Abstract
The Gene Ontology (GO) project (http://www.geneontology.org/) provides structured, controlled vocabularies and classifications that cover several domains of molecular and cellular biology and are freely available for community use in the annotation of genes, gene products and sequences. Many model organism databases and genome annotation groups use the GO and contribute their annotation sets to the GO resource. The GO database integrates the vocabularies and contributed annotations and provides full access to this information in several formats. Members of the GO Consortium continually work collectively, involving outside experts as needed, to expand and update the GO vocabularies. The GO Web resource also provides access to extensive documentation about the GO project and links to applications that use GO data for functional analyses.
INTRODUCTION
The era of genome-scale biology has seen the accumulation of vast amounts of biological data, accompanied by the widespread proliferation of biology-oriented databases. To make the best use of biological databases and the knowledge they contain, different kinds of information from different sources must be integrated in ways that make sense to biologists.
A major component of the integration effort is the development and use of annotation standards such as ontologies (1–4). Ontologies provide conceptualizations of domains of knowledge and facilitate both communication between researchers and the use of domain knowledge by computers for multiple purposes.
The Gene Ontology (GO) project is a collaborative effort to address two aspects of information integration: providing consistent descriptors for gene products, in different databases; and standardizing classifications for sequences and sequence features. The project began in 1998 as a collaboration between three model organism databases: FlyBase (Drosophila), the Saccharomyces Genome Database (SGD) and the Mouse Genome Informatics (MGI) project. Since then, the GO Consortium has grown to include many databases, including several of the world’s major repositories for plant, animal and microbial genomes (a current list of member organizations is included as Supplementary Material).
THE GO PROJECT
The GO project has three major goals: (i) to develop a set of controlled, structured vocabularies—known as ontologies—to describe key domains of molecular biology, including gene product attributes and biological sequences; (ii) to apply GO terms in the annotation of sequences, genes or gene products in biological databases; and (iii) to provide a centralized public resource allowing universal access to the ontologies, annotation data sets and software tools developed for use with GO data.
Ontologies
The GO project provides ontologies to describe attributes of gene products in three non-overlapping domains of molecular biology. Within each ontology, terms have free text definitions and stable unique identifiers. The vocabularies are structured in a classification that supports ‘is-a’ and ‘part-of’ relationships. The scope and structure of the GO vocabularies are described in more detail in references (5–7). In the current research environment, where new genome sequences are being rapidly generated, and where comparative genome analysis requires the integration of data from multiple sources, it is especially germane to provide rigorous ontologies that can be shared by the community.
Molecular Function (MF) describes activities, such as catalytic or binding activities, at the molecular level. GO molecular function terms represent activities rather than the entities (molecules or complexes) that perform the actions, and do not specify where, when or in what context the action takes place. Examples of individual molecular function terms are the broad concept ‘kinase activity’ and the more specific ‘6-phosphofructokinase activity’, which represents a subtype of kinase activity.
Biological Process (BP) describes biological goals accomplished by one or more ordered assemblies of molecular functions. High-level processes such as ‘cell death’ can have both subtypes, such as ‘apoptosis’, and subprocesses, such as ‘apoptotic chromosome condensation’.
Cellular Component (CC) describes locations, at the levels of subcellular structures and macromolecular complexes. Examples of cellular components include ‘nuclear inner membrane’, with the synonym ‘inner envelope’, and the ‘ubiquitin ligase complex’, with several subtypes of these complexes represented.
The recent development of the Sequence Ontology (SO) permits the classification and standard representation of sequence features. Defined sequence features include terms such as ‘exon’, whose meaning is widely accepted, and the more problematic term ‘pseudogene’, for which several different usages have yet to be resolved. Although the SO is a relatively new vocabulary, and is still undergoing refinement, it is already being used for genome annotation projects in Drosophila and Caenorhabditis elegans.
Annotations
Collaborating databases provide data sets comprising links between database objects and GO terms, with supporting documentation. Every annotation must be attributed to a source, which may be a literature reference, another database or a computational analysis; furthermore, the annotation must indicate the type of evidence the cited source provides to support the association between the gene product and the GO term. A standard set of evidence codes qualifies annotations with respect to different types of experimental determinations. For example, a direct assay to determine the function of the exact gene product being annotated is more reliable than a sequence architecture comparison.
