Abstract
Alternative splicing is widespread in mammalian gene expression, and variant splice patterns are often specific to different stages of development, particular tissues or a disease state. There is a need to systematically collect data on alternatively spliced exons, introns and splice isoforms, and to annotate this data. The Alternative Splicing Database consortium has been addressing this need, and is committed to maintaining and developing a value-added database of alternative splice events, and of experimentally verified regulatory mechanisms that mediate splice variants. In this paper we present two of the products from this project: namely, a database of computationally delineated alternative splice events as seen in alignments of EST/cDNA sequences with genome sequences, and a database of alternatively spliced exons collected from literature. The reported splice events are from nine different organisms and are annotated for various biological features including expression states and cross-species conservation. The data are presented on our ASD web pages (http://www.ebi.ac.uk/asd).
INTRODUCTION
Genome-wide analyses of alternative splicing have indicated, for human, that 40–60% of genes are alternatively spliced (1–6), suggesting that alternative splicing is a major contributor to the functional complexity of mammalian, and perhaps other, genomes. While most genes produce a modest number of transcript isoforms, some have banks of alternative exons that are combined to generate a large number of isoforms (7). Alternative splicing is regulated by a large assortment of splicing-associated proteins and factors (8–10). Regulation of alternative splicing may involve changes in the ratios of isoforms brought about by changes in the concentrations of general splicing factors (11,12). Defects in (alternative) splicing are increasingly recognized as causes of disease (13,14), and such disease states can have complicated inheritance patterns. Elucidation of the genetic mechanisms regulating alternative splicing is important for the development of novel diagnostic and therapeutic tools.
There is an increasing need to create value-added data bases on alternative splice events (reviewed in 15). To date, data on alternative splicing fall into two categories. (i) experimentally determined and characterized splice events from specific genes, as reported in bibliography databases such as MEDLINE, or in curated nucleotide and protein sequence databases such as EMBL (16) and SWISS-PROT (17). Efforts to create data sets based on these data include Alternative Exon Database (18), ASDB (19) and AsMamDB (20). (ii) computationally determined splice events observed through examination of alignments of EST/cDNA sequences with one another or with genomic DNA sequences—these include AltExtron (2), Asforms (21) and ASAP (22). These two approaches differ in the size, presentation and analysis of data and also in the extent, and accuracy, of annotation.
Computational approaches that use EST resources can be error prone, suffer from the limited gene coverage of available transcript resources and often do not have confidence attached to the predicted events. Further, while experimentally determined events often have annotation of biological properties (including regulatory mechanisms, expression states and functional changes in the encoded proteins), methods that can computationally generate such annotations are not readily available. Thus computational pipelines need to develop and incorporate methods that both associate confidence values with predicted events and also provide useful annotations that are indicative of biological properties.
The Alternative Splicing Database (ASD) consortium (see http://www.ebi.ac.uk/asd/asd-ec/index.html for details) is working to meet these challenges by combining and extending manually curated alternative exon databases with data generated through a computational pipeline. The ultimate product of the consortium is a unified ASD database that also implements RNA analysis tools, a literature agent and cross-species comparisons related to alternative splicing, as well as links to alternative exon-specific oligonucleotide arrays. We present here two products from the ASD consortium: (i) a large collection of computationally predicted data, and (ii) experimentally determined data collected from the full-text articles in peer-reviewed journals. These data are available from http://www.ebi.ac.uk/asd/.
DATABASES FROM THE ASD PROJECT
Manually curated database: AEDB (alternative exon database)
This database is an ongoing collection of experimental data relating to alternatively spliced exons as published in peer-reviewed journals (18). Collection of data is carried out using a defined assessment form. The collected data include not only the nucleotide sequences of alternatively spliced exons but also the reported biological properties, including tissue specificity, developmental regulation, alternative exon function and association with disease. An online submission system is in place for direct submission by the partners of the ASD consortium. The submission system will be developed further as a public repository of experimental data on alternative splicing. A web interface to the data is in place and allows querying through biological features, and provides hyperlinks to relevant MEDLINE abstracts and EMBL gene entries. The web server for AEDB was formally launched in December 2002 and currently contains around 1100 entries collected from nearly as many journal reports. Splice regulatory sequences reported in the literature are also collected and integrated into the database.
