Abstract
IMGT/GeneInfo is a user-friendly online information system that provides information on data resulting from the complex mechanisms of immunoglobulin (IG) and T cell receptor (TR) V(D)J recombinations. For the first time, it is possible to visualize all the rearrangement parameters on a single page. IMGT/GeneInfo is part of the international ImMunoGeneTics information system® (IMGT), a high-quality integrated knowledge resource specializing in IG, TR, major histocompatibility complex (MHC), and related proteins of the immune system of human and other vertebrate species. The IMGT/GeneInfo system was developed by the TIMC and ICH laboratories (with the collaboration of LIGM), and is the first example of an external system being incorporated into IMGT. In this paper, we report the first part of this work. IMGT/GeneInfo_TR deals with the human and mouse TRA/TRD and TRB loci of the TR. Data handling and visualization are complementary to the current data and tools in IMGT, and will subsequently allow the modelling of V(D)J gene use, and thus, to predict non-standard recombination profiles which may eventually be found in conditions such as leukaemias or lymphomas. Access to IMGT/GeneInfo is free and can be found at http://imgt.cines.fr/GeneInfo.
INTRODUCTION
The synthesis of the antigen receptors [immunoglobulins (IG) and T cell receptors (TR)] is complex and unique due to DNA molecular rearrangements in multiple loci, located on different chromosomes (1,2). This led to the creation in 1989 of the international ImMunoGeneTics information system® (‘IMGT’); a high-quality integrated knowledge resource specializing in IG, TR, major histocompatibility complex (MHC), and related proteins of the immune system of human and other vertebrate species (3). In vertebrates, the four TR loci, TRA, TRB, TRG and TRD, comprise variable (V), diversity (D) (for the TRB and TRD loci) and joining (J) genes, which rearrange in a combinatorial V(D)J way in order to encode, with a constant C gene, the α, β, γ and δ chains, respectively. The TRA/TRD locus organization is even more complex since the TRD locus is nestled within the TRA locus (2,4–6). The loci are shown in more detail in Table 1 (7). The human TRA locus spans 1000 kb and comprises 54 TRAV and 61 TRAJ (2), whereas the mouse TRA locus spans 1550 kb and comprises 98 TRAV and 60 TRAJ (6). Consequently, extensive work will be required to analyse all the possible TRA V-J combinations: 3294 (54 × 61) in human (2) and 5880 (98 × 60) in mouse (6). The TRB locus spans 620 kb in human and 700 kb in mouse, and comprises 67 and 35 TRBV genes, respectively, and two TRBD and 14 TRBJ genes [(2), and IMGT Repertoire http://imgt.cines.fr]. Analysis of the TRB loci will require the study of 1876 (67 × 2 × 14) and 980 (35 × 2 × 14) different TRB V-D-J combinations, respectively. The IMGT/GeneInfo information system is intended to give user-friendly and intuitive access to V(D)J recombination data in immunology. This information is complementary to that given in the IMGT/GENE-DB database, and the IMGT/GeneSearch, IMGT/GeneView and IMGT/LocusView tools (3). It is worth noting that IMGT/GeneInfo, developed by TIMC and ICH (also in collaboration with LIGM) is the first example of an external system being incorporated into IMGT. In this paper, we report the first part of this work: IMGT/GeneInfo_TR, which deals with human and mouse TRA/TRD and TRB loci. The IMGT/GeneInfo information system allows researchers working on VDJ recombination not only to decrease the work time on genomic analysis, but also to avoid the possibility of sequence errors, when V, D and J genes are manually extracted from raw data of up to 1550 kb loci. Results are obtained after a simple two-step process, allowing a practical visualization of all the rearrangement parameters within the same page: gene names, functionality, recombination signal (RS) sequences, locus positions, and sequences of exons and introns.
Table 1. T cell receptor V(D)J genes in IMGT/GeneInfo.
| TR V(D)J loci | TRDV | TRAV | TRAJ | TRBV | TRBD | TRBJ |
|---|---|---|---|---|---|---|
| Human | ||||||
| Total no. | 3 | 54 | 61 | 67 | 2 | 14 |
| Functional | 45 | 50 | 47 | 2 | 13 | |
| ORF | 1 | 8 | 6 | 0 | 1 | |
| Pseudo | 8 | 3 | 14 | 0 | 0 | |
| Locus size (kb) | TRAD: 1000 | TRB: 620 | ||||
| Sources | TRAD: AE000658–AE000662 | TRB: L36092 | ||||
| Mouse | ||||||
| Total no. | 6 | 98 | 60 | 35 | 2 | 14 |
| Functional | 79 | 38 | 21 | 2 | 11 | |
| ORF | 5 | 12 | 1 | 0 | 2 | |
| Pseudo | 14 | 10 | 13 | 0 | 1 | |
| Locus size (kb) | TRAD: 1550 | TRB: 700 | ||||
| Sources | TRAD: AE008683–AE008686 | TRB: AE00063–AE00065 | ||||
Sources: IMGT/LIGM-DB and GenBank.
