Abstract
MitoProteome is an object-relational mitochondrial protein sequence database and annotation system. The initial release contains 847 human mitochondrial protein sequences, derived from public sequence databases and mass spectrometric analysis of highly purified human heart mitochondria. Each sequence is manually annotated with primary function, subfunction and subcellular location, and extensively annotated in an automated process with data extracted from external databases, including gene information from LocusLink and Ensembl; disease information from OMIM; protein–protein interaction data from MINT and DIP; functional domain information from Pfam; protein fingerprints from PRINTS; protein family and family-specific signatures from InterPro; structure data from PDB; mutation data from PMD; BLAST homology data from NCBI NR; and proteins found to be related based on LocusLink and SWISS-PROT references and sequence and taxonomy data. By highly automating the processes of maintaining the MitoProteome Protein List and extracting relevant data from external databases, we are able to present a dynamic database, updated frequently to reflect changes in public resources. The MitoProteome database is publicly available at http://www.mitoproteome.org/. Users may browse and search MitoProteome, and access a complete compilation of data relevant to each protein of interest, cross-linked to external databases.
INTRODUCTION
Mitochondria are organelles found in most eukaryotic cells. Thought to have once been free-living prokaryotes, they are self-replicating and contain their own double-stranded, circular DNA which, in Homo sapiens, is 16 569 bp in length (1). A cell may contain only one mitochondrion, but hundreds, or even thousands of mitochondria are typically present in cells with substantial energy requirements.
In addition to their central role in energy metabolism, mitochondria are involved in many cellular processes and mitochondrial defects have been associated with apoptosis, aging, and a number of diseases, including Parkinson’s disease, diabetes mellitus and Alzheimer’s.
Only 13 proteins involved in oxidative phosphorylation are encoded by the mitochondrial genome; most of the estimated 1500 human mitochondrial proteins (2) involved in mitochondrial function are nuclear encoded, synthesized in the cytosol and targeted to mitochondria (3).
Data relevant to the study of mitochondrial proteins are dispersed among many heterogeneous public databases. A single database that consolidates and interlinks this data and couples it to powerful query, annotation and analysis systems would greatly facilitate mitochondria research. Such systems can permit biologists to pose questions that would otherwise require the understanding and manipulation of numerous disparate interfaces and be very time-intensive.
Existing mitochondrial protein sequence databases include the Organelle Genome Database [GOBASE (4), http://megasun.bch.umontreal.ca/gobase/], MITOP (5) (from the ‘Mitochondria Project’, http://mips.gsf.de/proj/medgen/mitop/), the Human Mitochondrial Protein Database (HMPDb, http://bioinfo.nist.gov:8080/examples/servlets/index.html) and MitoDrome (6) (http://bighost.area.ba.cnr.it/BIG/MitoDrome/). GOBASE is an organelle genome database and does not contain nuclear-encoded mitochondrial proteins. MITOP contains mitochondrial- and nuclear-encoded mitochondrial protein sequence information from five different species; however, only ∼300 human mitochondrial proteins are represented. The MITOP database may be browsed, but no search interface is available. HMPDb contains information extracted from public databases, with a web interface for searching. However, the HMPDb protein list is highly redundant. MitoDrome is a database of nuclear-encoded Drosophila melanogaster mitochondrial proteins. MitoDrome represents only 350 mitochondrial proteins and lacks extensive annotation.
We have developed MitoProteome, a mitochondrial protein sequence database containing, to our knowledge, the largest set of unique human mitochondrial protein sequences, with 847 in the initial MitoProteome Protein List. Many of the proteins in our protein list were obtained from mass spectrometric analysis of highly purified human heart mitochondria. Other mitochondrial proteins were retrieved from public databases by keyword search and from BLAST analysis. Each protein is manually curated with respect to function and subcellular location, and extensively annotated in an automated process that involves extracting and integrating relevant data from multiple external public databases. A query interface provides several options for browsing and searching the MitoProteome database.
