Diffraction-quality crystals of N. equitans neelaredoxin have been produced. The expression, purification and crystallization of the protein and preliminary X-ray crystallographic analysis of the crystals are reported.
Keywords: superoxide reductases, Nanoarchaeum equitans, oxidative stress, neelaredoxins
Abstract
Superoxide reductases (SORs) are the most recent oxygen-detoxification system to be identified in anaerobic and microaerobic bacteria and archaea. SORs are metalloproteins that are characterized by their possession of a catalytic nonhaem iron centre in the ferrous form coordinated by four histidine ligands and one cysteine ligand. Ignicoccus hospitalis, a hyperthermophilic crenarchaeon, is the only organism known to date to serve as a host for Nanoarchaeum equitans, a nanosized hyperthermophilic archaeon isolated from a submarine hot vent which completely depends on the presence of and contact with I. hospitalis cells for growth to occur. Similarly to I. hospitalis, N. equitans has a neelaredoxin (a 1Fe-type SOR) that keeps toxic oxygen species under control, catalysing the one-electron reduction of superoxide to hydrogen peroxide. Blue crystals of recombinant N. equitans SOR in the oxidized form (12.7 kDa, 109 residues) were obtained using polyethylene glycol (PEG 2000 MME) as precipitant. These crystals diffracted to 1.9 Å resolution at 100 K and belonged to the orthorhombic space group P212121, with unit-cell parameters a = 51.88, b = 82.01, c = 91.30 Å. Cell-content analysis suggested the presence of four monomers in the asymmetric unit. The Matthews coefficient (V M) was determined to be 1.9 Å3 Da−1, corresponding to an estimated solvent content of 36%. Self-rotation function and native Patterson calculations suggested a tetramer with 222 point-group symmetry, similar to other 1Fe-SORs. The three-dimensional structure will be determined by the molecular-replacement method.
1. Introduction
Superoxide reductases (SORs) constitute an enzyme family that is responsible for reduction of the superoxide anion (O2 ·−) to hydrogen peroxide (H2O2; Jenney et al., 1999 ▶; Pinto et al., 2010 ▶). This superoxide-scavenging enzyme family is present in prokaryotes, both anaerobic or microaerophilic, and represents an alternative strategy to the superoxide dismutase (SOD) family by operating through a distinct mechanism (McCord & Fridovich, 1969 ▶). SORs are only able to reduce the superoxide anion, whereas SODs couple its reduction and oxidation.
The reduction of O2 ·− is a one electron–two proton process that takes place in a highly conserved active site: a nonhaem ferrous ion (Fe2+) pentacoordinated by four equatorial histidines and one axial cysteine in a square-pyramidal geometry. The vacant axial coordination site of the Fe atom is the proposed binding site of the substrate in the ferrous state. In the ferric resting state, a glutamate residue or a water molecule acts as a sixth ligand, completing an octahedral coordination geometry (Pinto et al., 2010 ▶).
SORs can be distinguished by their number of metal centres. 1Fe-SORs or neelaredoxins have only one iron site that constitutes the active centre and is named centre II (Pinto et al., 2010 ▶). 2Fe-SORs or desulfoferrodoxins have an extra N-terminal domain harbouring a nonhaem iron coordinated by four cysteines, [Fe(Cys)4]3+, in a distorted tetrahedral geometry, designated as centre I (Archer et al., 1995 ▶). Other SORs, e.g. that from Treponema pallidum, were found to contain this extra N-terminal domain, although one of the cysteine ligands and the Fe atom are absent (Santos-Silva et al., 2006 ▶).
The first step in the catalytic reduction of O2 ·− is the binding of the substrate to the ferrous centre and the formation of an initial intermediate (T1), thought to be an Fe3+-hydroperoxo species which following a protonation step will decay and release the product, H2O2; finally, an oxidized H2O-bound or Glu-bound species is formed (Pinto et al., 2010 ▶). During substrate binding and product release, two amino-acid residues (Glu12 and Lys13; Archaeoglobus fulgidus 1Fe-SOR numbering) that are conserved in the majority of SORs have been proposed to play important roles in the mechanism (Yeh et al., 2000 ▶; Berthomieu et al., 2002 ▶). The positively charged Lys residue is thought to attract the substrate to the active centre, whereas the Glu residue is believed to be responsible for mediating the protonation step prior to its binding to the Fe atom. Interestingly, some mutagenesis studies in the A. fulgidus (Rodrigues et al., 2006 ▶), Desulfoarculus baarsii (Nivière et al., 2004 ▶) and D. vulgaris (Emerson et al., 2002 ▶) enzymes have shown that the absence of these two residues does not impair the overall catalytic cycle. Furthermore, the natural absence of the Glu residue in Nanoarchaeum equitans SOR has been shown not to impair the catalytic reduction of O2 ·− (Rodrigues et al., 2008 ▶). In fact, it was proposed that superoxide reduction by this enzyme also occurs via a two-step reaction involving the formation of a single transient (T1) as shown in Rodrigues et al. (2008 ▶), in analogy with other site-directed mutants lacking the active-site Glu residue.
