Abstract
Bacteria evolve their capacity to cause disease by acquiring virulence genes that are usually clustered in discrete genetic modules termed pathogenicity islands (PAI). Stable integration of PAIs into preexisting transcriptional networks coordinates expression from PAIs with ancestral genes in response to diverse environmental cues. Such transcriptional controls are evident in the regulation of the locus of enterocyte effacement (LEE), a PAI of enteropathogenic and enterohemorrhagic Escherichia coli. However, recent reports indicate that global posttranscriptional and posttranslational regulators, including CsrA, Hfq and ClpXP, fine-tune the transcriptional output from the LEE. Here, we highlight recent advances in understanding posttranscriptional and posttranslational regulation in attaching and effacing pathogens.
What are EPEC and EHEC?
Enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC) cause significant morbidity and mortality worldwide [1, 2]. EPEC is a waterborne pathogen that causes diarrhea, primarily among infants, in developing countries [1]. EHEC is spread via contaminated food and water and affects both infants and adults in developed countries [1]. Certain EHEC strains harbor Shiga toxins that cause bloody diarrhea and hemolytic uremic syndrome (HUS), a disease characterized by hemolytic anemia, thrombocytopenia, and acute renal failure [2].
Upon infection, EPEC and EHEC compete with the native microflora for limited nutrients, respond to stressors from the host immune system, and migrate to sites within the small or large intestine respectively, where they colonize and cause disease [1]. They belong to the attaching and effacing (A/E) family of pathogens because they adhere intimately to intestinal cells (attachment) and promote the destruction of microvilli (effacement). Upon attachment, EPEC and EHEC recruit host cytoskeletal proteins to form characteristic actin-filled membraneous protrusions, termed pedestals, beneath themselves (Figure 1). The locus of enterocyte effacement (LEE) is a 35–42 kb pathogenicity island that encodes transcriptional regulators, a functional type III secretion system (T3SS), and various exported translocators and effectors. Mutational analysis has revealed that the LEE is necessary for pedestal formation and disease [1] (Figure 2).
Figure 1.
Pedestal formation bv EPEC and EHEC. (a) Cartoon depiction of pedestal formation by EPEC and EHEC. Under inducible conditions the gatekeeper switch, composed of SepL and SepD, orchestrates the hierarchical secretion of translocators (EspA, EspB and EspD) over effectors (Tir and other effectors such as NleA, EspF and Map). While the EspADB translocon is being assembled, SepL directly binds to Tir and possibly other effectors and suppresses their export (i). After the maturation of the translocon that connects the bacterium to the host, effectors are trafficked into the host’s cytoplasm via the T3SS (i). Subsequently, Tir integrates into the host plasma membrane where it interacts with its ligand, intimin, located on the outer bacterial membrane. Tir-intimin interactions promote the clustering of Tir and initiate a signal transduction cascade that leads to the conscription and subsequent polymerization of actin to form membraneous protrusions, termed attaching and effacing (A/E) pedestals, underneath adherent bacteria (ii). (b) Scanning electron micrographic (SEM) image of EPEC forming pedestals on infected mammalian cells. The inset shows a pseudocolored image of two EPEC bacteria (green) infecting HeLa cells (purple). SEM image courtesy of Jorge Giron.
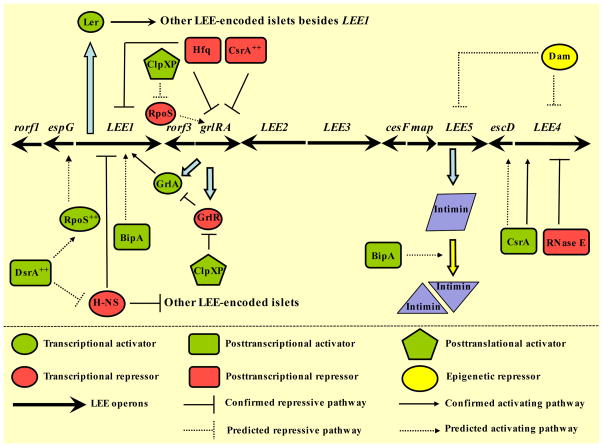
Figure 2.