Computer-generated databases: AltExtron and AltSplice
Methodology and data statistics. We first generate a high-quality data set of gene–transcript alignments that show more than one high scoring match between gene and transcript (EST/mRNA) sequences. Any transcript sequence that aligns with more than one gene or ambiguously with a single gene is discarded, and any gaps in the transcript sequence between matches are further examined (‘patched’) in an attempt to incorporate them into the scaffold of alignment data. Alternative splice events are then delineated through careful analysis of these alignments (details on methods is presented in 2). Alignment gaps on gene sequences are potential introns, with validation of these as transcript-confirmed introns being the crucial step in our work. Validation tools for this purpose are continually expanded with research activities on unusual or rare introns. A match in the alignment is taken as a confirmed exon if it is flanked on either side by a confirmed intron.
A comment on terminology that refers to constitutive splicing is in order here. Sometimes the word ‘constitutive’ is used to refer to an exon (or splice site or pattern) that is always used. This strict usage is problematic, as it is hard to know whether some splice pattern always occurs, or just does so for examined data. Thus we avoid the use of the term ‘constitutive form’, and instead use ‘normal form’ to refer to an exon or splice pattern that is usually adopted. Further, when it is necessary to say what the normal form is, this is (pragmatically) taken to be the annotated form.
Confirmed introns (or exons) that overlap with one another indicate alternative splicing events. Observed alternative events are described as: (i) exon/intron isoforms, where use of alternative donor or acceptor splice sites leads to truncation/extension of exons/introns; (ii) intron retention, where an intronic region is not spliced out; (iii) cryptic (or skipped) exons, where an entire alternative (or normal) exon is seen in some transcripts but not in others (i.e. a cassette exon); and (iv) alternating exon events (where the splice isoforms contain one or the other of a cryptic and skipped exon pair). The latter three events are further characterized as ‘complex’ or ‘simple’ events depending on whether the event also extends/truncates the flanking exons or not. Splice pattern(s) leading to expression of each observed event are derived by choosing representative(s) from the set of transcript sequences that confirm the event. Effects of alternative events on the coding frame of transcripts are determined and preliminary translations of alternative transcripts are derived.
The nomenclature used in our computer-generated databases differs from that used in AEDB in the case of exon extension or truncation events and in further categorization of ‘cassette exons’ as ‘skipped’ and ‘cryptic exons’. While the exon extension or truncation events are conventionally called alternative 5′ (or 3′) splice site events, we classify these as intron or exon isoforms. This nomenclature arises naturally in the way these events are identified from the alignment data. Categorization of cassette exons onto ‘cryptic’ or ‘skipped’ rather than just as alternative (cassette) exons has led to the identification of meaningful differences in splice characteristics (2).
Genes and transcript data along with observed introns, exons, splice patterns, alternative splice events and preliminary translations form the core of the presented data. These data are examined further for generating annotations.
At present two related computational pipelines are implemented on different gene datasets: AltExtron is the research and development pipeline, and is based around gene entries in the EMBL/GenBank sequence databases (16,23); AltSplice is the production pipeline, is tuned for integration with the other products of the ASD consortium, and is based on genes from the Ensembl genome annotation project (24). Both pipelines produce high-quality data, and until such time as AltSplice becomes a superset of AltExtron, we will present the data from both. Statistics on the generated data are given in Tables 1 and 2.
Table 1. ASD data (July 2003) statistics: AltExtron implementation for multiple species.
| No. of confirmed | No. of genes with at least one confirmed | No. of observed events | ||||||
|---|---|---|---|---|---|---|---|---|
| Organism | Introns | Exons | Intron/exon | Alternative event | Intron retention | Exon isoforms | Intron isoforms | Cassette exons |
| Human | 42922 | 29853 | 5584 | 2581 | 383 | 2046 | 3132 | 3844 |
| Mouse | 11461 | 8288 | 1766 | 658 | 127 | 440 | 718 | 541 |
| Fruit fly | 33241 | 20595 | 7881 | 1302 | 111 | 494 | 1294 | 797 |
| Caenorhabditis elegans | 41214 | 23043 | 9552 | 598 | 50 | 229 | 424 | 267 |
| Arabidopsis thaliana | 67455 | 51733 | 13302 | 758 | 156 | 372 | 633 | 70 |
| Rat | 1263 | 846 | 339 | 34 | 1 | 15 | 26 | 22 |
| Chicken | 1115 | 811 | 228 | 42 | 4 | 26 | 37 | 27 |
| Cow | 594 | 367 | 177 | 20 | 0 | 12 | 20 | 25 |
| Zebra fish | 1227 | 829 | 257 | 21 | 0 | 11 | 16 | 9 |
Table 2. ASD data (July 2003) statistics: AltSplice implementation for human.
