Sm23, a novel SGNH arylesterase from S. meliloti 1021, was crystallized in space group I4122 and diffraction data were collected to a resolution of 2.2 Å.
Keywords: Sm23, SGNH family, Sinorhizobium meliloti 1021, arylesterases
Abstract
Industrial demand for active biocatalysts with desirable biochemical properties is constantly increasing and the discovery and characterization of novel esterases is potentially useful for industrial processes. Here, X-ray crystallographic studies of an (R)-specific SGNH arylesterase (Sm23) from Sinorhizobium meliloti 1021 are reported. The recombinant protein was expressed in Escherichia coli with a His tag and purified to homogeneity. Sm23 was crystallized using 0.2 M magnesium formate as a precipitant and X-ray diffraction data were collected to a resolution of 2.2 Å with an R merge of 6.9%. The crystals of SM23 belonged to the I-centred tetragonal space group I4122, with unit-cell parameters a = b = 126.6, c = 190.9 Å. A molecular-replacement solution was obtained using the crystal structure of arylesterase from Mycobacterium smegmatis as a template.
1. Introduction
Microbacteria produce different classes of lipolytic enzymes, including esterases, lipases and phospholipases, which are generally involved in ester hydrolysis, transesterifications and condensations of small metabolites or nutrients (Arpigny & Jaeger, 1999 ▶; Bornscheuer, 2002 ▶; Kang et al., 2006 ▶). These enzymes are important biocatalysts with diverse biotechnological applications owing to their broad specificities, high stereoselectivities, stability in organic solvents and high processing yields. More specifically, microbial esterases are currently employed in a vast array of industrial processes, including detergent formulation, food processing, chemical synthesis, oil and lipid processing and water purification (Serdakowski & Dordick, 2008 ▶; Panda & Gowrishankar, 2005 ▶). Therefore, the isolation and engineering of biotechnologically relevant esterases is highly valuable in the pursuit of enzymes with better catalytic performance (Gupta & Khare, 2009 ▶; Burton et al., 2002 ▶).
Lipolytic enzymes have been classified into eight families based on their sequences and biological properties (Arpigny & Jaeger, 1999 ▶). Structural studies of lipolytic enzymes have revealed that many of them harbour a typical α/β-hydrolase fold with a common Sm-X-Nu-X-Sm (where Sm is a small residue, X is any residue and Nu is a nucleophile) motif around the active-site residue (Carr & Ollis, 2009 ▶; Nardini & Dijkstra, 1999 ▶). This motif is generally located in the tip of a loop connecting a β-strand and an α-helix in the centre of the structure, forming an extremely sharp turn referred to as a nucleophile elbow (Ollis et al., 1992 ▶). However, not all lipolytic enzymes harbour the α/β-hydrolase fold and the canonical active-site motif. For example, SGNH hydrolases contain a 8Gly-9Asp-10Ser-11Leu (GDSL) motif near the N-terminal region and their fold in crystal structures is clearly not a typical α/β-hydrolase fold (Akoh et al., 2004 ▶).
The SGNH hydrolases are characterized by a highly conserved four-residue sequence consisting of serine (S), glycine (G), asparagine (N) and histidine (H) in active sites or oxyanion holes. They are suggested to exhibit multifunctional properties and broad substrate specificities owing to the flexibility of their active sites (Mølgaard et al., 2000 ▶). Recently, several SGNH hydrolases have been identified, including arylesterase from Streptomyces coelicolor (Bielen et al., 2009 ▶) and rhamnogalacturonan acetylesterases from Bacillus subtilis (Martínez-Martínez et al., 2008 ▶) and B. halodurans (Navarro-Fernández et al., 2008 ▶). Although an increasing amount of molecular information regarding SGNH-family proteins is becoming available, knowledge concerning the reaction mechanisms and the structural identification of these proteins remains limited in comparison with other lipolytic enzymes.
In a previous study, we identified and characterized an oligomeric SGNH arylesterase from Sinorhizobium meliloti 1021, which was designated Sm23 (Hwang et al., 2010 ▶). Sm23 exhibited a substrate preference for short-chain fatty-acid esters such as C2 or C4 with an (R)-preference. Here, we report the purification and preliminary crystallographic analysis of Sm23 as an initial attempt to solve its crystal structure. Structural study of Sm23 will not only provide the molecular basis for the substrate-specificity and enantioselectivity of SGNH-family proteins, but will also provide a platform for the design and engineering of these enzymes for numerous industrial applications.