This 1Fe-SOR, a natural glutamate-lacking SOR mutant, is encoded in the genome of N. equitans, an anaerobic hyperthermophilic archaeon isolated from a marine hydrothermal vent with an optimal growth temperature of 363 K at pH 5.5 that is only capable of growing in the presence of Ignicoccus hospitalis, another hyperthermophilic archaeon, thus forming a unique intimate association (Huber et al., 2002 ▶; Jahn et al., 2008 ▶).
In this study, we describe the recombinant expression in Escherichia coli, purification, crystallization and preliminary X-ray crystallographic analysis of the 1Fe-SOR from N. equitans. This crystal structure (lacking the glutamate as the sixth ligand in the ferric state), together with the crystal structure of I. hospitalis SOR (which lacks the canonical lysine residue; Pinho et al., 2010 ▶), will further contribute to the understanding of the catalytic mechanism of SORs, showing how the active centre is disturbed by the absence of these previously believed to be key canonical residues.
2. Experimental procedures and results
2.1. Protein expression and purification
E. coli BL21 Gold (DE3) cells (Stratagene) containing the previously described plasmid pT7NNlr (Rodrigues et al., 2008 ▶) were grown aerobically at 310 K in M9 minimal medium (Ausubel et al., 1987 ▶) supplemented with 30 µg ml−1 kanamycin and enriched with 400 µM FeSO4·7H2O until the OD600nm reached 0.3. At this stage, 400 µM isopropyl β-d-1-thiogalactopyranoside was added, the temperature was lowered to 303 K and growth was continued for 20 h. The cells were then harvested by centrifugation at 10 000g for 10 min at 277 K.
Harvested cells were resuspended in a buffer consisting of 20 mM Tris–HCl pH 7.6, 1 mM phenylmethanesulfonyl fluoride (PMSF) and 20 µg ml−1 DNase (Sigma) and broken in a large-cell French press at 131 MPa. All subsequent purification steps were performed at pH 7.6 and 277 K. After ultracentrifugation at 186 000g for 1 h, the soluble extract was first dialyzed overnight against 20 mM Tris–HCl, 1 mM PMSF (buffer A) and then centrifuged at 48 384g for 15 min. The soluble fraction was subsequently loaded onto a Q-Sepharose Fast Flow column (XK 26/10; GE Healthcare) previously equilibrated with buffer A. The fraction containing N. equitans SOR was eluted in the flowthrough; it was then concentrated by ultrafiltration (Amicon, 10 kDa cutoff) and loaded onto a Superdex 75 column (XK 26/60; GE Healthcare) equilibrated with 20 mM Tris–HCl and 150 mM NaCl. The collected fractions were analyzed and judged to be pure on the basis of an SDS–PAGE gel.
In order to determine the correct molecular mass of the N. equitans SOR, the pure protein solution was loaded onto a size-exclusion column (Superdex 200 column, XK 10/300; GE Healthcare) using appropriate molecular-mass standards in parallel. The buffer used was 20 mM Tris–HCl pH 7.6, 150 mM NaCl with a flow rate of 0.5 ml min−1. The elution profile (Fig. 1 ▶ c) revealed a protein solution containing a tetramer with a molecular mass of around 46 kDa. In a final SDS–PAGE gel several bands were observed (Fig. 1 ▶ a) corresponding to different oligomeric forms as confirmed by the results of size-exclusion chromatography.
Figure 1.
(a) SDS–PAGE. Lane 1, low-molecular-weight markers; lane 2, N. equitans SOR protein sample eluted from the gel-filtration column. (b) UV–Vis absorption spectrum of pure N. equitans SOR; the region from 400 to 800 nm is amplified 5× in order to highlight the characteristic band with a maximum at ∼550 nm. (c) N. equitans SOR protein elution profile from a Superdex 200 (10/300) column. The 280 nm peak corresponds to a retention volume of 15.4 ml. The y axis corresponds to absorbance at 280 nm.