Transcriptional and extratranscriptional control of the LEE. The LEE is organized into five principal polycistronic operons (LEE1-5), the bicistronic operon grlRA, and numerous monocistronic genes. Under inducible conditions, the H-NS-mediated repression of LEE1 is relieved and transcription of ler is induced. Ler, an H-NS paralog, activates transcription from the other LEE operons primarily by counteracting the H-NS-mediated repression. Expression from the LEE leads to the synthesis and consequent assembly of a T3SS that connects the bacterial cytosol to the host cytoplasm and leads to pedestal formation and ensuing disease (see text for details). The posttranscriptional factors DsrA, BipA, Hfq, and CsrA, as well as the posttranslational factor ClpXP govern gene expression from the LEE by directly or indirectly influencing the abundance of the LEE-encoded regulator, Ler. Moreover, the regulators CsrA, BipA, Dam, and RNase E exert extra-transcriptional controls on the LEE-encoded lower tier genes within the Ler regulon (see text for details). Activators and repressors have been defined based on their capacity to globally induce or silence gene expression from the LEE respectively, and consequently affect pedestal formation. Thin filled lines with blunted ends or arrowheads depict regulatory pathways where the effect is direct and/or the mechanistic basis of gene expression has been defined, whereas thin dashed lines with blunted ends or arrowheads reflect presumptive circuits for which a detailed mechanism has not been established. ++ indicates overexpression of the gene product.
Coordinated regulation of virulence
During infection, A/E pathogens encounter changes in pH, osmolarity, ferric nitrate [Fe(NO3)3], Ca2+, temperature, quorum sensing, and bicarbonate (HCO3−), among others, and respond by coordinately regulating virulence gene expression in conjunction with other physiological processes [1, 3]. Whereas transcriptional control of the LEE has been rigorously characterized, recent evidence indicates that posttranscriptional and posttranslational factors play key roles in fine-tuning the transcriptional output from the LEE, and in addition might integrate virulence with other physiological processes. Here we discuss such regulatory mechanisms, and highlight the physiological and evolutionary benefits of posttranscriptional and posttranslational regulation as a complement to transcriptional control. Moreover, we suggest that coordinate regulation of ancestral traits such as motility and metabolism together with virulence traits is a key determinant of morbidity in A/E pathogens.
Intrinsic transcriptional control of the LEE
The LEE is organized into five major polycistronic operons (LEE1-LEE5), the bicistronic operon (grlR-grlA) and numerous monocistronic genes (grlA, escD, map, espG, cesF, rorf3) [1, 4–7] (Figure 2). Expression from the LEE1, LEE2, and LEE3 operons and escD results in synthesis and assembly of a T3SS, which traverses the inner and outer membranes, and the peptidoglycan layer [1, 5]. The LEE4 operon primarily encodes the extracellular components of the T3SS including EscF, EspA, EspB and EspD. EscF forms the needle tip of the T3SS, and EspA monomers polymerize to form a hollow filament that connects EscF to the host cell membrane [8–10]. Subsequently, EspB and EspD translocate through the EspA filament and integrate into the host plasma membrane thereby forming a contiguous pore between the bacterium and the host. Effectors, including Tir, enter the host cell through the portal and mediate the formation of pedestals [1, 11–15] (Figure 1a).
