 is furthermore involved in aromatic T-stacking (Tewari & Dubey, 2008 ▶) with Phe11 of the same protomer. His71, which adopts more diverse conformations in individual protomers, is found to make interactions with the side chains of the symmetry-related residues Arg17 and His71. Fig. 2 ▶ displays these interactions.
Figure 2.
Intra-protomer and inter-protomer interactions. Each C-terminus packs in a distinct conformation. However, the general trend is a horizontal/lateral placement of His70 and His71 with respect to each other, thereby mediating between three C-termini standing against each other (inter-protomer interaction) and the C-terminus packing against the Sm core (intra-protomer interaction). The initial part of the C-terminus interacts with the same region from an adjacent hexamer. As can be seen, the histidine pair (His70 and His71) mediates between the inter-protomer interaction of adjacent C-termini and the intra-protomer crystal contact that packs the C-terminus into the groove of the Sm core.
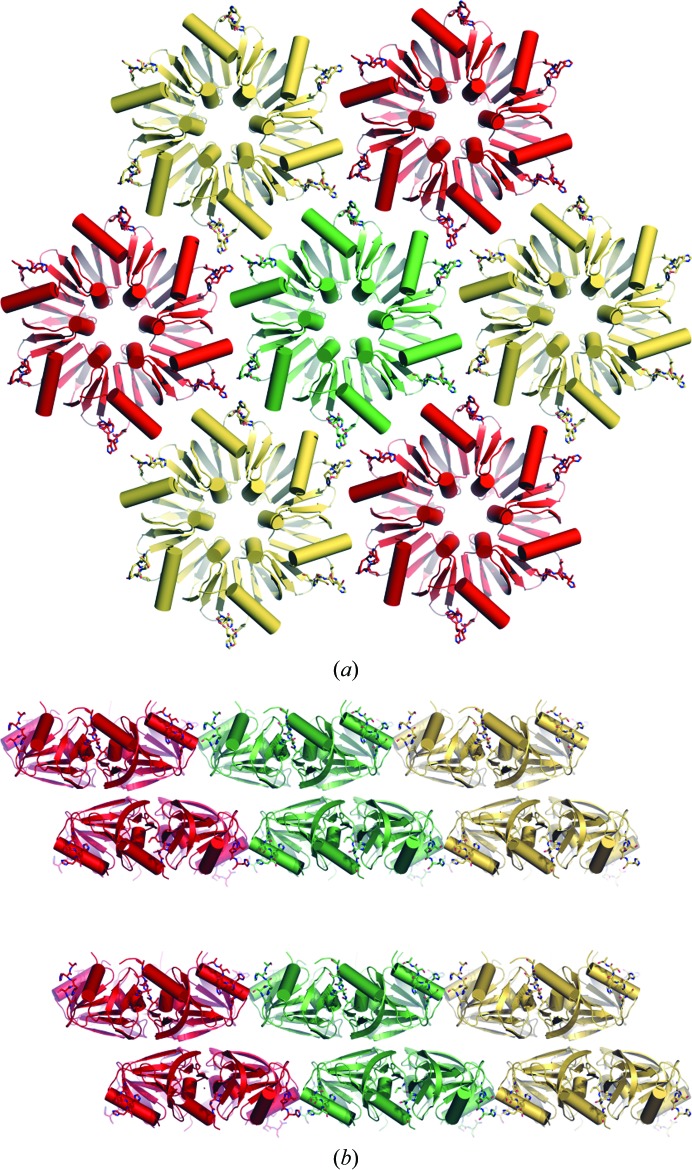
Similar to the structure of truncated Hfq (amino acids 1–72; PDB entry 1hk9; Sauter et al., 2003 ▶), HfqEc packs in layers with a ‘honeycomb’ pattern, with three C-termini standing against each other (Fig. 2 ▶) and interacting through favourable polar and hydrophobic interactions, stabilizing the HfqEc hexamers in hexagonal layers (Fig. 3 ▶). Each of the two HfqEc hexamers of the unit cell gives rise to a distinct ‘honeycomb’ layer (Fig. 3 ▶ a) and they pack against each other in a staggered manner with their distal sides facing to form a double layer (Fig. 3 ▶ b). In contrast to the structure of truncated E. coli Hfq (amino acids 1–72; PDB entry 1hk9; Sauter et al., 2003 ▶), in which the Hfq layers pack against each other in a ‘proximal-to-distal’ arrangement, the presence of the C-termini evidently causes the hexamers to favour packing via the more apolar distal side, with the C-termini of each double layer pointing towards the next double layer (Fig. 3 ▶ b). The double layers stack on top of each other and are separated by a distance of ∼30 Å along the c axis, therefore impairing any specific contacts between the conserved (L)Sm core of HfqEc subunits across adjacent double layers. On the other hand, this distance allows the accommodation of the unstructured C-terminal regions, which amount to about 30% of the mass of E. coli HfqEc.
