The transaldolase enzyme from T. acidophilum has been crystallized in two different space groups.
Keywords: Schiff bases, (α/β)8-barrel, fructose 6-phosphate, aldolases, pentose phosphate pathway
Abstract
The metabolic enzyme transaldolase from Thermoplasma acidophilum was recombinantly expressed in Escherichia coli and could be crystallized in two polymorphic forms. Crystals were grown by the hanging-drop vapour-diffusion method using PEG 6000 as precipitant. Native data sets for crystal forms 1 and 2 were collected in-house to resolutions of 3.0 and 2.7 Å, respectively. Crystal form 1 belonged to the orthorhombic space group C2221 with five monomers per asymmetric unit and crystal form 2 belonged to the monoclinic space group P21 with ten monomers per asymmetric unit.
1. Introduction
Transaldolase (TA) is a ubiquitous enzyme of carbon metabolism that catalyzes the reversible transfer of a three-carbon dihydroxyacetone unit between sugar phosphates as part of the nonoxidative part of the pentose phosphate pathway (Samland & Sprenger, 2009 ▶). In the cellular context, it catalyzes the interconversion of the ketose substrates fructose 6-phosphate (F6P) and sedoheptulose 7-phosphate (S7P) in an equilibrium reaction with the aldose phosphates erythrose 4-phosphate (E4P) and glyceraldehyde 3-phosphate (G3P) as alternative acceptor substrates,
TA does not contain a cofactor but utilizes an active-centre lysine that forms Schiff-base adducts with donor ketose substrates in a multistep reaction sequence involving carbinolamine-type, iminium-type and enamine-type intermediates. Structural analysis of TAs from different organisms by X-ray diffraction revealed that all hitherto characterized TA proteins fold into an (α/β)8-barrel (TIM barrel) consisting of alternating α-helices and β-strands in a doughnut-shaped structure, the central core of which is constituted of eight parallel β-strands (Jia, Huang et al., 1996 ▶; Thorell et al., 2000 ▶). Despite the overall conserved subunit topology and fold, different quaternary assemblies are observed for TAs from different domains of life (Samland & Sprenger, 2009 ▶). While TAs from bacteria and eukarya form homodimers, the archaeal enzymes exist as homodecamers made up of two pentameric discs (a dimer of two pentamers) in a face-to-face orientation.
A molecular reaction mechanism for TA has been proposed on the basis of mutagenesis studies and structural analysis of a borohydride-reduced dihydroxyacetone Schiff-base adduct trapped at the active site of TA from Escherichia coli (Jia et al., 1997 ▶; Schörken et al., 2001 ▶). The steady-state kinetic data and structural information implicated two conserved acidic residues, a Glu and an Asp, as functioning as general acid/base catalysts at different stages of the reaction cycle. The conserved Glu has been proposed to first activate the essential lysine via a bridging catalytic water and then to facilitate dehydration of the transiently formed carbinolamine intermediate. Breakdown of the scissile C3—C4 bond of the Schiff base thus formed is believed to be initiated by ionization of the C4 hydroxyl group catalyzed by the conserved Asp residue. However, this functional assignment needs to be confirmed by structural analysis of genuine reaction intermediates trapped in the active site of a TA.
Here, we report the crystallization and preliminary X-ray diffraction analysis of TA from Thermoplasma acidophilum (TacTA) as part of a study towards understanding the molecular reaction mechanism of TA by structural analysis of authentic on-pathway intermediates.
2. Materials and methods
2.1. Cloning of Ta0616, protein expression and purification
An operon encoding transketolase I (Ta0618), transketolase II (Ta0617) and transaldolase (Ta0616) from T. acidophilum was amplified from chromosomal DNA of T. acidophilum using PCR. The whole operon was then cloned into pET28a vector (Novagen) using NdeI and HindIII restriction sites and appropriate primers (MWG Biotech). A double stop codon was inserted at the end of gene Ta0616 encoding TacTA (residues 1–223) to prevent expression of the C-terminal His tag. The correctness of the sequence was confirmed by complete sequencing of the gene.