The protein concentration and total iron content were determined using the bicinchoninic acid protein assay (Pierce; Smith et al., 1985 ▶) and the 2,4,6-tripyridyl-S-triazine method (Fischer & Price, 1964 ▶), respectively. The final protein sample had an iron content of 0.7 mol iron per monomer and the protein was concentrated to 15 mg ml−1 using a 10 kDa cutoff Centricon (Vivaspin).
2.2. Crystallization and cryoprotection
Preliminary crystallization screening was carried out with protein concentrated to 15 mg ml−1 in 20 mM Tris–HCl pH 7.6, 150 mM NaCl using the vapour-diffusion technique. Nanolitre-scale drops were prepared with the commercially available Structure I & II kit (Molecular Dimensions) using a Cartesian Crystallization Robot Dispensing System (Genomics Solutions) and polystyrene round-bottom Greiner 96-well CrystalQuick plates (Greiner Bio-One). One crystallization drop was prepared per screened condition by mixing 100 nl protein solution with 100 nl reservoir solution. The drops were equilibrated against 100 µl reservoir solution. After 4 d, nearly all of the drops were still clear, suggesting that N. equitans SOR was not sufficiently concentrated for crystallization. The protein concentration was therefore increased to 30 mg ml−1 and two additional 96-well plate screenings were performed using the Structure I & II and PEG II screens (Quiagen). Crystals were now observed in five different crystallization conditions (Structure I & II condition No. 94 and PEG II screen conditions No. 7, 21, 55 and 58) containing different types of polyethylene glycol as precipitant. However, these crystals could not be manually reproduced in 24-well plates (Hampton Research Cryschem plates made of optically clear polystyrene) using either the hanging-drop or sitting-drop vapour-diffusion methods. A systematic screening around the initial hits was performed by modifying the precipitant and salt concentrations and trying different proportions of protein to reservoir solution in the drop (1:2, 1:1 and 2:1) in final drop volumes of 1, 2 and 3 µl, but no crystals were ever observed. As a last attempt, crystallization drops were prepared on a Greiner 96-well CrystalQuick plate and finally two blue crystals (Fig. 1 ▶) were successfully obtained in condition No. 21 of PEG II screen (Qiagen) consisting of 20% PEG 2000 MME only. The polystyrene used in manufacturing the Greiner plates was likely to have played a key role in the crystallization of N. equitans SOR. The crystals were grown using a 2 µl drop obtained by mixing 1 µl protein solution and 1 µl reservoir solution. Their dimensions were 100 × 80 × 40 µm (Fig. 2 ▶) and they could be harvested, cryocooled and used for data collection.
Figure 2.
Blue crystals of N. equitans superoxide reductase (neelaredoxin) grown in 20%(v/v) PEG 2000 MME. The largest crystal dimensions are 0.1 × 0.08 × 0.04 mm.
2.3. Data collection and processing
The strategy for this data collection was to solve the structure by molecular replacement since several homologous structures are available in the PDB. However, it is noteworthy mentioning that no anomalous signal was detected in the measured data at the wavelength used (0.97930 Å).