Figure 3.
Crystal structure. (a) Similar to the crystal structure of truncated HfqEc (amino acids 1–72; PDB entry 1hk9; Sauter et al., 2003 ▶), the hexameric molecules pack in a hexagonal ‘honeycomb’ pattern inside the crystal. (b) In the crystal of full-length HfqEc the hexamers pack in staggered double layers interspaced by an ∼30 Å void, which leaves room for the unstructured termini. Interestingly, the coordination between the double layers is unstaggered. Hexamers from each unit cell are coloured red, gold and green.
Inspection of the diffraction images showed streaked and diffuse diffraction spots in some crystal orientations. Furthermore, the native Patterson map displayed several rather strong maxima (6–8% of the origin peak; 5.5–8.8σ) close to the origin as well as peaks related to each other by sixfold rotational symmetry at w = 0. These observations may be indicative of lattice-translocation disorder (Howells & Perutz, 1954 ▶; Bragg & Howells, 1954 ▶; Trame & McKay, 2001 ▶; Wang, Kamtekar et al., 2005 ▶; Wang, Rho et al., 2005 ▶; Hwang et al., 2006 ▶; Tanaka et al., 2008 ▶; Zhu et al., 2008 ▶). Lattice translocation refers to situations in which layers or groups of molecules in a crystal are displaced relative to each other in a stochastic manner by a discrete set of translation vectors and can occur in crystals of homo-multimeric as well as monomeric macromolecules. A textbook example of lattice translocation, manifesting itself as thick layers of space between layers of molecules, is the crystal structure of the carboxysome shell protein CsoS1C (Tsai et al., 2009 ▶), in which the interlayer space accommodates another layer of protein molecules for which the electron density is uninterpretable. In our case, the interlayer space hosts the intrinsically unstructured C-termini of HfqEc.
Crystal packing that lacks lattice-stabilizing contacts in one direction in combination with lattice-translocation defects could be the basis of the anisotropic diffraction that hampers high-resolution data collection from this crystal form.
4. Conclusions
In the crystal structure of HfqEc reported here we observe the initial segment of the C-terminal region to be bound to the body of the hexamer; at the same time, its involvement in intermolecular contacts constrains the remaining C-terminal portion, which is not involved in contacts, to project towards the solvent on the proximal side of the hexamer (Fig. 3 ▶ b).
A certain variation in the conformations of the Hfq C-termini, with the best fitting conformations extending laterally away from the hexameric core, was observed by small-angle X-ray scattering (Beich-Frandsen et al., 2011 ▶). At variance with this, the crystallization process apparently selects conformers with the C-termini on the proximal face, indicating this conformation to be one of the low-energy states of the molecule, with no significant structural differences at the subunit and quarternary structure level compared with truncated HfqEc or other Hfq structures.
Note added in proof: Since the acceptance of this article the following new Hfq structures have been deposited: S. aureus (PDB entry 3qsu), E. coli (2y90, 3qo3) and P. aeruginosa (3qui).