PROTEIN LIST
An initial set of human mitochondrial protein sequences was generated by including the human subset of mitochondrial protein sequences from MITOP and sequences from SWISS-PROT (7), selected based on the ‘subcellular location’ field of protein records. BLAST (8) searches were then performed using the initial set of protein sequences as query sequences and GenBank (9) and SWISS-PROT as target databases. Only full-length sequences with E-values < 1e-5 and sequence identity > 90% were considered for inclusion in the MitoProteome Protein List and annotations were checked manually. This set of mitochondrial protein sequences acquired from public resources was clustered at 90% sequence similarity using CD-HIT (10)—a program for clustering large protein databases at different sequence identity levels—to remove redundant sequences. A group of 615 proteins detected experimentally by mass spectrometry (LC/MS/MS) at MitoKor was added to the above protein list. A unique protein list was generated by sorting based on GenBank GI and the final set of proteins was clustered at 98% to remove redundant proteins, resulting in an initial MitoProteome Protein List of 847 proteins.
For each sequence in the MitoProteome Protein List, protein sequence and protein name were obtained from GenBank records. RefSeq ID and SWISS-PROT accession were parsed from FASTA headers and enzymes were manually assigned EC numbers using the ENZYME (11) nomenclature database. Molecular weight calculations were performed with a Perl program adapted from the Oscore.pm module (12) and isoelectric point values were calculated with the IEP program from the EMBOSS suite (13). Annotation of primary function, subfunction and subcellular location was accomplished manually with the help of databases such as MITOP, LocusLink (14) and PROLYSIS (http://delphi.phys.univ-tours.fr/Prolysis/ec.html).
PROTEIN ANNOTATION
Data extracted from various public databases can provide clues as to the function of a given protein. The MitoProteome Protein List was enriched with additional data derived from a variety of sources, including: Pfam (15) (functional domains); InterPro (16) (protein family and family-specific signatures); PRINTS (17) (fingerprints); DIP (18) and MINT (19) (protein–protein interactions); OMIM (20) (disease information); PDB (21) (structural information); PMD (22) (mutations); LocusLink, Ensembl and SDSC COMBNR (23), a non-redundant compilation of protein sequences from GenBank NR, SWISS-PROT and TrEMBL databases (gene information); NCBI NR (24) (top BLAST hits); and related proteins derived from LocusLink and SWISS-PROT.
To retrieve relevant data from the DIP, MINT, OMIM, PDB, InterPro and PMD databases, we ran BLAST searches using the protein list as query sequences, and developed Perl scripts to select hits with full-length sequences, E-values < 1e-5, and sequence identity > 90%. Perl scripts were also developed to extract gene information from the LocusLink and ENSEMBL databases and to find related proteins from LocusLink and SWISS-PROT. HMMPfam (25) was run with an E-value threshold of 1e-5 to obtain domain information from Pfam and all MitoProteome proteins were queried against PRINTS using the FingerPRINTScan program (26).
RESULTS AND DATABASE IMPLEMENTATION
MitoProteome is implemented as an object-relational database, using Oracle9i Enterprise Edition Release 9.2.0.2.0. Schema diagrams and materials describing the initial database results and implementation are available online (see Table 1).
Table 1. Additional material available online.
| MitoProteome Database: Results and Implementation | http://www.mitoproteome.org/NAR/2004_database/results_and_implementation.html |
| MitoProteome Database Schema Diagrams | http://www.mitoproteome.org/NAR/2004_database/mitoproteome_schema.pdf |
| http://www.mitoproteome.org/NAR/2004_database/mitoproteome_schema.tif |
DATABASE UPDATES
The process of updating the MitoProteome database begins with updating the MitoProteome Protein List. For each sequence in the MitoProteome Protein List, we first query SDSC COMBNR to see whether the defining database record for that protein is still current or if it has expired. For each expired record, we step through a set of rules looking for a current protein record for that protein.
We first look for a current protein record with an identical GenBank accession and updated GenBank version. Failing to find such a record, we next look for any current records with identical sequences (and consistent taxonomy classification). Often, several current records will be found to have sequences identical to that of the expired record. In such cases, SWISS-PROT records are given preference, followed by curated RefSeq protein records, records from other databases such as DDBJ, EMBL, PIR, and lastly RefSeq model protein records. If a current record is found in COMBNR, the corresponding entry in the MitoProtoeme Protein List is updated (with the Mito ID remaining unchanged), flagged as ‘updated’, and the GenBank GI for both the updated MitoProteome record and its corresponding current record are logged. If no corresponding current record is found in COMBNR, the MitoProteome record is flagged as ‘expired’.