Under refractory conditions, the global regulator H-NS binds to cis-regulatory sequences located upstream of LEE1-3, rorf3, grlR, grlA and LEE5 and inhibits transcription initiation [1, 16–18] (Figure 2). H-NS-dependent transcriptional regulation is governed by diverse environmental cues including temperature and HCO3− ions, and it appears to be the most prominent repressor of the LEE [1, 19] (Figure 2). Under conditions conducive for pedestal formation, H-NS-mediated repression of LEE1 is relieved and transcription of ler is induced [1, 19]. Consequently, Ler activates transcription from the other LEE-encoded transcriptional units, including grlRA, LEE2, LEE3, LEE4, LEE5, map and espG, primarily by competing with H-NS for overlapping binding sites [1, 6, 16, 18, 20, 21] (Figure 2). The grlRA operon encodes a global transcriptional repressor of the LEE, GrlR, and an associated activator, GrlA [5]. Ler, GrlR, and GrlA are components of an autoregulatory loop in which GrlA further enhances transcription of ler [16, 17, 22], whereas GrlR binds to and inactivates GrlA, thereby limiting transcription of ler [17, 23].
Besides transcriptional control, several recent studies have highlighted the importance of posttranscriptional and posttranslational mechanisms in refining gene expression from the LEE [24–33]. Importantly, many of these mechanisms were originally identified in E. coli [34–38], but are ubiquitously used by other bacterial pathogens including EPEC and EHEC [35, 36, 39].
Why control the LEE at the posttranscriptional and posttranslational levels?
There are several reasons why posttranscriptional and posttranslational controls might have evolved to complement transcriptional control. First, the operon organization of bacterial genes limits the capacity of transcription factors to differentially modulate genes within the same transcription unit. For instance, Ler activates the transcription of all the genes encoded within LEE2, LEE3, LEE5 and LEE4 operons without selectively affecting the expression of genes within individual transcription units [1, 40]. Moreover, such a genetic organization is particularly constraining when gene products required in different stoichiometric ratios are encoded on the same transcript. For instance, the LEE4-encoded polycistronic transcript encodes a regulator (SepL), the structural components of a mature T3SS (EspA, EspD, EspB, and EscF), chaperones (CesD2 and L0017), and an effector (EspF) [29]. While most of the genes in the transcript contribute to T3SS in general, they are required in different concentrations; the translocators EspA, EspB and EspD are made in excess relative to the regulator SepL [29]. In EHEC, this is accomplished by posttranscriptional processing within the intracistronic segment of sepL, followed by the selective degradation of sepL and the concomitant stabilization of espA, espD, espB [29, 31].
Second, posttranscriptional and posttranslational mechanisms allow bacteria to control gene expression over a wide dynamic range, degrading some transcripts or proteins when they are no longer needed, or retaining others in abeyance for rapid mobilization at a later time [41]. For instance, the RNA-binding protein CsrA represses translation from some transcripts by promoting their degradation (e.g. pgaABCD) [42] or without affecting transcript stability (e.g. hfq) [43]. Likewise, the adaptor protein RssB can target the alternative stationary phase sigma factor, RpoS, for degradation by the ClpXP protease or simply bind to and sequester it, consequently repressing the expression of RpoS-activated genes [44]. Thus, by affecting substrate activity, stability and/or abundance [41, 44–48], posttranscriptional and posttranslational mechanisms fine-tune gene expression in a way not easily accomplished by transcriptional controls alone.
Third, posttranscriptional and posttranslational controls of gene regulation provide a means to rapidly and globally adapt to diverse environmental stimuli. The use of global regulators of ancestral processes for this purpose allows bacterial pathogens to coordinate virulence with other physiological processes. Examples of such global regulatory factors include CsrA, Hfq, DsrA, and ClpXP, whose activities are described in detail below.
Lastly, posttranscriptional regulation is energetically efficient because small RNAs (sRNAs) governing this process, such as DsrA and CsrB, or small proteins such as CsrA and Hfq, which exert much of the posttranscriptional control, can be synthesized quickly and with less energy compared to the relatively larger transcription factors [41]. In summary, posttranscriptional and posttranslational mechanisms provide a complement to transcriptional control for highly plastic regulatory responses to diverse environmental stimuli [41].