High-quality GO annotations, normally based on curatorial review of published literature and supported by experimental evidence, are now available for gene products in many model organisms. In addition, large sets of annotations made using automated methods cover both model organisms and less experimentally tractable organisms, including human. A number of different automatic methods have been applied (e.g. 8–12), all of which are represented by the evidence code IEA (‘inferred from electronic annotation’). Table 1 provides a snapshot of current annotations in the GO database; a more detailed table is maintained on the web at http://www.geneontology.org/doc/GO.current.annotations.shtml. Additional information on GO annotations can be found in references (5–8) and (13).
Table 1. Status of the GO vocabularies.
| Totals | July 1, 2000 | July 1, 2003 |
|---|---|---|
| All valid termsa | 4493 | 13412 |
| Terms with definitions | 250 | 11105 |
| Terms with synonyms | 301 | 2813 |
| Terms with db cross-references | 1042 | 12317 |
| Associationsb | 30654 | 7781954 |
| Gene products | 13016 | 1549236 |
| Sequences | 0 | 21916 |
| Pathsc | 30941 | 314886 |
aExcludes obsolete terms.
bIndividual associations between any gene product and any GO term.
cParent–child relationships traced from any GO term to the root (molecular function, biological process or cellular component).
The SO is being used by the collaborating databases for genomic feature annotation. Like GO annotations, SO annotations are curated using both manual work by experts and purely computational methodologies.
GO slims
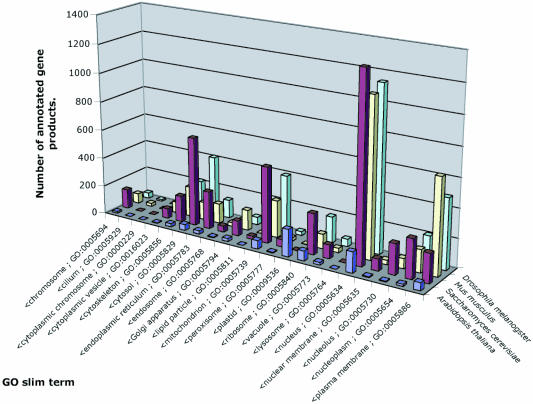
For many purposes, in particular reporting the results of GO annotation of a genome or cDNA collection, it is very useful to have a high-level view of each of the three ontologies. These subsets of the GO have become known as ‘GO slims’, the first of which was constructed for the annotation of the Drosophila genome (13). An example of a GO slim analysis is shown in Figure 1.
Figure 1.
Application of a GO slim set in genome annotation. The number of gene products annotated to each term in each of four model organism genomes is shown for a GO slim set taken from the cellular component ontology (data as of August 1, 2003).
The shared use of GO slims makes comparisons of summary GO term distributions very easy. Different applications, however, may require different GO slim sets tailored to the specific needs of an analysis. To address this, the GO Consortium makes both generic and specific GO slim files available. The generic GO slim file is kept up to date with respect to the full ontologies, and specific GO slim files that have been used in particular publications or analyses are archived.
THE GO DATABASE
The GO database consists of a MySQL database that captures GO content and a Perl object model and Application Programmer Interface (API) to simplify database access and help programmers write tools that use the GO data. The GO relational database is released monthly in several versions: termdb includes the ontologies, definitions and cross-references to other databases; assocdb includes all data in termdb plus associations to gene products; and seqdb adds protein sequences for annotated gene products (where available). A fourth version, seqdblite, is equivalent to seqdb without the IEA-based associations; this version is used by the AmiGO browser (see below).
The GO database schema models generic graphs, including the GO structure (a directed acyclic graph, or DAG) relationally. At the core of the schema are two relational tables for capturing all terms (also called nodes) and term–term relationships (arcs). The two relationship types, ‘is-a’ and ‘part-of,’ are represented as a ‘relationship type’ attribute in the relationship table.