New records are manually verified to be localized to mitochondria and annotated with primary function, subfunction, subcellular location and (if the protein is an enzyme) EC number. The remaining automated process of extracting and integrating data from external databases, and updating the MitoProteome database then continues.
QUERY INTERFACE
MitoProteome is publicly accessible at http://www.mitoproteome.org/. We have developed query forms for browsing and searching the MitoProteome Protein List, Disease Catalogue, Domain and Motifs, Gene Catalogue, Protein–protein interactions and Structure data. One can browse and search using any of these data categories as a starting point, and access all available MitoProteome data corresponding to proteins selected from the query results. For each data category, a ‘Browse’ button, when pressed, executes a query to select all records containing data in that category. A ‘Basic Search’ form allows the user to search for a keyword or expression contained in any of the fields selected from the fields relevant to that data set. The query results page presents a sortable list of proteins matching the query criteria, along with appropriate summary information.
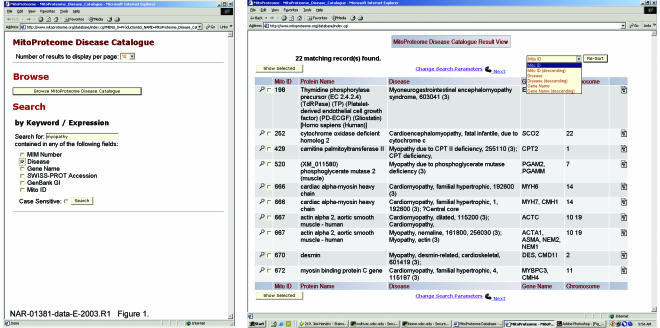
A sample query and results page from the MitoProteome Disease Catalogue are shown in Figure 1. In this example, the search is for all MitoProteome proteins containing the keyword ‘myopathy’ in the disease field of the MitoProteome Disease Catalogue. The query returned 22 matching records. Summary information for each protein in the Disease Catalogue query results page includes protein name, disease, gene name and chromosome. Table 2 summarizes the query forms and results shown for each of the MitoProteome data catalogues.
Figure 1.
Sample database query and result view from the MitoProteome Disease Catalogue.
Table 2. Query forms and results shown for each of the MitoProteome data catalogues.
| Data category | Search fields | Query results fields |
|---|---|---|
| Protein List | Database IDs (Mito ID, GenBank GI, SWISS-PROT Accession, RefSeq ID, EC number) | Mito ID, protein name, EC number, primary function, subfunction, subcellular location, molecular weight, isoelectric point, sequence length |
| Protein name, description, primary function, subfunction, subcellular location | ||
| Physical properties (molecular weight, isoelectric point, sequence length) | ||
| Disease | OMIM number | Mito ID, protein name, disease, gene symbol, chromosome |
| Disease | ||
| Gene symbol | ||
| SWISS-PROT accession | ||
| GenBank GI | ||
| Mito ID | ||
| Domain | Pfam model name | Mito ID, GenBank GI, protein name, model name, domain start, domain end, score, E-value |
| GenBank GI | ||
| Mito ID | ||
| Gene | Locus ID | Mito ID, protein name, gene name, gene symbol, chromosome |
| Official gene symbol | ||
| Official gene name | ||
| Alias gene name | ||
| Alias symbol | ||
| Locus type | ||
| Product name | ||
| Chromosome | ||
| Chromosome location | ||
| Unigene ID | ||
| RefSeq nucleic accession | ||
| RefSeq protein accession | ||
| Variant description | ||
| RefSeq Assembly | ||
| Strain | ||
| Mito ID | ||
| Interaction | Mito ID | Mito ID, protein name, percent identity, description DIP node A (BLAST hit), description DIP node B (interaction partner), method |
| Domain involved | ||
| Method of detection | ||
| For both BLAST hit and interaction partner: DIP node, PIR entry, SWISS-PROT accession, GenBank GI, description, organism | ||
| Structure | PDB ID | Mito ID, protein name, PDB ID, GenBank GI, percent identity, E-value |
| Mito ID | ||
| GenBank GI |
MitoProteome may be searched for keywords contained in any of the listed search fields. The user may select the number of records to show in query results and may sort query results based on any of the query results fields. A ‘Browse’ button is also available from within each query form. Pressing the ‘Browse’ button from within any given data category is equivalent to constructing a query to search for all records containing data in that category. Selecting a protein from the query results page yields the complete set of data relevant to that protein collected in all MitoProtoeme data catalogues.