| Event | Number of occurrences |
|---|---|
| Intron isoforms | 9757 |
| Exon isoforms | 5214 |
| Cassette exons | 12470 |
| as simple event | 9042 |
| as complex event | 1086 |
| as simple and complex | 2342 |
| Intron retention | 3441 |
| as simple event | 2605 |
| as complex event | 332 |
| as simple and complex | 504 |
| Mutually exclusive exons | 1056 |
| as simple event | 854 |
| as complex event | 111 |
| as simple and complex | 91 |
| No. of genes showing at least one alternative splice event | 8314 |
| No. of genes showing at least one confirmed intron/exon | 15550 |
| No. of confirmed introns | 162328 |
| No. of confirmed exons | 122499 |
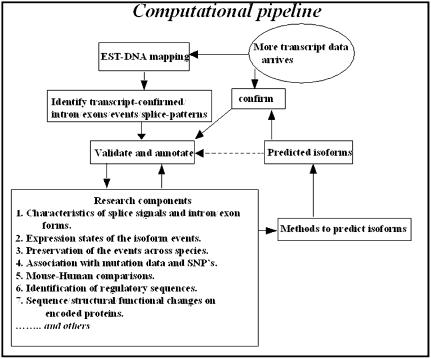
Derived annotations. Data generation and annotation activities that are planned for the computational pipeline are shown in Figure 1. The annotation activities that are already carried out are mentioned below.
Figure 1.
Schematic representation of the pipeline for computational generation of alternative splice data and annotation.
Characteristics of splice signals and intron/exon forms. Introns are grouped as strong or weak GT-AG introns (depending on the score for the donor site), U12 GT-AG introns, GC-AG introns and AG-dependent introns. The strong GT-AG introns are further categorized, depending on the observed nucleotide at position +3 of the donor splice site, as G3 introns, A3 introns and Y3 introns. This type of categorization revealed biologically important characteristics (2,25). Donor and acceptor sites are scored for their strength; putative polypyrimidine tracts and branch points are identified and scored. Weight matrices that characterize the different intron/exon types are presented.
Expression states of the isoform events. Expression state for intron/exon features and the isoform events are delineated by examining the annotations of EST libraries from which the confirming transcript sequences are derived. Controlled vocabularies are implemented by adopting the eVOC ontologies (26). We associate expression specificity to four major domains: namely, pathology, developmental stage, anatomical site and cell type. These annotations enable large-scale expression mining.
Preservation of the splice events across species. An important part of our pipeline is to generate evolutionary profiles of gene splicing patterns. Methods have been standardized to delineate conserved intron/exon features, alternative events and modulations (such as use of alternative frame of translation or alternative stop codon) between human and mouse (27).
Association with mutation data and SNPs. We have developed methods (as documented in our ASD web pages) to delineate allele specificity of observed alternative splice events and transcript isoforms, and data relating to allele specificity of transcript isoforms are included in AltSplice.
Data and access
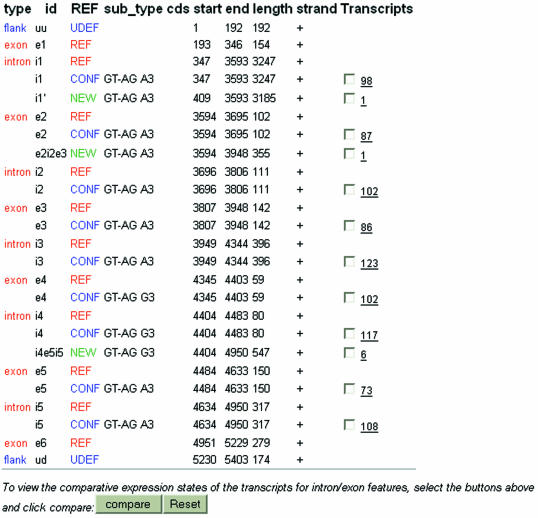
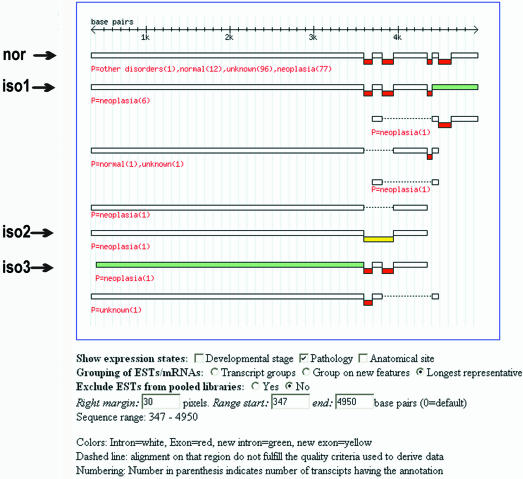
Data and documentation are available from the ASD web pages at http://www.ebi.ac.uk/asd/. Data can be downloaded as flat files or queried through web interfaces. Currently the interface is implemented for AltExtron data. Genes can be queried through types of splice events, protein keywords, and database cross-references [such as EMBL and SWISS-PROT accession numbers, HUGO gene symbols (28), Gene Ontology identifiers (29,30) and protein identifiers]. Browsers that enable one to select from eVOC standard vocabularies and GO classifiers facilitate querying through expression states and protein function/process/location. An output page lists all the available database cross-references; an important aspect being hyperlinks to homologous genes from other organisms (as extracted from AltExtron and Ensembl). Observed events are listed and are hyperlinked to a page that gives detailed intron–exon organization, and expression states for each of the isoforms. The output page further shows the splice structures (an example is presented in Fig. 2) where introns and exons are annotated for their relationship to a reference structure (as presented in the CDS feature of the EMBL entry). The query output page hyperlinks to a splice pattern viewer that gives a visual presentation of the observed isoform transcript structures and their expression states (Fig. 3).
