Abstract
The Saccharomyces Genome Database (SGD; http://www.yeastgenome.org/), a scientific database of the molecular biology and genetics of the yeast Saccharomyces cerevisiae, has recently developed several new resources that allow the comparison and integration of information on a genome-wide scale, enabling the user not only to find detailed information about individual genes, but also to make connections across groups of genes with common features and across different species. The Fungal Alignment Viewer displays alignments of sequences from multiple fungal genomes, while the Sequence Similarity Query tool displays PSI-BLAST alignments of each S.cerevisiae protein with similar proteins from any species whose sequences are contained in the non-redundant (nr) protein data set at NCBI. The Yeast Biochemical Pathways tool integrates groups of genes by their common roles in metabolism and displays the metabolic pathways in a graphical form. Finally, the Find Chromosomal Features search interface provides a versatile tool for querying multiple types of information in SGD.
INTRODUCTION
The Saccharomyces Genome Database (SGD) collects and organizes biological information about genes and proteins of the yeast Saccharomyces cerevisiae from the scientific literature, and presents this information on individual Locus Pages for each yeast gene. In addition to collecting detailed information about S.cerevisiae genes and proteins, SGD is continuing to develop new ways for users to visualize the relationships of yeast genes to one another and to similar genes and proteins from different species. The new tools described below help place individual genes in a larger biological context, using flexible interfaces that allow users to choose the format in which results are presented, or to use the results generated by one tool as a starting point for analysis with another tool.
FUNGAL ALIGNMENT VIEWER
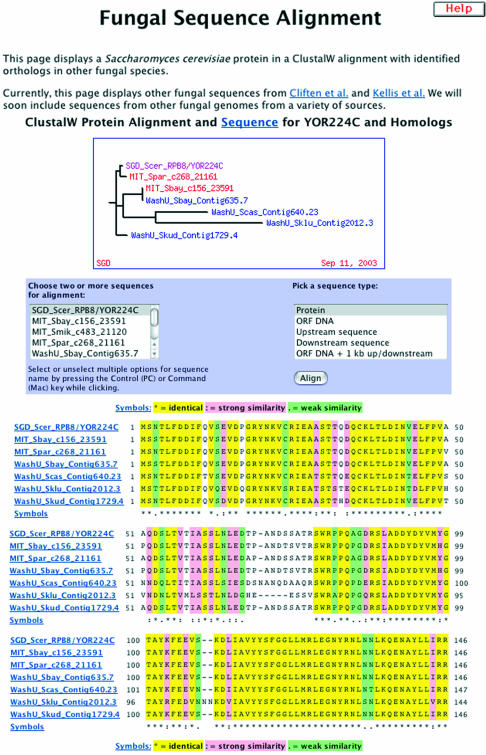
The Fungal Alignment Viewer (Fig. 1), accessible from the Comparison Resources pulldown menu in the right-hand column of the Locus Page, displays available sequences from other fungal genomes that have similarity to a predicted protein sequence from S.cerevisiae. At the top of the viewer is a dendrogram showing the relationships between the sequences, as determined using ClustalW (1). The next portion of the viewer displays the ClustalW alignment itself, with a control menu. The default display option is to include all available fungal protein sequences in the alignment. Colored highlighting provides easy visualization of the degree of sequence conservation: amino acid residues identical in all sequences are highlighted in yellow; those sharing strong similarity in pink; and those sharing weak similarity in green. All sequences are displayed at the bottom of the page. Each is also available on an individual page, in either FASTA or GCG format, by clicking on the name of that sequence in the alignment.
Figure 1.
The Fungal Alignment Viewer. The Fungal Alignment Viewer displays available fungal sequences related to an S.cerevisiae sequence of interest. At the top of the display is a dendrogram showing the relationships between all included sequences. The available sequences are aligned via ClustalW with color coding to indicate conservation (yellow = identical, pink = strong similarity, green = weak similarity). Above the ClustalW alignment is a control menu that allows selection of sequences and type of alignment (protein, ORF DNA, upstream sequence, downstream sequence, ORF DNA + 1 kb up/downstream). When the ‘Align’ button is pressed, the alignment is recomputed according to the user’s selections. Individual sequences present in the alignment are displayed below the ClustalW alignment (not shown here). In the alignment shown, the partial sequence from S.mikatae, available from the sequence selection menu, has not been included so that sequence similarity between the C-terminal portions of the other available sequences can be visualized by ClustalW.
The selection control menu above the alignment allows the user to regenerate the alignment using custom parameters. The left side of the menu allows selection from the available sequences, while the right side of the menu allows selection of various alignment types: protein, DNA from the open reading frame (ORF), the upstream flanking DNA, the downstream flanking DNA, or the ORF DNA plus upstream and downstream flanking regions. This provides a quick and easy way to customize the alignment for specific sequences or regions of interest, or to remove partial sequences from the alignment.