For each record selected from the query results page, all MitoProteome data relevant to that protein are displayed. Database IDs are linked to external databases, and sequences may be imported to the Biology Workbench (27) (http://workbench.sdsc.edu), where one can perform BLAST searches against other databases, sequence alignments, structure predictions, motif and profile extractions as well as numerous other analyses.
DISCUSSION
MitoProteome is a dynamic database, combining an accurate and comprehensive human mitochondrial protein list with expert annotation, extensive value-added information, a powerful query interface, extensive cross-links to external databases and the ability to import sequences to the Biology Workbench (http://workbench.sdsc.edu) suite of sequence analysis tools.
The MitoProteome Protein List was derived from three major sources: known mitochondrial proteins from MITOP and SWISS-PROT; proteins with high sequence homology to MITOP- and SWISS-PROT-derived mitochondrial proteins, based on BLAST scores; and proteins detected by mass spectrometric analysis of highly purified human heart tissue. It is possible for a protein to have very high sequence homology to a known mitochondrial protein, but no existing annotation or experimental evidence pointing to mitochondrial localization. There could also be a low degree of error associated with the assignment of subcellular localization based on mass spectrometric analysis. To account for these possibilities, we carefully examined available experimental evidence for each protein included in the MitoProteome Protein List. Each MitoProteome protein was also manually annotated with respect to primary function, subfunction, subcellular location and EC number.
We are currently investigating methods to automate the manual annotation steps of assigning function, subcellular location and EC number. With these exceptions, the process of generating the MitoProteome Protein List, extracting and integrating value-added information, and updating the MitoProteome Protein List and database is entirely automated. A major advantage of such automation is that we are able to perform frequent MitoProteome database updates to reflect updates in other public databases.
Future work will also include the development of additional query interfaces, enhancements to existing interfaces, and the development of mitoproteome interaction and signaling networks. We will also incorporate tools for finding new mitochondrial proteins in public databases, including mitochondrial proteins from other species. To this end, our group has developed a new method for the prediction of nuclear-encoded mitochondrial proteins in eukaryotic species (C. Guda, E. Fahy and S. Subramaniam, submitted). This method predicts mitochondria-targeted proteins based on the patterns of Pfam domain occurrence and the amino acid compositional differences between mitochondrial and non-mitochondrial sequences.
The MitoProteome Protein List may be downloaded as a flat file database and MitoProteome protein sequences may be downloaded in FASTA format. The MitoProteome Protein List and FASTA sequences are also available from within the Biology Workbench.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to acknowledge Chittibabu Guda, a member of the SDSC MitoProteome team, and thank Brian Saunders for his help in finding gene information and related proteins. The MitoProteome query interface was developed with the help of DataScope, written by Roger Unwin. This work was supported by Life Sciences Informatics (LS:IT) Program Grant L99-1077.