Supplementary Material
PDB reference: Hfq, 3qhs
Acknowledgments
The work in the KD-C and UB laboratories was supported by grants F1722 (KD-C) and F1720 (UB) in the framework of the Special Research Program (SFB17) on ‘Modulators of RNA fate and function’ from the Austrian Science Fund. We thank the beamline scientists of the ID23-2 microfocus beamline of the European Synchrotron Radiation Facility for their assistance during data collection and Irina Grishkovskaya (Department of Structural and Computational Biology, University of Vienna) for fruitful discussions and support.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Arluison, V., Folichon, M., Marco, S., Derreumaux, P., Pellegrini, O., Seguin, J., Hajnsdorf, E. & Regnier, P. (2004). Eur. J. Biochem. 271, 1258–1265. [DOI] [PubMed]
- Arluison, V., Hohng, S., Roy, R., Pellegrini, O., Régnier, P. & Ha, T. (2007). Nucleic Acids Res. 35, 999–1006. [DOI] [PMC free article] [PubMed]
- Baba, S., Someya, T., Kawai, G., Nakamura, K. & Kumasaka, T. (2010). Acta Cryst. F66, 563–566. [DOI] [PMC free article] [PubMed]
- Beich-Frandsen, M., Vecerek, B., Konarev, P. V., Sjöblom, B., Kloiber, K., Hämmerle, H., Rajkowitsch, L., Miles, A. J., Kontaxis, G., Wallace, B. A., Svergun, D. I., Konrat, R., Bläsi, U. & Djinović-Carugo, K. (2011). Nucleic Acids Res. 10.1093/nar/gkq1346. [DOI] [PMC free article] [PubMed]
- Bøggild, A., Overgaard, M., Valentin-Hansen, P. & Brodersen, D. E. (2009). FEBS J. 276, 3904–3915. [DOI] [PubMed]
- Bragg, W. L. & Howells, E. R. (1954). Acta Cryst. 7, 409–411.
- Chao, Y. & Vogel, J. (2010). Curr. Opin. Microbiol. 13, 24–33. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2007). PyMOL. http://www.pymol.org.
- Ding, Y., Davis, B. M. & Waldor, M. K. (2004). Mol. Microbiol. 53, 345–354. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Franze de Fernandez, M. T., Hayward, W. S. & August, J. T. (1972). J. Biol. Chem. 247, 824–831. [PubMed]
- Howells, E. R. & Perutz, M. F. (1954). Proc. R. Soc. London Ser. A, 225, 315–329.
- Hwang, W. C., Lin, Y., Santelli, E., Sui, J., Jaroszewski, L., Stec, B., Farzan, M., Marasco, W. A. & Liddington, R. C. (2006). J. Biol. Chem. 281, 34610–34616. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Lease, R. A. & Belfort, M. (2000). Mol. Microbiol. 38, 667–672. [DOI] [PubMed]
- Lease, R. A. & Woodson, S. A. (2004). J. Mol. Biol. 344, 1211–1223. [DOI] [PubMed]
- Link, T. M., Valentin-Hansen, P. & Brennan, R. G. (2009). Proc. Natl Acad. Sci. USA, 106, 19292–19297. [DOI] [PMC free article] [PubMed]
- Moll, I., Leitsch, D., Steinhauser, T. & Bläsi, U. (2003). EMBO Rep. 4, 284–289. [DOI] [PMC free article] [PubMed]
- Møller, T., Franch, T., Højrup, P., Keene, D. R., Bächinger, H. P., Brennan, R. G. & Valentin-Hansen, P. (2002). Mol. Cell, 9, 23–30. [DOI] [PubMed]