2. Experimental procedures and results
2.1. Protein expression and purification
The gene encoding Sm23 was cloned into the bacterial expression vector pQE30 (Qiagen, USA) and the protein was overexpressed in Escherichia coli as previously described (Hwang et al., 2010 ▶). Transformed E. coli cells were grown in LB medium containing 100 µg ml−1 ampicillin at 310 K. 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added when the optical density at 600 nm reached 0.6. The culture was allowed to grow at 310 K for 4 h before the cells were harvested via centrifugation (8000g, 20 min, 279 K). For purification, the cell pellet was resuspended in buffer A consisting of 50 mM sodium phosphate pH 8.0, 300 mM NaCl and 10 mM imidazole, followed by sonication. The cell lysate was centrifuged at 15 000 rev min−1 for 20 min and the supernatant was loaded onto a HiTrap nickel-chelating column (GE Healthcare, USA) pre-equilibrated with buffer A. After 1 h of incubation, the protein was washed twice with buffer A containing 20 mM imidazole. The protein was then eluted in the same buffer with 250 mM imidazole and desalted using a PD-10 column (GE Healthcare, USA) with 20 mM Tris–HCl pH 8.0. The purity of the Sm23 was verified via SDS–PAGE and mass-spectrometric analysis. The purified protein was concentrated to 5 mg ml−1 using Vivaspin 30K (Sartorius-Stedim Biotech, Germany) for crystallization. Protein concentrations were determined with a Bio-Rad protein-assay kit (Bio-Rad Laboratories, USA) using bovine serum albumin (BSA) as a standard (Bradford, 1976 ▶). Sm23 was produced using E. coli XL1-Blue harbouring the pQE30 plasmid (Qiagen, USA). The N-terminal His tag was not removed and Sm23 thus contained an additional 12 amino acids within its N-terminus: MRGSHHHHHHGS.
2.2. Crystallization
Crystallization trials of Sm23 were carried out via the microbatch method (Chayen et al., 1990 ▶) under Al’s Oil using Crystal Screen and Crystal Screen 2 (Hampton Research, USA) crystal screening solutions at 285 K. A drop consisting of 1 µl screening solution and 1 µl protein solution was placed into each well of a Nunc 96-well Mini Tray (Nalge Nunc International, USA). Crystals appeared in various screening conditions. However, the rectangular rod crystals grown in condition No. 44 (0.2 M magnesium formate dehydrate) of Crystal Screen were employed for further diffraction experiments since crystallization was well reproduced in this condition and the crystals obtained using this condition showed the best diffraction quality.
2.3. X-ray data collection and data processing
Crystals were mounted in cryoloops (Hampton Research, USA) and 15% glycerol was used as a cryoprotectant. Data were collected at a wavelength of 1.000 Å using an ADSC Quantum 315 CCD detector on beamline PAL 4A at Pohang Accelerator Laboratory (PAL), Republic of Korea from a crystal that was flash-cooled in a cold nitrogen-gas stream at 100 K. The diffraction data were processed with the HKL-2000 package (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
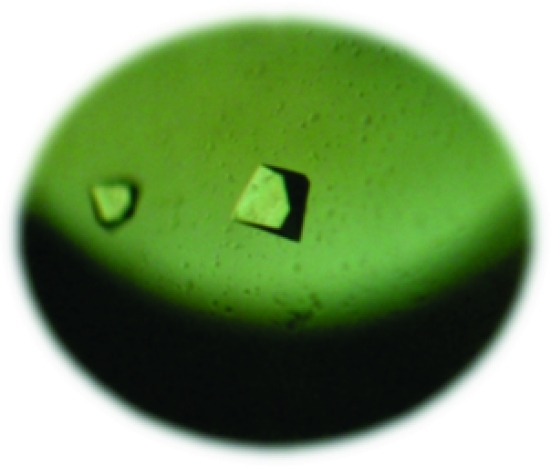
In a previous study, a new type of (R)-specific SGNH arylesterase was overexpressed and purified (Hwang et al., 2010 ▶). An amino-acid sequence-homology search using BLAST identified Sm23 as an SGNH esterase. Further sequence comparisons with other SGNH-family members confirmed the presence of a catalytic triad consisting of Ser10, Asp187 and His190. Sm23 was expressed in E. coli XL1-Blue cells and purified to homogeneity for structural studies. The purification of Sm23 was straightforward, requiring only one Ni–NTA metal-affinity step. The protein was concentrated to 5 mg ml−1 for crystallization. The molecular weight and purity of Sm23 were ∼24 kDa and ∼98%, respectively, as judged from SDS–PAGE. This value was in agreement with the calculated molecular mass of 24 315 Da including the N-terminal His tag. The initial crystallization conditions for Sm23 were screened using the microbatch method under Al’s Oil. Diffraction-quality crystals were obtained using 0.2 M magnesium formate dehydrate as a precipitant. The crystals grew to final dimensions of approximately 0.1 × 0.1 × 0.1 mm within 4 d (Fig. 1 ▶). The crystals diffracted to a resolution of 2.0 Å. The diffraction data set was processed at 2.2 Å resolution with 100% completeness and an R merge of 6.9%. Higher resolution reflections were not processed since R merge in the last resolution bin was higher than 30%. Data-collection statistics are summarized in Table 1 ▶.
Figure 1.
Sm23 crystals were obtained from 0.2 M magnesium formate dihydrate. Crystal dimensions are 0.1 × 0.1 × 0.1 mm.