TacTA was heterologously expressed in E. coli BL21 Star (DE3) (Invitrogen). Cells were grown for 14–18 h at 310 K after induction of gene expression with 0.5 mM IPTG. The protein was initially enriched by fractional ammonium sulfate precipitation, in which the protein precipitated in the fraction containing 50–80%(w/v) ammonium sulfate. This fraction was desalted by exhaustive dialysis against 20 mM Tris–HCl pH 7.5 at 279 K and was then further purified using a Fractogel EMD TMAE (S) (Merck KgaA, Germany) anion-exchange step (elution buffer: 20 mM Tris pH 7.5 supplemented with 1 M NaCl) and subsequent heat precipitation (20 min at 333 K). The homogeneity was judged by SDS–PAGE analysis. Finally, the buffer was exchanged to 20 mM glycylglycine pH 7.5 and the protein was stored at 273–279 K until use.
2.2. Steady-state kinetic analysis
Transaldolase activity (conversion of the donor F6P and the acceptor E4P into the products G3P and S7P) was analyzed in a coupled spectrophotometric steady-state assay using the auxiliary enzymes triosephosphate isomerase (TIM) and sn-glycerol-3-phosphate:NAD+ 2-oxidoreductase (G3PDH) to detect the formation of G3P, which is derived from the cleavage of F6P. The concomitant oxidation of NADH was followed spectrophotometrically at 340 nm in 20 mM glycylglycine pH 7.5 supplemented with 50 U ml−1 TIM, 5 U ml−1 G3PDH and varying concentrations of F6P (up to 15 mM) and E4P (up to 2 mM) at 303 K. Potential F6P or fructose 1,6-bisphosphate (FBP) aldolase activity was tested with the same assay but using F6P or FBP as the sole substrate (in the absence of any acceptor substrate).
2.3. Crystallization and data collection
Crystals of TacTA were grown by the hanging-drop vapour-diffusion method at 291 K using a modification of the protocol that was established for the crystallization of TA from E. coli (Jia, Lindqvist et al., 1996 ▶). A 3 µl droplet of 16 mg ml−1 TacTA in a two-component buffer (6 mM Tris and 14 mM glycylglycine pH 7.5) was mixed with 3 µl reservoir solution consisting of 0.1 M sodium citrate pH 4, 10%(w/v) PEG 6000 and 25%(v/v) glycerol and equilibrated against 500 µl reservoir solution. The pH of the resulting crystallization mixture was determined to be 4.5.
Crystals of TacTA were mounted in cryoloops (Hampton Research, USA), flash-cooled by direct immersion in liquid nitrogen and placed on the goniometer head. Diffraction data for both crystal forms were collected in-house with an R-AXIS IV++ imaging-plate system (Rigaku–MSC, Japan) using Cu Kα radiation with a wavelength of 1.5418 Å generated by a rotating-anode generator (Rigaku MicroMax-007). Data sets were collected under cryogenic conditions at 100 K (XSTREAM2000, Rigaku–MSC, Japan). Data sets were processed and scaled using XDS (Kabsch, 2010 ▶).
3. Results and discussion
TacTA could be recombinantly produced in E. coli strain BL21 (DE3) Star, for which the highest expression yields were obtained amongst a set of different tested strains. Starting from 10 g cell material, a total of 15–20 mg protein could be purified to homogeneity by fractional salt precipitation, anion-exchange chromatography and heat precipitation. Purified TacTA exhibited a single band at 26 kDa on SDS–PAGE, which is in reasonable agreement with the calculated molecular mass of 24 437 Da. The enzyme consists of 223 amino-acid residues and thus belongs to the transaldolase-like proteins, which are approximately 100 residues shorter in length than the classical transaldolases (Samland & Sprenger, 2009 ▶). Steady-state kinetic analysis confirmed that TacTA exhibits transaldolase activity, as predicted from sequence similarity to TAs from other organisms, with a k cat of 16.61 ± 1.11 s−1 (303 K, pH 7.5) at saturating concentrations of the substrates F6P and E4P. In contrast, no F6P or fructose 1,6-bisphosphate aldolase activity could be detected.