MATERIALS AND METHODS
IMGT/GeneInfo data extraction
The following references (from GenBank and IMGT/LIGM-DB) were used for data extraction: human (Homo sapiens) TRA/TRD (AE000658–AE000662) and TRB (L36092) loci, and mouse (Mus musculus) TRA/TRD (AE008683–AE008686) and TRB (AE00063, AE00064, AE00065) loci. Extracted data included the following information for each V, D and J gene: its functionality (functional, pseudogene, ORF), positions of the first and last nucleotide for the gene, V-intron and exon(s) and for the three parts of the recombination signals RS (heptamer, spacer, nonamer). The positions of the V, D and J genes in the TRA/TRD and TRB loci were determined from the first nucleotide of the TRAC and TRBC2 genes, respectively. Data manually extracted from the files were collected for each gene of the six loci. A program automatically extracts nucleotide sequences using the positions of the various elements [gene, V-intron, exon(s), heptamer, spacer, nonamer].
IMGT/GeneInfo query
IMGT/GeneInfo is currently available for the TRA/TRD and TRB loci of human and mouse. The IMGT/GeneInfo query is a two-step process.
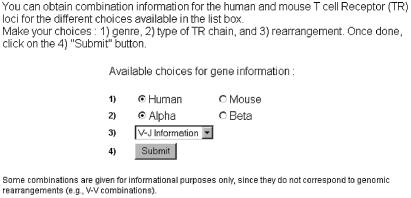
Step one: on the first page (Fig. 1), the user selects the species (human or mouse), the locus TRA/TRD (α) or TRB (β) and the gene combinations (V-V, V-J, V-D-J). Some combinations are given for informational purposes only, since they do not correspond to genomic rearrangements (e.g. V-V combinations).
Figure 1.
IMGT/GeneInfo query page.
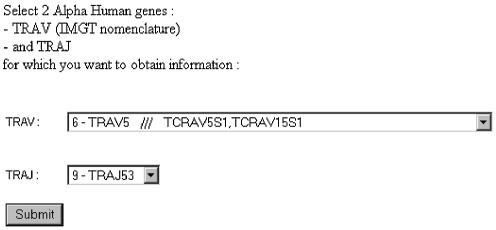
Step two: The second page is generated automatically, and the user then chooses the genes (V, D, J) for which information is required (Fig. 2). Gene choice can be made either according to the gene name [official IMGT nomenclature or previous ones (2,6)], or the relative position of the gene within the locus (e.g. on the TRA locus, position number 1 for the V gene is the most in 5′, and position 1 for the J gene is the most in 3′). All combinations are available, for example, TRAV5 and TRAJ53 (Fig. 2).
Figure 2.
IMGT/GeneInfo gene choice page.
IMGT/GeneInfo results
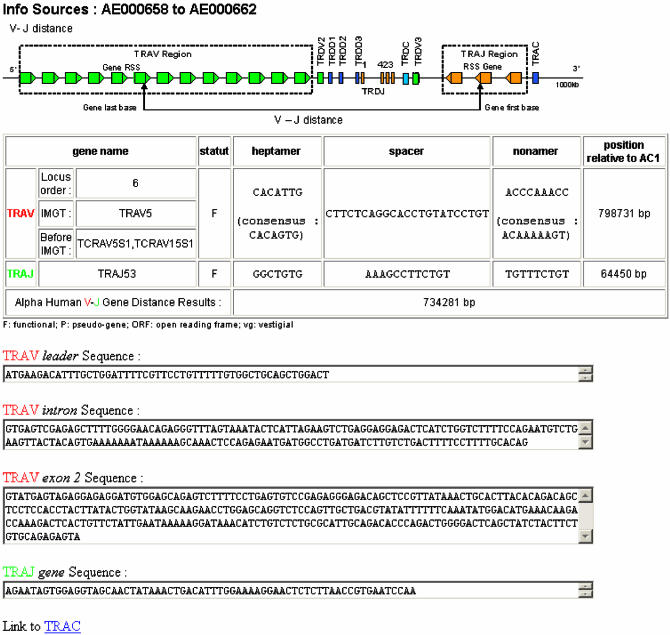
The IMGT/GeneInfo results page is divided into four parts. Reading from top to bottom: Part one is the source from which information was collected (e.g. AE000658 for the human TRA/TRD locus). Part two is an image that corresponds to the selected combination of genes and that explains visually which gene types are concerned, how the genes and the RS are oriented, and how distances between genes were computed. Part three is a table that contains a summary, for each gene, with the gene name, the functionality (functional, pseudogene, ORF) and the nucleotide sequences for each RS part (heptamer, spacer, nonamer). It also contains the corresponding consensus sequence when it exists; the position relative to TRAC for the TRA/TRD loci and to TRBC2 for the TRB loci; and the genomic distance in base pairs between the genes of the selected combination, in their germline configuration. Part four corresponds to the sequences of the gene and, for a V gene, to its various parts (leader, V-intron, exon 2). These sequences can be selected for copy and paste. A colour code is associated with all information originating from the same gene to make it easier to see and remember. A link is provided to the constant gene (e.g. TRAC) from which distances are calculated.