GO RESOURCES
Access to ontologies and annotations in all formats
The output of the GO project—vocabularies, annotations, database and accompanying tools—are in the public domain and are readily accessible via the GO web pages at http://www.geneontology.org/. The GO Consortium gives permission for any of its products to be used without license, in accordance with its redistribution and citation policy. Highlights of that policy are:
(i) that the Gene Ontology Consortium is clearly acknowledged as the source of the product;
(ii) that any GO Consortium file(s) displayed publicly include the revision number(s) and/or date(s) of the relevant GO file(s);
(iii) that neither the content of a GO file(s) nor the logical relationships embedded within the GO file(s) be altered in any way.
The full GO Redistribution and Citation Policy document is available online at http://www.geneontology.org/doc/GO.cite.html. A list of useful URLs and addresses is included in the Supplementary Material.
The MySQL database described above can be downloaded locally, and Perl APIs are provided. The GO Consortium’s ontologies and annotations are also available as flat files (the most frequently updated format at the time of writing) and as RDF XML; the latter is available with or without annotation data included. The MySQL and XML formats are released monthly. The flat files are updated continally, and monthly snapshots are archived. Current and archival releases of all three formats can be downloaded from the GO web site.
Documentation
The GO web resource includes an extensive set of documentation pages (see http://www.geneontology.org/doc/GO.contents.doc.html). Topics include an overview of the GO project and the ontologies, guides to editorial style, file formats and annotation practices, and frequently asked questions (FAQ).
Software/tools
A variety of browsers that provide visualization and query capabilities for the GO are available. For example, the AmiGO browser (developed by the GO software group at Berkeley; see http://www.godatabase.org/cgi-bin/go.cgi) provides a web interface for searching and displaying the ontologies, term definitions and associated annotated gene products for the entire spectrum of contributing organism databases represented in the GO database. AmiGO easily allows users to browse a tree-like view of the GO structure and to search for terms using a variety of different keys such as a name, synonym, definition, numerical identifier or cross-referenced entry in an external database. The summary view presents the list of gene products associated with each term. The results may be constrained by the evidence code used in the association or by the organization that submitted the association. Representative amino acid sequences are available for most genes, and these can be selected and downloaded as FASTA files. Using GOst, the GO BLAST server, users may submit a query sequence and retrieve the sequences and GO annotations of all similar gene products in the GO database.
The GO software group has also developed DAG-Edit, a tool that provides a graphical interface to browse, query and edit GO or any other vocabulary that has a DAG data structure. GO curators use DAG-Edit to manage the GO vocabularies. The tool has also been used by other groups to build ontologies for a wide range of biological subjects, such as anatomies and developmental timelines for several model organisms, human diseases and plant growth environment. DAG-Edit is an open source Java application that is installed locally. A user guide is available within the application and on the web (http://www.geneontology.org/doc/dagedit_userguide/dagedit.html).
DAG-Edit is updated regularly to add features and improve performance; the current version can be downloaded from http://sourceforge.net/project/showfiles.php?group_id=36855.
The GO Software web page (http://www.geneontology.org/doc/GO.tools.html) provides a catalogue of GO-related tools developed by members of the GO Consortium or by GO users. In addition to AmiGO, there are several more applications for browsing and searching the GO vocabularies and annotations. Other available software includes applications for correlating data from the GO project and other sources (including, but not limited to, microarray data), as well as tools that are not specific to, but can be used in conjunction with, GO data.
Other resources
Literature collection. The GO project maintains a bibliography of peer-reviewed publications (124 as of August 2003) relevant to the development and use of the GO vocabularies and annotation sets at http://www.geneontology.org/doc/GO.biblio.html. Many of the publications document the curation and display of GO annotations within a wide variety of databases, whereas others make use of GO terms and gene product annotations in the interpretation of large-scale experimental results. Still other papers describe novel uses of GO terms (e.g. in text mining), software that uses GO data and integration of the GO with other ontological resources.