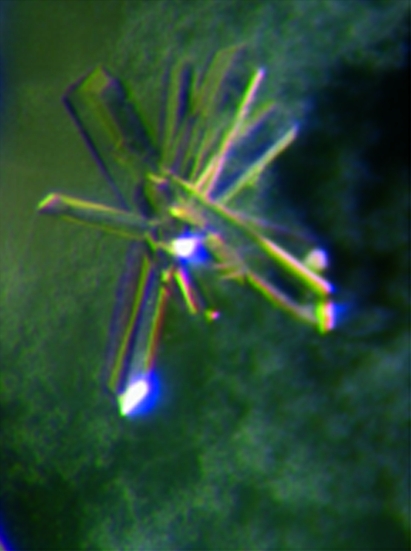
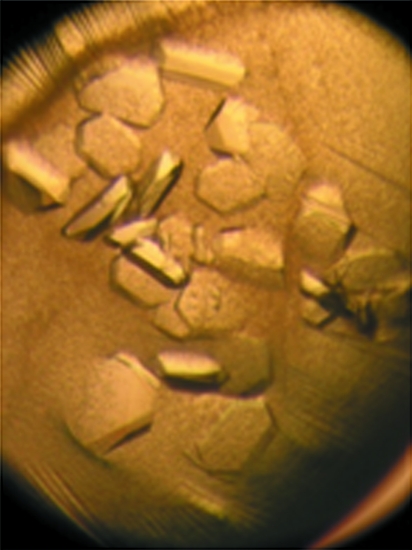
Two polymorphic crystal forms of TacTA were obtained under identical chemical conditions at 291 K. It turned out that the temperature conditions of crystallization setup prior to crystallization were important in determining which crystal form was preferentially grown. When crystallization setup was carried out at 303 K, TacTA crystallized in form 1 with approximate crystal dimensions of 0.6 × 0.1 × 0.1 mm (Fig. 1 ▶). In contrast, a second crystal form (form 2) with dimensions of 0.4 × 0.4 × 0.1 mm (Fig. 2 ▶) was obtained when crystallization was set up at 291 K. Occasionally, crystals of both forms grew in the same drop. However, when crystals of form 2 appeared first it was rarely observed that the other crystal form grew in the same drop.
Figure 1.
Representative crystal (0.6 × 0.1 × 0.1 mm) of TacTA in the orthorhombic space group C2221.
Figure 2.
Representative crystal (0.4 × 0.4 × 0.1 mm) of TacTA in the monoclinic space group P21.
Native data sets from crystal forms 1 and 2 were collected in-house under cryogenic conditions to resolutions of 3.0 and 2.7 Å, respectively (Table 1 ▶). The space group of form 1 was identified as the orthorhombic space group C2221, while form 2 belonged to the monoclinic space group P21. Initial molecular-replacement phasing using the structure of TA from Thermus thermophilus HB8 (PDB entry 1wx0; Y. Asada & N. Kunishima, unpublished work) as a search model with Phaser (Winn et al., 2011 ▶) was successful and revealed the presence of either five (form 1) or ten (form 2) monomers in the asymmetric unit. The Matthews coefficients were 2.63 Å3 Da−1 (form 1) and 2.56 Å3 Da−1 (form 2), indicating a similar solvent content of ∼52% for both crystal forms, which falls within the typical range for protein crystals. Initial trials showed that both crystal forms could withstand soaking with substrate solution.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Crystal form 1 | Crystal form 2 | |
|---|---|---|
| Space group | C2221 | P21 |
| Unit-cell parameters (Å, °) | a = 147.10, b = 171.30, c = 98.90, α = β = γ = 90.0 | a = 98.98, b = 129.17, c = 100.10, α = γ = 90.0, β = 108.2 |
| Resolution range (Å) | 29.63–3.00 (3.10–3.00) | 29.40–2.70 (2.80–2.70) |
| Mosaicity (°) | 0.39 | 0.23 |
| No. of observations | 138527 (13021) | 474815 (36419) |
| No. of unique reflections | 24858 (2303) | 65081 (6176) |
| 〈I/σ(I)〉 | 20.8 (4.2) | 21.0 (5.3) |
| Completeness (%) | 97.9 (99.8) | 98.9 (91.3) |
| Multiplicity | 5.6 (5.6) | 7.3 (5.9) |
| Rmerge† (%) | 7.8 (44.9) | 8.1 (33.5) |
| Rmeas‡ (%) | 8.6 (49.5) | 8.7 (36.8) |
| B factor from Wilson plot (Å2) | 54.5 | 49.3 |
| Matthews coefficient (Å3 Da−1) | 2.63 | 2.56 |