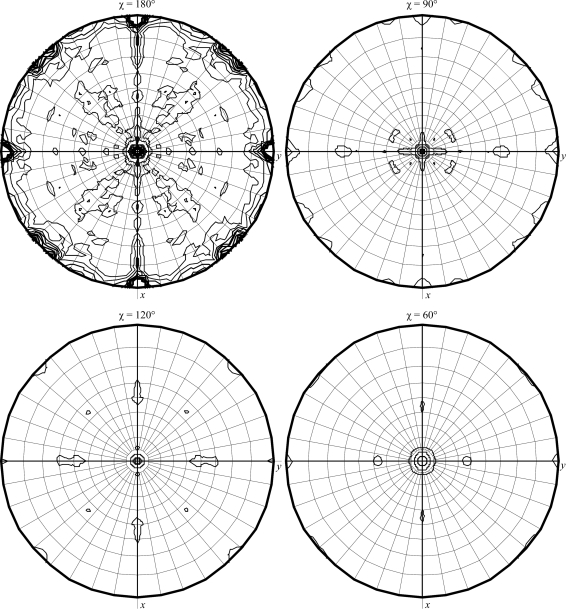
Diffraction data from a crystal of N. equitans SOR flash-cooled using 40%(v/v) PEG 2000 MME as a cryoprotecting solution were collected to 1.88 Å resolution on beamline ID23-1 of the European Synchrotron Radiation Facility (ESRF) in Grenoble using an ADSC Quantum Q315r CCD detector (Fig. 3 ▶). The crystal belonged to the orthorhombic space group P212121, with unit-cell parameters a = 51.88, b = 82.01, c = 91.30 Å. The data were integrated and scaled with XDS (Kabsch, 2010 ▶). The diffracted intensities obtained with XDS were subsequently merged with SCALA and converted to structure factors with CTRUNCATE from the CCP4 suite (Winn et al., 2011 ▶). Data-collection and processing statistics are presented in Table 1 ▶. Matthews coefficient calculations (Matthews, 1968 ▶) indicated the presence of four molecules in the asymmetric unit, with a corresponding V M value of 1.9 Å3 Da−1 and a predicted solvent content of 36%. A self-rotation Patterson function calculation (Fig. 4 ▶) revealed strong peaks corresponding to twofold noncrystallographic rotation axes, suggesting the presence of a tetramer in the asymmetric unit with 222 point symmetry as in other known 1Fe-SOR crystal structures. Finally, a native Patterson map calculation (Fig. 5 ▶) showed a strong peak at (0, 0.17, 0.5) indicative of a twofold noncrystallographic rotation axis parallel to, but not coincident with, the crystallographic c axis.
Figure 3.
Diffraction images collected on ESRF beamline ID23-1 from an N. equitans 1Fe-SOR single crystal (inset). A composite image of the first (top half, frame 1) and the last (bottom half, frame 241) diffraction images collected is shown, demonstrating the effect of crystal mosaicity on spot shape. The green scale bars in the inset are 100 µm in length. The mosaicity range of the collected data was 0.249–0.642°.
Table 1. Diffraction data-collection and processing parameters.
Values in parentheses are for the highest resolution shell.
| Beamline | ESRF ID23-1 |
| Detector | ADSC Quantum Q315r |
| Wavelength (Å) | 0.97930 |
| No. of frames | 214 |
| Oscillation range (°) | 0.5 |
| Data-processing program | XDS |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 51.88, b = 82.01, c = 91.30 |
| Resolution (Å) | 45.1–1.88 (1.95–1.88) |
| No. of observations | 129895 (8825) |
| Unique reflections | 31984 (2864) |
| Completeness (%) | 98.6 (91.6) |
| Multiplicity | 4.1 (3.1) |
| Rmerge† (%) | 5.4 (38.3) |
| Rp.i.m.‡ (%) | 3.0 (23.7) |
| Rmeas§ (%) | 6.2 (45.3) |
| 〈I/σ(I)〉 | 14.1 (2.6) |
| Wilson plot B factor (Å2) | 28 |
| Z¶ | 4 |
| VM (Å3 Da−1) | 1.9 |
| Estimated solvent content (%) | 36 |
The merging R factor R
merge = 
 × 100, where Ii(hkl) is the observed intensity, 〈I(hkl)〉 is the average intensity of multiple observations from symmetry-related reflections.
× 100, where Ii(hkl) is the observed intensity, 〈I(hkl)〉 is the average intensity of multiple observations from symmetry-related reflections.
The precision-independent R factor R
p.i.m. = 
 × 100; N is the multiplicity (Diederichs & Karplus, 1997 ▶).
× 100; N is the multiplicity (Diederichs & Karplus, 1997 ▶).
The redundancy-independent R factor R
meas = 
 × 100 (Diederichs & Karplus, 1997 ▶).
× 100 (Diederichs & Karplus, 1997 ▶).
No. of monomers in the asymmetric unit according to the Matthews coefficient (Matthews, 1968 ▶).
Figure 4.
MOLREP self-rotation function calculation showing the presence of twofold NCS axes perpendicular to the crystallographic c axis. The resolution range of the self-rotation search was 15–3 Å.
Figure 5.
w = 1/2 section of the native Patterson map showing a large non-origin peak corresponding to a noncrystallographic twofold axis parallel to the crystallographic c axis. The map was calculated using all data and contoured at 5σ intervals starting at 3σ. The origin peak is 133σ and the NCS peak is 38σ.