Figure 2.
Example output of gene intron–exon structures. The human C2F gene is presented here. Introns and exons from the annotated splice form are labelled as ‘REF’. The transcript-confirmed introns and exons are labelled as either ‘CONF’ (if they conform to those in annotated splice form) or ‘NEW’ (if they are alternatives to those in annotated splice form). The new intron i1′ represents an intron isoform, the new exon e2i2e3 represents intron retention, and the new intron i4e5i5 represents a cassette exon event. The splice patterns are shown graphically in Figure 3.
Figure 3.
Visualization of splice isoforms for the human C2F gene. Transcript sequences are grouped onto splice patterns, which are then displayed (color coding distinguishes introns from exons, and normal from alternative forms). The first pattern corresponds to the normal form of splicing as reported in the EMBL Database; patterns that are labelled iso1–3 correspond to the observed alternative splice patterns (relating to the three alternative events seen in Fig. 2). The splice pattern labelled iso1 illustrates a skipped exon event, iso2 illustrates an intron retention event and iso3 illustrates an intron isoform event. Note that, for an exon to be shown in the view, both ends must have been confirmed. Transcript sequences can be grouped in three different ways. Each of the derived splice patterns is annotated for the expression state, with the annotation for a splice pattern obtained by consolidating those of its component multiple transcripts.
CONCLUSIONS
We present data on alternative splicing in human and other species; such data are derived through two means: (i) through computational delineation using available transcript sequences; and (ii) through collecting experimentally determined splice events as reported in peer-reviewed journals. The reported splice events and isoform transcripts are annotated for various biological features. Generation and characterization of data are carried out under the direction of the ASD consortium. Further updates will contain the results of microarray-based experimental validations and characterizations of alternative splicing in disease-related genes.
Acknowledgments
ACKNOWLEDGEMENTS
The work of Susan Lilley (UQ) in her project to rationalize the AltExtron pipeline is acknowledged. T.A.T. and S.S. acknowledge the European Commission for the grant (QLK3-CT-2002-02062) that supports this work.