| Monomers in asymmetric unit | 5 | 10 |
R
merge = 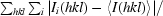
 ;
;
R
meas = 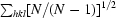
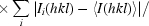
 , where Ii(hkl) is the intensity of the ith observation of reflection hkl and 〈I(hkl)〉 is the mean value of Ii(hkl) for all measurements; N is the multiplicity.
, where Ii(hkl) is the intensity of the ith observation of reflection hkl and 〈I(hkl)〉 is the mean value of Ii(hkl) for all measurements; N is the multiplicity.
We are now aiming at collecting data sets at higher resolution using synchrotron radiation and additionally solving the structures of reaction intermediates after soaking TacTA crystals with different physiological substrates in order to obtain further insights into the enzymatic reaction mechanism. In this context, we also wish to analyze whether or not similar catalytic principles govern the processing of the sugar substrates and derived intermediates to those that have recently been proposed for transketolase (Asztalos et al., 2007 ▶; Tittmann & Wille, 2009 ▶). As opposed to TA, transketolase transfers two-carbon rather than three-carbon units and contains the bioorganic cofactor thiamine diphosphate. Since the molecular origins of the different bond-fission specificity in the two enzymes are not understood, a comparative structural analysis of reaction intermediates will provide insight.
Acknowledgments
This work was supported by the Friedrich-Naumann-Stiftung (stipend to AL-L) and the Göttingen Graduate School for Neurosciences and Molecular Biosciences funded by the Deutsche Forschungsgemeinschaft (to KT).
References
- Asztalos, P., Parthier, C., Golbik, R., Kleinschmidt, M., Hübner, G., Weiss, M. S., Friedemann, R., Wille, G. & Tittmann, K. (2007). Biochemistry, 46, 12037–12052. [DOI] [PubMed]
- Jia, J., Huang, W., Schörken, U., Sahm, H., Sprenger, G. A., Lindqvist, Y. & Schneider, G. (1996). Structure, 4, 715–724. [DOI] [PubMed]
- Jia, J., Lindqvist, Y., Schneider, G., Schörken, U., Sahm, H. & Sprenger, G. A. (1996). Acta Cryst. D52, 192–193. [DOI] [PubMed]
- Jia, J., Schörken, U., Lindqvist, Y., Sprenger, G. A. & Schneider, G. (1997). Protein Sci. 6, 119–124. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Samland, A. K. & Sprenger, G. A. (2009). Int. J. Biochem. Cell Biol. 41, 1482–1494. [DOI] [PubMed]
- Schörken, U., Thorell, S., Schürmann, M., Jia, J., Sprenger, G. A. & Schneider, G. (2001). Eur. J. Biochem. 268, 2408–2415. [DOI] [PubMed]
- Thorell, S., Gergely, P., Banki, K., Perl, A. & Schneider, G. (2000). FEBS Lett. 475, 205–208. [DOI] [PubMed]
- Tittmann, K. & Wille, G. (2009). J. Mol. Cat. B Enzym. 61, 93–99.
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.