- Moskaleva, O., Melnik, B., Gabdulkhakov, A., Garber, M., Nikonov, S., Stolboushkina, E. & Nikulin, A. (2010). Acta Cryst. F66, 760–764. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Nielsen, J. S., Bøggild, A., Andersen, C. B., Nielsen, G., Boysen, A., Brodersen, D. E. & Valentin-Hansen, P. (2007). RNA, 13, 2213–2223. [DOI] [PMC free article] [PubMed]
- Nikulin, A., Stolboushkina, E., Perederina, A., Vassilieva, I., Blaesi, U., Moll, I., Kachalova, G., Yokoyama, S., Vassylyev, D., Garber, M. & Nikonov, S. (2005). Acta Cryst. D61, 141–146. [DOI] [PubMed]
- Rolle, K., Zywicki, M., Wyszko, E., Barciszewska, M. Z. & Barciszewski, J. (2006). J. Biochem. 139, 431–438. [DOI] [PubMed]
- Sauter, C., Basquin, J. & Suck, D. (2003). Nucleic Acids Res. 31, 4091–4098. [DOI] [PMC free article] [PubMed]
- Schumacher, M. A., Pearson, R. F., Møller, T., Valentin-Hansen, P. & Brennan, R. G. (2002). EMBO J. 21, 3546–3556. [DOI] [PMC free article] [PubMed]
- Sittka, A., Lucchini, S., Papenfort, K., Sharma, C. M., Rolle, K., Binnewies, T. T., Hinton, J. C. & Vogel, J. (2008). PLoS Genet. 4, e1000163. [DOI] [PMC free article] [PubMed]
- Sledjeski, D. D., Whitman, C. & Zhang, A. (2001). J. Bacteriol. 183, 1997–2005. [DOI] [PMC free article] [PubMed]
- Sonnleitner, E., Hagens, S., Rosenau, F., Wilhelm, S., Habel, A., Jäger, K. E. & Bläsi, U. (2003). Microb. Pathog. 35, 217–228. [DOI] [PubMed]
- Tanaka, S., Kerfeld, C. A., Sawaya, M. R., Cai, F., Heinhorst, S., Cannon, G. C. & Yeates, T. O. (2008). Science, 319, 1083–1086. [DOI] [PubMed]
- Tewari, A. K. & Dubey, R. (2008). Bioorg. Med. Chem. 16, 126–143. [DOI] [PubMed]
- Trame, C. B. & McKay, D. B. (2001). Acta Cryst. D57, 1079–1090. [DOI] [PubMed]
- Tsai, Y., Sawaya, M. R. & Yeates, T. O. (2009). Acta Cryst. D65, 980–988. [DOI] [PubMed]
- Vaguine, A. A., Richelle, J. & Wodak, S. J. (1999). Acta Cryst. D55, 191–205. [DOI] [PubMed]
- Vecerek, B., Moll, I., Afonyushkin, T., Kaberdin, V. & Bläsi, U. (2003). Mol. Microbiol. 50, 897–909. [DOI] [PubMed]
- Vecerek, B., Rajkowitsch, L., Sonnleitner, E., Schroeder, R. & Bläsi, U. (2008). Nucleic Acids Res. 36, 133–143. [DOI] [PMC free article] [PubMed]
- Walter, T. S. et al. (2005). Acta Cryst. D61, 651–657. [DOI] [PMC free article] [PubMed]
- Wang, J., Kamtekar, S., Berman, A. J. & Steitz, T. A. (2005). Acta Cryst. D61, 67–74. [DOI] [PubMed]
- Wang, J., Rho, S.-H., Park, H. H. & Eom, S. H. (2005). Acta Cryst. D61, 932–941. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Zhang, A., Wassarman, K. M., Ortega, J., Steven, A. C. & Storz, G. (2002). Mol. Cell, 9, 11–22. [DOI] [PubMed]
- Zhu, X., Xu, X. & Wilson, I. A. (2008). Acta Cryst. D64, 843–850. [DOI] [PMC free article] [PubMed]
- Zwart, P. H., Grosse-Kunstleve, R. W. & Adams, P. D. (2005). CCP4 Newsl. 43, contribution 7.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Hfq, 3qhs