Community input. The GO effort is greatly enriched by input from its user community. Several routes are available for users to comment on various aspects of the GO. Comments and suggestions for changes and updates to the ontologies can be submitted via a GO project page at the SourceForge site (http://sourceforge.net/projects/geneontology), whereupon each suggestion is evaluated by GO Consortium members. Different ‘trackers’ available from the SourceForge site allow GO users to report problems or request features for the AmiGO browser, and to submit suggestions for additions and changes to the ontologies; items can be assigned to individuals or groups within the GO Consortium who have relevant expertise. This system allows the submitter to track the status of a suggestion, both online and by email, allows other users to see what changes are currently under consideration, and archives all entries and associated communications.
Mailing lists. GO also has several mailing lists, covering general questions and comments, the GO database and software, and summaries of changes to the ontologies. The lists are described at http://www.geneontology.org/GO_contacts.html. Any questions about contributing to the GO project should be directed to the main GO mailing list at go@geneontology.org.
SUMMARY
The GO project provides an ongoing example of community development of bioinformatics standards. Combining the expertise of biologists from multiple sub-disciplines, the computational expertise of artificial intelligence researchers, and input from multiple users of the system, the GO Consortium continues to develop and expand these classification systems for molecular biology.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The Gene Ontology Consortium is supported by NIH/NHGRI grant HG02273, and by grants from the European Union RTD Programme ‘Quality of Life and Management of Living Resources’ (QLRI-CT-2001-00981 and QLRI-CT-2001-00015).
REFERENCES
- 1.Gruber T.R. (1993) A translational approach to portable ontologies. Knowl. Acq., 5, 199–220. [Google Scholar]
- 2.Jones D.M. and Paton,R.C. (1999) Toward principles for the representation of hierarchical knowledge in formal ontologies. Data Knowl. Eng., 31, 102–105. [Google Scholar]
- 3.Schulze-Kremer S. (1998) Ontologies for molecular biology. Pac. Symp. Biocomput., 3, 695–706. [PubMed] [Google Scholar]
- 4.Stevens R., Goble,C.A., and Bechhofer.S. (2000) Ontology-based knowledge representation for bioinformatics. Brief. Bioinform., 1, 398–414. [DOI] [PubMed] [Google Scholar]
- 5.Blake J.A. and Harris,M. (2003) The Gene Ontology Project: Structured vocabularies for molecular biology and their application to genome and expression analysis. In Baxevanis,A.D., Davison,D.B., Page,R., Stormo,G. and Stein,L. (eds), Current Protocols in Bioinformatics. Wiley and Sons, Inc., New York. [Google Scholar]
- 6.The Gene Ontology Consortium (2001) Creating the gene ontology resource: design and implementation. Genome Res., 11, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Gene Ontology Consortium (2000) Gene Ontology: tool for the unification of biology. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camon E., Magrane,M., Barrell,D., Binns,D., Fleischmann,W., Kersey,P., Mulder,N., Oinn,T., Maslen,J., Cox,A. et al. (2003) The Gene Ontology Annotation (GOA) Project: Implementation of GO in SWISS-PROT, TrEMBL, and InterPro. Genome Res., 13, 662–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mi H., Vandergriff,J., Campbell,M., Narechania,A., Majoros,W., Lewis,S., Thomas,P.D. and Ashburner,M. (2003) Assessment of genome-wide protein function classification for Drosophila melanogaster. Genome Res., 13, 2118–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pouliot Y., Gao,J., Su,Q.J., Liu,G.G. and Ling,X.B. (2001) DIAN: a novel algorithm for genome ontological classification. Genome Res., 11, 1766–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okazaki Y., Furuno,M., Kasukawa,T., Adachi,J., Bono,H., Kondo,S., Nikaido,I., Osato,N., Saito,R. and Suzuki,H. et al. (2002) Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature, 420, 563–573. [DOI] [PubMed] [Google Scholar]
- 12.Xie H., Wasserman,A., Levine,Z., Novik,A., Grebinskiy,V., Shoshan,A. and Mintz,L. (2002) Large scale protein annotation through Gene Ontology. Genome Res., 12, 785–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams M.D., Celniker,S.E., Holt,R.A., Evans,C.A., Gocayne,J.D., Amanatides,P.G., Scherer,S.E., Li,P.W., Hoskins,R.A., Galle,R.F. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.