RNA-binding proteins regulate the LEE
Carbon storage regulator A (CsrA), also called RsmA, was originally identified as a repressor of glycogen biosynthesis and biofilm formation in nonpathogenic E. coli [49]. Since then, CsrA has been found to regulate diverse physiological processes including carbon homeostasis, quorum sensing, peptide uptake, morphogenesis of flagella, and T3SS [34, 35].
CsrA is highly conserved across diverse bacteria but has been most extensively characterized in E. coli [34, 35]. CsrA exists as a homodimer and binds to single-stranded tracts in the leader segment of transcripts containing a core motif, AGGA or ANGGA and influences messenger RNA (mRNA) stability and/or translation [34, 35, 50, 51]. The regulatory sRNAs CsrB and CsrC contain repetitive AGGA/ANGGA tracts that sequester multiple CsrA molecules and reduce the effective concentration and activity of CsrA [51–53]. Orthologs of the Csr genes are highly conserved in A/E pathogens [24].
In EPEC, CsrA acts as an activator or repressor of the LEE in a manner that depends upon its concentration [24] (Figure 2). CsrA binds to two ANGGA motifs in the untranslated leader segment of the LEE4 transcript, and increases the steady-state levels, likely by stabilizing the message. Additionally, CsrA also activates the expression of the inner membrane protein of the T3SS, EscD, through an intermediate regulator. As a consequence, CsrA facilitates pedestal formation [24]. By contrast, high concentrations of CsrA globally inhibit gene expression from the LEE by reducing grlRA transcript levels [24]. Moreover, CsrA binds to as many as three sites within the grlRA transcript, one of which is in close vicinity of the Shine-Dalgarno sequence, a topological feature generally conserved in repressed transcripts [34, 35]. Taken together, these observations suggest that the CsrA-mediated repression of grlRA might result from reduced transcript stability [24].
Besides CsrA, the highly conserved RNA-chaperone Hfq has recently been identified as an important posttranscriptional regulator of the LEE in EPEC and EHEC [25, 26]. Inactivation of hfq renders E. coli hypersensitive to a plethora of environmental stressors [54]. Hfq has since been recognized to play a key role in stress responses and virulence of diverse bacteria [55]. Hfq exists as a homohexamer and assumes a toroidal conformation that contains distinct proximal and distal RNA-binding surfaces [56, 57]. Structural studies with Hfq from E. coli reveal that the proximal face binds A/U rich sequences, whereas the distal face simultaneously recognizes tandem poly-(A-R-E) tracts [56–58]. This enables Hfq to stabilize base-pairing between sRNAs and target mRNAs, which have limited complementarity, and affect transcript stability and/or translation [57].
In EHEC, Hfq represses the LEE via two pathways and consequently affects pedestal formation [25, 26] (Figure 2). In the exponential phase, Hfq destabilizes the grlRA transcript resulting in reduced expression of GrlA. This, in turn, causes a reduction in levels of Ler, and global silencing of the LEE [25]. By contrast, in the stationary phase, Hfq-mediated repression of the LEE remains largely independent of grlRA and instead occurs by translational repression of Ler [25, 26] (Figure 2). In both EPEC and EHEC the leader segment of the grlRA transcript possesses a canonical poly-(A-R-E) motif (5’-AGA AAA AGA AAG-3’), raising the possibility that the transcript binds to the distal face of Hfq. It remains to be determined whether ler is directly regulated by Hfq.