Currently the Fungal Alignment Viewer provides access to sequences from several other Saccharomyces genomes, including those of four closely related sensu stricto species (Saccharomyces paradoxus, Saccharomyces mikatae, Saccharomyces kudriavzevii and Saccharomyces bayanus) and two more distantly related sensu lato species (Saccharomyces castellii and Saccharomyces kluyveri). The genomes of S.paradoxus, S.mikatae and S.bayanus were sequenced at 6–8× coverage by the Fungal Genome Initiative at the Whitehead Institute (2). The genomes of S.mikatae, S.bayanus, S.kudriavzevii, S.castellii and S.kluyveri were sequenced at 2–4× coverage by Mark Johnston’s group at Washington University (3). The availability of sequence data from the closely related sensu stricto species allows the evaluation of conservation in non-protein coding features such as regulatory elements, while data from the more distantly related sensu lato species are valuable in the analysis of sequence conservation in protein sequences and domains. The Fungal Alignment Viewer will be a dynamic resource not limited to the data currently available. Current plans include the addition of sequences from Schizosaccharomyces pombe, Candida albicans and other fungi as sequence information becomes available.
An additional tool, the Synteny Viewer (not shown; also accessible from the Comparison Resources pulldown menu), visualizes the conserved order of genes amongst the three sensu stricto species sequenced by the Fungal Genome Initiative (2), indicating orthologs and paralogs where determined, and showing the contigs from each sequenced genome aligned with the chromosomal sequence of S.cerevisiae, as held by SGD. These fairly complete genomes from species closely related to S.cerevisiae provide detailed information about individual gene sequences and about genome organization and evolution as a whole. Individual features shown for the sensu stricto species are hyperlinked to the Fungal Alignment Viewer when related sequences are available.
This combination of sequence data from both sensu stricto and sensu lato Saccharomyces species is a powerful resource for investigation of the Saccharomyces genome, providing information about genome organization as well as identifying residues and regions crucial for the function of specific proteins. The Fungal Alignment Viewer and the Synteny Viewer provide users with versatile web interfaces for analyzing these data.
SEQUENCE SIMILARITY QUERY TOOL
The Sequence Similarity Query tool displays sequences from any species that are related to a given protein from S.cerevisiae. Each protein sequence in SGD is used as a query for the PSI-BLAST (Position-Specific Iterated BLAST) program (4) against the most recent version of the non-redundant (nr) protein sequence data set (5) at NCBI (6). This data set contains the sum of all unique entries from all species for which sequence data exist, compiled from the Swiss-Prot, PIR, PDB and GenPept Databases. The PSI-BLAST program identifies families of related proteins using an iterative BLAST procedure that groups related hits into families. An initial pass using BLAST identifies the strongest hits to the query sequence. Subsequent iterations use information from both the initial query sequence and all accumulated hits to generate a query for BLAST to find additional related sequences. The process can be allowed to run until no new additional sequences are identified. This iterative procedure often finds protein relationships that are not identified in single-pass methods (4), making it highly effective for identifying protein families. In the first run of this analysis at SGD, ∼60% of S.cerevisiae proteins identified similar proteins from Homo sapiens, Mus musculus, Arabidopsis thaliana, S.pombe, Drosophila melanogaster, Caenorhabditis elegans and Neurospora crassa. The data in this tool will be updated quarterly by running PSI-BLAST against NCBI’s most up-to-date version of the nr protein sequence database.
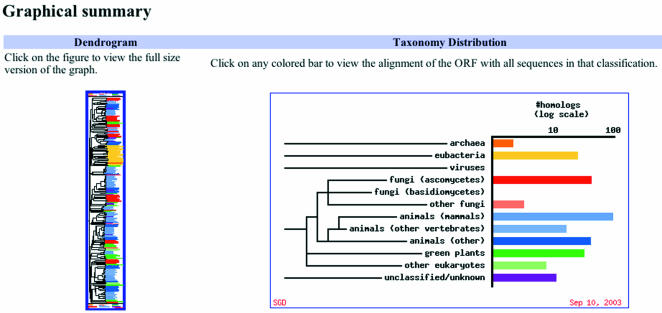
Accessible from the Comparison Resources pulldown menu in the right-hand column of the Locus Page, the Sequence Similarity Query resource produces a graphical summary of results (Fig. 2), as well as options for generating alignments for any sequence with more than one hit in the nr protein data set. The taxonomic distribution of all hits is presented as a color-coded bar graph with the following major taxonomic categories: Archaea, Eubacteria, Viruses, Fungi (Ascomycetes), Fungi (Basidiomycetes), Fungi (other), Animals (mammals), Animals (other vertebrates), Animals (other), Green plants, Other eukaryotes and Unclassified/unknown. The relationships between the sequences, including the initial query sequence, are diagrammed in a dendrogram with the same color coding as that in the bar graph.
Figure 2.
The Sequence Similarity Query Tool. In the graphical summary of the Sequence Similarity Query tool, a dendrogram shows the relationships between the sequences in a protein family. A bar graph shows the distribution of members of the protein family within broad taxonomic groupings. The color coding used in the bar graph is also used for the names of the sequences within the dendrogram to indicate the taxonomic grouping.