Table 1. X-ray data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| Space group | I4122 |
| Unit-cell parameters (Å) | a = b = 126.6, c = 190.9 |
| Wavelength (Å) | 1.000 |
| Resolution (Å) | 50.00–2.20 (2.28–2.20) |
| Unique reflections | 39412 (3870) |
| Completeness (%) | 100 (100) |
| Multiplicity | 14.2 (14.0) |
| Rmerge† (%) | 6.9 (25.4) |
| Mean I/σ(I) | 57.0 (11.7) |
R
merge = 
 .
.
The diffraction data were indexed in the I-centred tetragonal space group I422, with unit-cell parameters a = b = 126.6, c = 190.9 Å. Given the systematic absence of reflections along (00l), the true space group of the SGNH arylesterase crystals was determined to be I4122. The structural model of Sm23 was obtained by molecular replacement using the crystal structure of arylesterase from Mycobacterium smegmatis (PDB code 2q0q; Mathews et al., 2007 ▶), which shares 40% sequence identity with Sm23, as the search model. Using the program MOLREP (Vagin & Teplyakov, 2010 ▶), the best molecular-replacement solution was obtained with an R factor of 42% and a correlation coefficient of 71.4%. Three Sm23 molecules can be packed into one asymmetric unit, corresponding to a solvent content of 55.6% and a calculated Matthews coefficient (V M) of 2.77 Å3 Da−1 (Matthews, 1968 ▶).
Modelling and refinement are currently under way. The crystal structure of Sm23 will contribute to our understanding of the reaction mechanisms of SGNH arylesterases as well as their R-stereospecific characteristics.
Acknowledgments
TDK is supported by an Ajou University Research Grant (Ajou–General Subjects) and by the Korea Research Foundation Grant funded by the Korean Government (KRF-2009-0089832). KKK is supported by the Korea Healthcare Technology R&D Project (A092006).
References
- Akoh, C. C., Lee, G.-C., Liaw, Y.-C., Huang, T.-H. & Shaw, J.-F. (2004). Prog. Lipid Res. 43, 534–552. [DOI] [PubMed]
- Arpigny, J. L. & Jaeger, K.-E. (1999). Biochem. J. 343, 177–183. [PMC free article] [PubMed]
- Bielen, A., Cetković, H., Long, P. F., Schwab, H., Abramić, M. & Vujaklija, D. (2009). Biochimie, 91, 390–400. [DOI] [PubMed]
- Bornscheuer, U. T. (2002). FEMS Microbiol. Rev. 26, 73–81. [DOI] [PubMed]
- Bradford, M. M. (1976). Anal. Biochem. 7, 248–254. [DOI] [PubMed]
- Burton, S. G., Cowan, D. A. & Woodley, J. M. (2002). Nature Biotechnol. 20, 37–45. [DOI] [PubMed]
- Carr, P. D. & Ollis, D. L. (2009). Protein Pept. Lett. 16, 1137–1148. [DOI] [PubMed]
- Chayen, N. E., Shaw Stewart, P. D., Maeder, D. L. & Blow, D. M. (1990). J. Appl. Cryst. 23, 297–302.
- Gupta, A. & Khare, S. K. (2009). Crit. Rev. Biotechnol. 29, 44–54. [DOI] [PubMed]
- Hwang, H., Kim, S., Yoon, S., Ryu, Y., Lee, S. Y. & Kim, T. D. (2010). Int. J. Biol. Macromol. 46, 145–152. [DOI] [PubMed]
- Kang, H.-Y., Kim, J. F., Kim, M. H., Park, S.-H., Oh, T.-K. & Hur, C.-G. (2006). FEBS Lett. 580, 2736–2740. [DOI] [PubMed]
- Martínez-Martínez, I., Navarro-Fernández, J., Lozada-Ramírez, J. D., García-Carmona, F. & Sánchez-Ferrer, A. (2008). Proteins, 71, 379–388. [DOI] [PubMed]
- Mathews, I., Soltis, M., Saldajeno, M., Ganshaw, G., Sala, R., Weyler, W., Cervin, M. A., Whited, G. & Bott, R. (2007). Biochemistry, 46, 8969–8979. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Mølgaard, A., Kauppinen, S. & Larsen, S. (2000). Structure, 8, 373–383. [DOI] [PubMed]
- Nardini, M. & Dijkstra, B. W. (1999). Curr. Opin. Struct. Biol. 9, 732–737. [DOI] [PubMed]
- Navarro-Fernández, J., Martínez-Martínez, I., Montoro-García, S., García-Carmona, F., Takami, H. & Sánchez-Ferrer, A. (2008). J. Bacteriol. 190, 1375–1382. [DOI] [PMC free article] [PubMed]
- Ollis, D. L., Cheah, E., Cygler, M., Dijkstra, B., Frolow, F., Franken, S. M., Harel, M., Remington, S. J., Silman, I. & Schrag, J. (1992). Protein Eng. 5, 197–211. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Panda, T. & Gowrishankar, B. S. (2005). Appl. Microbiol. Biotechnol. 67, 160–169. [DOI] [PubMed]
- Serdakowski, A. L. & Dordick, J. S. (2008). Trends Biotechnol. 26, 48–54. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]