Besides non-catalytic RNA-binding proteins, the single-strand specific endoribonuclease RNase E also controls gene expression from the LEE of EHEC [29]. RNase E is a component of the bacterial degradosome, a multiprotein complex involved in the maturation or degradation of heterogeneous RNA species [37]. In E. coli, RNase E displays relaxed sequence specificity with a preference for A/U-rich transcripts [36]. Consequently, RNase E and Hfq co-regulate several transcripts antagonistically [36]. RNase E is responsible for the posttranscriptional processing of the LEE4-encoded sepLespADB transcript in EHEC [29] (Figure 2). The 5’ fragment containing sepL undergoes rapid degradation rendering SepL undetectable. By contrast, the 3’ segment spanning espA and the downstream genes remains stable [29]. Additionally, the leader segment of sepL possesses a noncanonical ribosome-binding site (RBS) that is highly divergent from the near-consensus RBS observed in the leader segment of espA. Lodato and Kaper propose that inefficient recruitment of the ribosome to the RBS of sepL renders the sepL segment of the transcript sensitive to RNase E; by contrast, proficient binding of the ribosome to the espA segment and ensuing translation might sterically hinder binding by ribonucleases [29]. Such a mechanism accounts for the stoichiometric difference between the abundance of the regulator SepL and the translocators (EspA, EspB and EspD), and might also promote the transition from the export of translocators, which connect the T3SS to the host cell, to that of the effectors (e.g. Tir) [48]. Thus, in response to appropriate environmental cues, degradation of the sepL transcript and reduction in SepL levels could permit the secretion of effectors, which would be otherwise sequestered by SepL [29, 48]. However, it remains to be determined whether posttranscriptional processing of sepLespADB occurs in other A/E pathogens. Furthermore, the physiological relevance of maturation of the LEE4 transcript during synthesis of the T3SS needs to be evaluated by selective mutation of the RNase E cleavage sites in sepL.
sRNA-mediated regulation of the LEE
Regulatory sRNAs are ubiquitously conserved in the three taxonomic domains of life where they affect all steps of gene regulation [59]. In E. coli, DsrA exists as an 87-nucleotide, untranslated transcript that modulates gene expression by antisense base-pairing with its target mRNAs in the presence of the RNA-chaperone Hfq [60, 61]. DsrA binds to hns and rpoS mRNAs, destabilizing the former but promoting translation of the latter [62, 63]. Riboregulation of these global regulators enables DsrA to regulate responses to an array of environmental stressors [61]. In EHEC, overexpression of dsrA activates ler in an hns- and rpoS-dependent manner [27] (Figure 2). While details of this regulation have not been elucidated, it is likely that the observed phenotype results from a mechanism similar to that observed in E. coli. Thus, high levels of DsrA could result in loss of H-NS-mediated repression of the LEE and, consequently, activation of Ler. DsrA-mediated activation of Ler also requires a functional rpoS allele. Paradoxically, RpoS has also been recognized as a repressor of Ler, when DsrA is expressed in single copy, and RpoS levels are low [27, 28]. However, increasing DsrA concentrations could promote translation of RpoS and lead to activation of Ler [27]. Thus, RpoS could act as a dose-dependent repressor or activator of the LEE, in a manner that depends on DsrA levels (Figure 2).
Intriguingly, the effects of DsrA are pathovar-specific, as overexpression has negligible effects on LEE gene expression in EPEC [27]. One possible explanation is that the basal levels of the DsrA message might be inherently higher in EPEC as compared to EHEC [27]. Thus, DsrA could contribute to the higher overall level of expression from the LEE of EPEC as compared to EHEC. A confounding factor in studies on EPEC and EHEC is that inactivation of dsrA was without effect. In this regard, riboregulators, including DsrA, often exhibit functional redundancy [41, 64]. In E. coli, inactivation of dsrA, rprA, and arcZ is required to substantially reduce expression of rpoS under nutrient deprivation conditions [64], and the same might be necessary in EPEC and EHEC to observe physiologically relevant changes in gene expression from the LEE.