3. Concluding remarks
The crystallographic structure of the N. equitans 1Fe-SOR will be solved by molecular replacement using its closest structural homologue in the PDB as a search model. Knowledge of this structure will contribute to understanding the reaction mechanism of SOR with superoxide, since this protein may be considered a ‘natural mutant’ of this family that lacks the canonical glutamate residue that has so far been considered to be a key residue for catalysis of the reaction.
Acknowledgments
This work was supported by research grants PTDC/BIA-PRO/111940/2009, PTDC/BIA-PRO/67263/2006 and PTDC/BIA-PRO/70429/2006 funded by Fundação para a Ciência e Tecnologia (FCT), Ministério da Ciência e Ensino Superior, Portugal and the European Union FEDER program. ESRF support for data collection on ID23-1 is also acknowledged. FP is the recipient of IBET fellowship 21/07/09 CB IBET. AFP is the recipient of FCT PhD grant SFRH/BD/41355/2007. LCP is the recipient of a technician fellowship within FCT grant PTDC/BIA-PRO/70429/2006.
References
- Archer, M., Huber, R., Tavares, P., Moura, I., Moura, J. J., Carrondo, M. A., Sieker, L. C., LeGall, J. & Romão, M. J. (1995). J. Mol. Biol. 251, 690–702. [DOI] [PubMed]
- Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (1987). Editors. Current Protocols in Molecular Biology. New York: Greene Publishing Associates/Wiley Interscience.
- Berthomieu, C., Dupeyrat, F., Fontecave, M., Verméglio, A. & Nivière, V. (2002). Biochemistry, 41, 10360–10368. [DOI] [PubMed]
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol. 4, 269–275. [DOI] [PubMed]
- Emerson, J. P., Coulter, E. D., Cabelli, D. E., Phillips, R. S. & Kurtz, D. M. (2002). Biochemistry, 41, 4348–4357. [DOI] [PubMed]
- Fischer, D. S. & Price, D. C. (1964). Clin. Chem. 10, 21–31. [PubMed]
- Huber, H., Hohn, M. J., Rachel, R., Fuchs, T., Wimmer, V. C. & Stetter, K. O. (2002). Nature (London), 417, 63–67. [DOI] [PubMed]
- Jahn, U., Gallenberger, M., Paper, W., Junglas, B., Eisenreich, W., Stetter, K. O., Rachel, R. & Huber, H. (2008). J. Bacteriol. 190, 1743–1750. [DOI] [PMC free article] [PubMed]
- Jenney, F. E., Verhagen, M. F., Cui, X. & Adams, M. W. (1999). Science, 286, 306–309. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCord, J. M. & Fridovich, I. (1969). J. Biol. Chem. 244, 6049–6055. [PubMed]
- Nivière, V., Asso, M., Weill, C. O., Lombard, M., Guigliarelli, B., Favaudon, V. & Houée-Levin, C. (2004). Biochemistry, 43, 808–818. [DOI] [PubMed]
- Pinho, F. G., Romão, C. V., Pinto, A. F., Saraiva, L. M., Huber, H., Matias, P. M., Teixeira, M. & Bandeiras, T. M. (2010). Acta Cryst. F66, 605–607. [DOI] [PMC free article] [PubMed]
- Pinto, A. F., Rodrigues, J. V. & Teixeira, M. (2010). Biochim. Biophys. Acta, 1804, 285–297. [DOI] [PubMed]
- Rodrigues, J. V., Abreu, I. A., Cabelli, D. & Teixeira, M. (2006). Biochemistry, 45, 9266–9278. [DOI] [PubMed]
- Rodrigues, J. V., Victor, B. L., Huber, H., Saraiva, L. M., Soares, C. M., Cabelli, D. E. & Teixeira, M. (2008). J. Biol. Inorg. Chem. 13, 219–228. [DOI] [PubMed]
- Santos-Silva, T., Trincão, J., Carvalho, A. L., Bonifácio, C., Auchère, F., Raleiras, P., Moura, I., Moura, J. J. & Romão, M. J. (2006). J. Biol. Inorg. Chem. 11, 548–558. [DOI] [PubMed]
- Smith, P. K., Krohn, R. I., Hermanson, G. T., Mallia, A. K., Gartner, F. H., Provenzano, M. D., Fujimoto, E. K., Goeke, N. M., Olson, B. J. & Klenk, D. C. (1985). Anal. Biochem. 150, 76–85. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yeh, A. P., Hu, Y., Jenney, F. E., Adams, M. W. & Rees, D. C. (2000). Biochemistry, 39, 2499–2508. [DOI] [PubMed]