REFERENCES
- 1.Anderson S., Bankier,A.T., Barrell,B.G., de Bruijn,M.H., Coulson,A.R., Drouin,J., Eperon,I.C., Nierlich,D.P., Roe,B.A., Sanger,F. et al. (1981) Sequence and organization of the human mitochondrial genome. Nature, 290, 457–465. [DOI] [PubMed] [Google Scholar]
- 2.Taylor S.W., Fahy,E., Zhang,B., Glenn,G.M., Warnock,D.E., Wiley,S., Murphy,A.N., Gaucher,S.P., Capaldi,R.A., Gibson,B.W. et al. (2003) Characterization of the human heart mitochondrial proteome. Nat. Biotechnol., 21, 281–286. [DOI] [PubMed] [Google Scholar]
- 3.Lang B.F., Gray,M.W. and Burger,G. (1999) Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet., 33, 351–397. [DOI] [PubMed] [Google Scholar]
- 4.O’Brien E.A., Badidi,E., Barbasiewicz,A., deSousa,C., Franz Lang,B. and Burger,G. (2003) GOBASE—a database of mitochondrial and chloroplast information. Nucleic Acids Res., 31, 176–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scharfe C., Zaccaria,P., Hoertnagel,K., Jaksch,M., Klopstock,T., Dembowski,M., Lill,R., Prokisch,H., Gerbitz,K.D., Neupert,W. et al. (2000) MITOP, the mitochondrial proteome database. Nucleic Acids Res., 28, 155–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sardiellom M., Licciulli,F., Catalano,D., Attimonelli,M. and Caggese,C. (2003) MitoDrome: a database of Drosophila melanogaster nuclear genes encoding proteins targeted to the mitochondrion. Nucleic Acids Res., 31. 322–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O’Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 9.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J. and Wheeler,D.L. (2003) GenBank. Nucleic Acids Res., 31, 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W., Jaroszewski,L and Godzik,A. (2001) Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics, 17, 282–283. [DOI] [PubMed] [Google Scholar]
- 11.Bairoch A. (2000) The ENZYME database in 2000. Nucleic Acids Res., 28, 304–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore R.E., Young,M.K. and Lee,T.D. (2002) Qscore: an algorithm for evaluating SEQUEST database search results. J. Am. Soc. Mass Spectrom., 13, 378–386. [DOI] [PubMed] [Google Scholar]
- 13.Rice P., Longden,I. and Bleasby,A. (2000) EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet., 16, 276–277. [DOI] [PubMed] [Google Scholar]
- 14.Pruitt K.D., Katz,K.S., Sicotte,H. and Maglott,D.R. (2000) Introducing RefSeq and LocusLink: curated human genome resources at the NCBI. Trends Genet., 16, 44–47. [DOI] [PubMed] [Google Scholar]
- 15.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L.L. (2002) The Pfam Protein Families Database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulder N.J, Apweiler,R., Attwood,T.K., Bairoch,A., Barrell,D., Bateman,A., Binns,D., Biswas,M., Bradley,P., Bork,P. et al. (2003) The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res., 31, 315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Attwood T.K., Bradley,P., Flower,D.R., Gaulton,A., Maudling,N., Mitchell,A., Moulton,G., Nordle,A., Paine,K., Taylor,P. et al. (2003) PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res., 31, 400–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xenarios I., Salwínski,L., Duan,X,J., Higney,P., Kim,S. and Eisenberg,D. (2002) DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res., 30, 303–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zanzonia A., Montecchi-Palazzi,L., Quondam,M., Ausiello,G., Helmer-Citterich,M. and Cesareni,G. (2002) MINT: a Molecular INTeraction database. FEBS Lett., 513, 135–140. [DOI] [PubMed] [Google Scholar]
- 20.Hamosh A., Scott,A.F., Amberger,J., Bocchini,C., Valle,D. and McKusick,V.A. (2002) Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res., 30, 52–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawabata T., Ota,M. and Nishikawa,K. (1999) The Protein Mutant Database. Nucleic Acids Res., 27, 355–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J., Ning,Y., Hedley,W., Saunders,B., Chen,Y., Tindill,N., Hannay,T. and Subramaniam,S. (2002) The Molecule Pages database. Nature, 420, 716–717. [DOI] [PubMed] [Google Scholar]
- 24.Benson D.A., Karsch-Mizrachi,I, Lipman,D.J., Ostell,J. and Wheeler,D.L. (2003) GenBank. Nucleic Acids Res., 31, 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eddy S.R. (1998) Profile hidden Markov models. Bioinformatics, 14, 755–763. [DOI] [PubMed] [Google Scholar]
- 26.Scordis P., Flower,D.R. and Attwood,T.K. (1999) FingerPRINTScan: Intelligent searching of the PRINTS motif database. Bioinformatics, 15, 799–806. [DOI] [PubMed] [Google Scholar]
- 27.Subramaniam S. (1998) The Biology Workbench—a seamless database and analysis environment for the biologist. Proteins, 32, 1–2. [PubMed] [Google Scholar]