REFERENCES
- 1.Brett D., Hanke,J., Lehmann,G., Haase,S., Delbruck,S., Krueger,S., Reich,J. and Bork,P. (2000) EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett., 474, 83–86. [DOI] [PubMed] [Google Scholar]
- 2.Clark F. and Thanaraj,T.A. (2002) Categorization and characterization of transcript-confirmed constitutively and alternatively spliced introns and exons from human. Hum. Mol. Genet., 11, 451–464. [DOI] [PubMed] [Google Scholar]
- 3.Kan Z., Rouchka,E.C., Gish,W.R. and States,D.J. (2001) Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res., 11, 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 5.Mironov A.A., Fickett,J.W. and Gelfand,M.S. (1999) Frequent alternative splicing of human genes. Genome Res., 9, 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Modrek B., Resch,A., Grasso,C. and Lee,C. (2001) Genome-wide analysis of alternative splicing using human expressed sequence data. Nucleic Acids Res., 29, 2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmucker D., Clemens,J.C., Shu,H., Worby,C.A., Xiao.J., Muda,M., Dixon,J.E. and Zipursky,S.L.L. (2000) Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell, 101, 671–684. [DOI] [PubMed] [Google Scholar]
- 8.Smith C.W.J. and Valcárcel,J. (2000) Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biol. Sci., 25, 381–388. [DOI] [PubMed] [Google Scholar]
- 9.Stamm S. (2002) Signals and their transduction pathways regulating alternative splicing: a new dimension of the human genome. Hum. Mol. Genet., 11, 2409–2416. [DOI] [PubMed] [Google Scholar]
- 10.Philips A.V. and Cooper, T.A. (2000) RNA processing and human disease. Cell. Mol. Life Sci., 57, 235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez A.J. (1998) Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet., 32, 279–305. [DOI] [PubMed] [Google Scholar]
- 12.Black D.L. (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu Rev. Biochem., 72, 291–336. [DOI] [PubMed] [Google Scholar]
- 13.Faustino N.A. and Cooper,T.A. (2003) Pre-mRNA splicing and human disease. Genes Dev., 17, 419–437. [DOI] [PubMed] [Google Scholar]
- 14.Stoilov P., Meshorer,E., Gencheva,M., Glick,D., Soreq, H. and Stamm,S. (2002) Defects in pre-mRNA processing as causes of and predisposition to diseases. DNA Cell Biol., 21, 803–818. [DOI] [PubMed] [Google Scholar]
- 15.Thanaraj T.A. and Stamm,S. (2003) Prediction and statistical analysis of alternatively spliced exons. Prog. Mol. Sub. Biol., 31, 1–31. [DOI] [PubMed] [Google Scholar]
- 16.Stoesser G., Baker,G., van den Broek,A., Garcia-Pastor,M., Kanz,C., Kulikova,T., Leinonen,R., Lin,Q., Lombard,V., Lopez,R. et al. (2003) The EMBL nucleotide sequence database: major new developments. Nucleic Acids Res., 31, 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.-C., Estreicher,A., Gasteiger,A., Martin,M.J., Michoud,K., O’Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stamm S., Zhu,J., Nakai,K., Stoilov,P. Stoss,O. and Zhang,M.Q. (2000) An alternative-exon database and its statistical analysis. DNA Cell Biol., 19, 739–756. [DOI] [PubMed] [Google Scholar]
- 19.Dralyuk I., Brudno,M., Gelfand,M.S., Zorn,M. and Dubchak,I. (2000) ASDB: database of alternatively spliced genes. Nucleic Acids Res., 28, 296–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji H., Zhou,Q., Wen,F., Xia,H., Lu,X. and Li,Y. (2001) AsMamDB: an alternative splice database of mammals. Nucleic Acids Res., 29, 260–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brett D., Pospisil,H., Valcárcel,J., Reich,J. and Bork,P. (2001) Alternative splicing and genome complexity. Nature Genet., 30, 29–30. [DOI] [PubMed] [Google Scholar]
- 22.Lee C., Atanelov,L., Modrek,B. and Xing,Y. (2003) ASAP: the Alternative Splicing Annotation Project. Nucleic Acids Res., 31, 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J. and Wheeler,D.L. (2003) GenBank. Nucleic Acids Res., 31, 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clamp M., Andrews,D., Barker,D., Bevan,P., Cameron,G., Chen,Y., Clark,L., Cox,T., Cuff,J., Curwen,V. et al. (2003) Ensembl 2002: accommodating comparative genomics. Nucleic Acids Res., 31, 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thanaraj T.A. and Clark,F. (2001) Human GC-AG alternative intron isoforms with weak donor sites show enhanced consensus at acceptor exon positions. Nucleic Acids Res., 29, 2581–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelso J., Visagie,J., Theiler,G., Christoffels,A., Bardien,S., Smedley,D., Otgaar,D., Greyling,G., Jongeneel,C.V., McCarthy,M.I. et al. (2003) eVOC: A controlled vocabulary for unifying gene expression data. Genome Res., 13, 1222–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thanaraj T.A., Clark,F. and Muilu,J. (2003) Conservation of human alternative splice events in mouse. Nucleic Acids Res., 31, 2544–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wain H.M., Lush,M., Ducluzeau,F. and Povey,S. (2002) Genew: the Human Gene Nomenclature Database. Nucleic Acids Res., 30, 169–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwisht,S.S., Eppig,J.T. et al. (2000) Gene Ontology: tool for the unification of biology. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashburner M., Ball,C.A., Blake,J.A., Butler,H., Cherry,J.M., Corradi,J., Dolinski,K., Janan T., Eppig, J.T., Harris,M. et al. (2001) Creating the Gene Ontology resource: design and implementation. Genome Res., 11, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]