Posttranslational control of the LEE
Information on how posttranslational factors affect virulence of A/E pathogens is more limited than for posttranscriptional regulators. The ATP-dependent protease ClpXP is one of the few posttranslational regulators whose role in the virulence of EHEC is well established [28, 30]. Substrates bind to ClpX in an ATP-dependent manner and are subsequently hydrolyzed in the inner cavity of the catalytic component ClpP [65]. In EHEC, ClpXP activates the transcription of ler in an rpoS- and grlR-dependent manner [28, 30] (Figure 2). In E. coli K-12, the adaptor protein RssB is sufficient to target RpoS for degradation by ClpXP [44]. RssB is highly conserved in A/E pathogens suggesting that clpXP-mediated activation of the LEE might result in part from the direct degradation of RpoS by ClpXP. ClpXP effects also appear to be mediated via GrlR because inactivation of grlR completely bypasses the requirement for functional clpXP [28]. The observation that the stability and abundance of GrlR is elevated in a clpXP mutant of EHEC raises the possibility that GrlR could be a direct substrate of ClpXP [28]. This is perhaps not surprising as intrinsic regulators of horizontally acquired genes frequently integrate into ancestral regulatory circuits, including those acting at the posttranscriptional and posttranslational level [35, 55, 66].
Other extra-transcriptional mechanisms affecting the LEE
Posttranscriptional regulation has also been implicated in the phenotypic plasticity of EspADB translocons evident in different EHEC strains [31]. High secretors exhibit more T3SSs on their surface than low secretors. Paradoxically, high-secretors export more translocator proteins but contain less mRNA in comparison to low-secretor strains [31]. The regulatory factors and the corresponding networks mediating this response have yet to be elucidated, except in so far as the effect appears to be at the posttranscriptional level. In EHEC, the EspA translocon also functions as an adhesin [1], and evidence in other systems suggests that adhesin levels can contribute to tissue tropism amongst related strains [66]. Such strain-specific effects raise the possibility that rewiring of posttranscriptional regulatory networks amongst EHEC strains might confer alternate sites of attachment within the intestinal tract and/or facilitate colonization of different tissues within the host.
A variety of other extra-transcriptional mechanisms have been described that regulate LEE expression, though little detailed mechanistic information is available. For example, the ribosome binding GTPase BipA induces gene expression from the LEE of EPEC and EHEC by promoting the steady-state transcript levels of ler [32]. Moreover, BipA is also required for the proteolysis of intimin in EPEC [32] (Figure 2). In addition, epigenetic control via the DNA-modifying enzyme DNA adenine methyltransferase (Dam) has also been implicated in regulation of the LEE in EHEC [67]. The physiological role of Dam is to methylate the adenine residue located within the tetranucleotide tract, GATC [68]. The resulting methylation pattern globally affects numerous cellular processes including general gene expression, chromosomal replication, and methyl-directed mismatch repair [67]. In EHEC, inactivation of dam induces gene expression from the LEE and promotes bacterial adherence and pedestal formation [33] (Figure 2). This effect appears to be mediated at the translational or posttranslational level as the abundance of the LEE-encoded proteins, but not transcripts, is dramatically elevated in the mutant [33]. It is likely that this effect on the LEE is regulated through an intermediary because Dam specifically modifies DNA [67, 68].
Elucidating the posttranscriptional and posttranslational ‘virulence regulome’ of A/E pathogens
The majority of posttranscriptional and posttranslational regulators that govern the LEE are ancestral factors shared between nonpathogenic E. coli and A/E pathogens. Such factors affect a plethora of physiological processes. For instance, CsrA, Hfq and ClpXP control motility, metabolism and adaptation to stress in nonpathogenic and pathogenic E. coli [24, 25, 35, 55, 69]. The flagellum of EPEC and EHEC possesses adhesive properties and has been shown to facilitate bacterial colonization [1, 70]. Moreover, the ability of EHEC to switch between glycolytic and gluconeogenic substrates is critical to its pathogenicity in vivo [71]. Thus, the ability of A/E pathogens to cause disease is not exclusively the result of traits conferred by horizontally acquired PAIs. Rather, a successful infection requires the contribution of both ancestral and newly acquired traits, acting in a coordinated spatiotemporal manner. Techniques that permit the genome-wide identification of regulons and their corresponding traits are therefore important for elucidating the ’virulence regulome’ of A/E pathogens. In this regard, RNA immunoprecipitation (RIP) of RNA-binding proteins, such as CsrA and Hfq, coupled to the sequencing of the bound transcripts will be instrumental in identifying the direct targets of posttranscriptional factors. Moreover, investigating transcriptome and proteome profiles of these regulators would facilitate discrimination between direct and indirect regulatory targets. At the posttranslational level, catalytically inactive variants might prove useful. For example, ClpXP variants that trap but do not degrade substrates would permit identification of direct targets. Likewise, indirect targets of ClpXP could be inferred by pairing substrate trapping with proteomic analysis. Such approaches have been successfully employed to identify regulons of posttranscriptional and posttranslational factors in nonpathogenic E. coli and other bacteria [55, 72]. Furthermore, comparative metatranscriptomic and metaproteomic analyses could also shed light on the evolution of posttranscriptional and posttranslational regulatory networks in different A/E pathogens, as has been done for orthologous transcriptional networks in Yersinia pestis and Salmonella Typhimurium [66, 73].