Implementation
IMGT/GeneInfo is deployed in the IMGT information system using Java Servlet technology. The interface uses HTML, JavaScript and CSS.
DISCUSSION AND CONCLUSION
Large genome sequencing allows us to analyse complex loci over few hundred kilobases and to accurately determine their regulation mechanisms. However, raw data utilization in all genetic fields is difficult, and needs a substantial background expertise. This complexity is greatly increased in the IG and TR loci, because of the potential rearrangements of any given V, D and J gene (5). To date, immunologists working on these loci need to manually copy and paste all the potential combinations from sequence databases. The system presented here is the fruit of a collaboration between three laboratories offering complementary backgrounds in immunology, genomics and biocomputing. The IMGT/GeneInfo system allows researchers who work on V(D)J recombinations to greatly decrease the genomic work time as well as to avoid the possibility of sequence errors, working on loci manually shortened to 1550 kb rather than on large raw data. Only two steps are needed to obtain all rearrangement parameters (i.e. gene names, functionality, gene positions, RS, exon and V-intron sequences). The IMGT/GeneInfo information system facilitates easy data archiving. Moreover, because of its ease of use, we expect that this information system will be used as a teaching tool on V(D)J recombination mechanisms.
CITING IMGT/GeneInfo
Authors who use IMGT/GeneInfo are strongly encouraged to cite this article and the IMGT/GeneInfo home page URL, at http://imgt.cines.fr/GeneInfo.
ACCESS AND CONTACT
IMGT/GeneInfo home page: http://imgt.cines.fr/GeneInfo
IMGT/GeneInfo Contact: tpbaum@imag.fr
IMGT home page: http://imgt.cines.fr
IMGT contact: lefranc@ligm.igh.cnrs.fr
TIMC contact: tpbaum@imag.fr
ICH contact: patrice.marche@cea.fr
LIGM contact: lefranc@ligm.igh.cnrs.fr
Figure 3.
IMGT/GeneInfo results page.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Matthew U’Ren-Gerente for his help editing in English. IMGT/GeneInfo is funded by institutional grants from the Institut National de la Recherche Médicale (INSERM), the Commissariat à l’Energie Atomique (CEA) and a specific grant from ‘Thématiques Prioritaires de la Région Rhône-Alpes’. The IMGT is funded by the EU 5th PCRDT (QLG2-2000-01287) programme, the Centre National de la Recherche Scientifique (CNRS), and the Ministère de la Recherche et de l’Education Nationale.
REFERENCES
- 1.Lefranc M.-P. and Lefranc,G. (2001) The Immunoglobulin FactsBook. Academic Press, London, UK, 458 pp. [Google Scholar]
- 2.Lefranc M.-P. and Lefranc,G. (2001) The T Cell Receptor FactsBook. Academic Press, London, UK, 398 pp. [Google Scholar]
- 3.Lefranc M.-P. (2003) IMGT, the international ImMunoGeneTics database®, http://imgt.cines.fr. Nucleic Acids Res., 31, 307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glusman G., Rowen,L., Lee,I., Boysen,C., Roach,J.C., Smit,A.F., Wang,K., Koop,B.F. and Hood,L. (2001) Comparative genomics of the human and mouse T cell receptor loci. Immunity, 15, 337–349. [DOI] [PubMed] [Google Scholar]
- 5.Pasqual N., Gallagher,M., Aude-Garcia,C., Loiodice,M., Thuderoz,F., Demongeot,J., Ceredig,R., Marche,P. and Jouvin-Marche,E. (2002) Quantitative and qualitative changes in V–J α rearrangements during mouse thymocytes differentiation: implication for a limited T cell receptor α chain repertoire. J. Exp. Med., 196, 1163–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosc N. and Lefranc,M.-P. (2003) The mouse (Mus musculus) T cell receptor α (TRA) and δ (TRD) variable genes. Dev. Comp. Immunol., 27, 465–497. [DOI] [PubMed] [Google Scholar]
- 7.Gallagher M., Obeïd,P., Marche,P.N. and Jouvin-Marche,E. (2001) Both TCRα and TCRδ chain diversity are regulated during thymic ontogeny. J. Immunol., 167, 1447–1453. [DOI] [PubMed] [Google Scholar]