The Sequence Similarity Query resource provides multiple options for viewing alignments of hits to the query sequence. In the ‘Align by Best Hits’ section, one can select sequences of interest from the top 10 hits (i.e. lowest E-values). The ‘Align by Species’ section allows one or more groups of sequences to be selected by species. Both sections provide options for constructing either a multiple alignment of all selected sequences or pairwise alignments of each sequence against the query from S.cerevisiae. Pages displaying the alignment of selected sequences include links to download the GenBank accession numbers of all sequences included.
OTHER NEW TOOLS
Two other major new features at SGD provide new types of information about S.cerevisiae genes and proteins, and facilitate more detailed searches of the database. The Metabolic Pathways tool visualizes S.cerevisiae metabolic pathways and the genes involved in them. The Chromosomal Features Search is a versatile tool that provides many advanced options for querying the database.
Using the MetaCyc Database of metabolic reactions (7) and the Pathway Tools software created by Peter Karp’s group (8), SGD has generated metabolic pathway information for S.cerevisiae. Metabolic pathways are represented graphically and can be viewed at multiple levels of detail, from general summaries to detailed diagrams showing the chemical structures of each compound. Enzymatic activities shown on each pathway diagram are linked to the SGD Locus Pages of the proteins involved, and conversely, SGD Locus Pages are linked to all relevant pathway diagrams. The pathways are being curated at SGD to provide an up-to-date resource specific to the metabolic pathways present in S.cerevisiae.
The Find Chromosomal Features search provides advanced options for finding a gene or genes based on one or more criteria, including feature type (e.g. ORF, tRNA); protein molecular weight, length or pI; chromosomal location; the presence of introns; and associated Gene Ontology terms (9,10). The results page indicates numbers of hits for each step of the query to help the user refine the query, if necessary, and displays all results in a tabular form with links to the individual Locus Pages of all genes that satisfy the query. The list of results can be analyzed directly with other tools, including the GO Term Finder, the GO Term Mapper or Expression Connection, or it can be downloaded as a tab-delimited file.
SUMMARY
SGD has developed several new tools to increase ease of access to information about genes and proteins from S.cerevisiae and related proteins of other species. Two new tools provide information about related sequences. The Fungal Alignment Viewer displays alignments of S.cerevisiae protein or DNA sequences with those of other fungi. The Sequence Similarity Query tool provides information about all currently available protein sequences similar to a given protein sequence from S.cerevisiae. Visualization of metabolic pathways has been added, providing a unified source of information about the biochemical pathways in yeast. Finally, the Find Chromosomal Features search provides greater search capabilities of the basic gene and protein information already available at SGD. These tools will facilitate the use of S.cerevisiae as a model organism and reference for comparison with other species.
REFERENCES
- 1.Chenna R., Sugawara, H., Koike,T., Lopez,R., Gibson,T.J., Higgins,D.G. and Thompson,J.D. (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res., 31, 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kellis M., Patterson,N., Endrizzi,M., Birren,B. and Lander,E.S. (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature, 423, 241–254. [DOI] [PubMed] [Google Scholar]
- 3.Cliften P., Sudarsanam,P., Desikan,A., Fulton,L., Fulton,B., Majors,J., Waterston,R., Cohen,B.A. and Johnston,M. (2003) Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science, 301, 71–76. [DOI] [PubMed] [Google Scholar]
- 4.Altschul S.F. and Koonin,E.V. (1998) Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem. Sci., 23, 444–447. [DOI] [PubMed] [Google Scholar]
- 5.Altschul S.F., Boguski,M.S., Gish,W. and Wootton,J.C. (1994) Issues in searching molecular sequence databases. Nature Genet., 6, 119–129. [DOI] [PubMed] [Google Scholar]
- 6.Wheeler D.L., Church,D.M., Federhen,S., Lash,A.E., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Sequeira,E., Tatusova,T.A. and Wagner L. (2003) Database resources of the National Center for Biotechnology. Nucleic Acids Res., 31, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karp P.D., Riley,M., Paley,S.M. and Pellegrini-Toole,A. (2002) The MetaCyc Database. Nucleic Acids Res., 30, 59–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karp P.D., Paley,S. and Romero,P. (2002) The Pathway Tools software. Bioinformatics, 18, S225–S232. [DOI] [PubMed] [Google Scholar]
- 9.The Gene Ontology Consortium. (2001) Creating the gene ontology resource: design and implementation. Genome Res., 11, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dwight S.S., Harris,M.A., Dolinski,K., Ball, C.A., Binkley,G., Christie,K.R., Fisk,D.G., Issel-Tarver,L., Schroeder,M., Sherlock,G., Sethuraman,A., Weng,S., Botstein,D. and Cherry,J.M. (2002) Saccharomyces Genome Database (SGD) provides secondary gene annotation using the Gene Ontology (GO). Nucleic Acids Res., 30, 69–72. [DOI] [PMC free article] [PubMed] [Google Scholar]