Lastly, even though trans-acting mutations are useful in identifying regulators of pathogenesis, such mutations affect several biological pathways, of which only some contribute to virulence. Therefore, construction of cis-regulatory mutations, for example in the binding sites of posttranscriptional and posttranslational regulators, to selectively affect particular pathways, will provide a better understanding of the relative contributions of these pathways in bacterial colonization and disease. Moreover, such analysis of trans- and cis-acting mutations should not be limited to in vitro studies, because predictions based on in vitro assays might not translate in vivo. This is particularly true for posttranscriptional regulators. For example, inactivation of hfq induces the expression of the T3SS and virulence-associated effectors in Shigella sonnei, Vibrio cholerae, and Pseudomonas aeruginosa [26, 74–76]. However, the hfq mutant of each of these pathogens is dramatically attenuated in vivo [55, 74, 75, 77]. The in vivo pathogenicity profile of the hfq mutants is not unexpected because hfq mediates adaptation to a number of different stressors [55]. Thus, despite enhanced virulence in vitro, hfq mutants of EPEC and EHEC will likely be attenuated in vivo due to enhanced sensitivity to host stressors.
Concluding remarks
Posttranscriptional and posttranslational regulation significantly expand the regulatory flexibility of A/E pathogens and provide additional checkpoints to refine transcriptional output. However, our current knowledge of such regulatory mechanisms, which might be both specific to A/E pathogens and more generally applicable, is still in its infancy. Nonetheless, such modes of regulation could be key factors coordinating the expression of newly acquired virulence genes with ancient metabolic processes. Finally, a comprehensive understanding of the extra-transcriptional honing of the transcriptional output might prove essential for the development of novel therapeutic measures to effectively combat A/E pathogens (Box 1).
Box 1. Outstanding questions.
How do environmental cues affect extra-transcriptional regulators of the LEE?
What genes comprise the virulence regulon of extra-transcriptional factors and what is the contribution of such regulation to the morbidity associated with A/E pathogens in vivo?
What is the hierarchical organization of the different posttranscriptional and posttranslational regulators that govern the LEE?
Can highly conserved extra-transcriptional factors serve as targets for broad-spectrum antibiotics?
How have ancestral extra-transcriptional networks evolved to coordinate expression of ancient processes with recently acquired pathogenic traits during development of virulence in A/E pathogens?
Are there posttranscriptional and posttranslational virulence factors that are specific to A/E pathogens, and do such regulators affect ancestral genes just as ancestral regulators modulate virulence genes?
Do orthologs of extra-transcriptional regulators with established roles in the virulence of other pathogens function similarly in A/E pathogens?
Acknowledgments
We regret that space limitations precluded citation of all the work in this area. We are grateful to the members of the Kalman and Romeo labs for helpful discussions. Work in our lab is supported by grants from the National Institute of Health - R01DK074731-01 and R01A1056067-01 (to D.K.) and R01-GM059969 (to T.R.). S.B is a recipient of the National Science Foundation award 0450303 subaward I-66-606-63 to Emory University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mellies JL, et al. Enteropathogenic and enterohemorrhagic Escherichia coli virulence gene regulation. Infect Immun. 2007;75:4199–4210. doi: 10.1128/IAI.01927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karch H, et al. Enterohaemorrhagic Escherichia coli in human medicine. Int J Med Microbiol. 2005;295:405–418. doi: 10.1016/j.ijmm.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Kenny B, Abe Akio, Stein Markkus, Finlay B Brett. Enteropathogenic Escherichia coli protein cecretion is induced in response to conditions similar to those in the gastrointestinal tract. Infect and Immun. 1997;65:2606–2612. doi: 10.1128/iai.65.7.2606-2612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mellies JL, et al. The Per regulon of enteropathogenic Escherichia coli : identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler) Mol Microbiol. 1999;33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- 5.Deng W, et al. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc Natl Acad Sci U S A. 2004;101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchez-SanMartin C, et al. Transcriptional regulation of the orf19 gene and the tir-cesT-eae operon of enteropathogenic Escherichia coli. J Bacteriol. 2001;183:2823–2833. doi: 10.1128/JB.183.9.2823-2833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tauschek M, et al. Transcriptional analysis of the grlRA virulence operon from Citrobacter rodentium. J Bacteriol. 2010;192:3722–3734. doi: 10.1128/JB.01540-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sekiya K, et al. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc Natl Acad Sci U S A. 2001;98:11638–11643. doi: 10.1073/pnas.191378598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson RK, et al. Role of EscF, a putative needle complex protein, in the type III protein translocation system of enteropathogenic Escherichia coli. Cell Microbiol. 2001;3:753–762. doi: 10.1046/j.1462-5822.2001.00159.x. [DOI] [PubMed] [Google Scholar]
- 10.Daniell SJ, et al. The filamentous type III secretion translocon of enteropathogenic Escherichia coli. Cell Microbiol. 2001;3:865–871. doi: 10.1046/j.1462-5822.2001.00168.x. [DOI] [PubMed] [Google Scholar]
- 11.Shaw RK, et al. EspA filament-mediated protein translocation into red blood cells. Cell Microbiol. 2001;3:213–222. doi: 10.1046/j.1462-5822.2001.00105.x. [DOI] [PubMed] [Google Scholar]
- 12.Wachter C, et al. Insertion of EspD into epithelial target cell membranes by infecting enteropathogenic Escherichia coli. Mol Microbiol. 1999;31:1695–1707. doi: 10.1046/j.1365-2958.1999.01303.x. [DOI] [PubMed] [Google Scholar]
- 13.Wolff C, et al. Protein translocation into host epithelial cells by infecting enteropathogenic Escherichia coli. Mol Microbiol. 1998;28:143–155. doi: 10.1046/j.1365-2958.1998.00782.x. [DOI] [PubMed] [Google Scholar]
- 14.Ide T, et al. Characterization of translocation pores inserted into plasma membranes by type III-secreted Esp proteins of enteropathogenic Escherichia coli. Cell Microbiol. 2001;3:669–679. doi: 10.1046/j.1462-5822.2001.00146.x. [DOI] [PubMed] [Google Scholar]
- 15.Kenny B, et al. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1999;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 16.Barba J, et al. A positive regulatory loop controls expression of the locus of enterocyte effacement-encoded regulators Ler and GrlA. J Bacteriol. 2005;187:7918–7930. doi: 10.1128/JB.187.23.7918-7930.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jimenez R, et al. Molecular characterization of GrlA, a specific positive regulator of ler expression in enteropathogenic Escherichia coli. J Bacteriol. 2010;192:4627–4642. doi: 10.1128/JB.00307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haack KR, et al. Interaction of Ler at the LEE5 (tir) operon of enteropathogenic Escherichia coli. Infect Immun. 2003;71:384–392. doi: 10.1128/IAI.71.1.384-392.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Umanski T, et al. Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology. 2002;148:2735–2744. doi: 10.1099/00221287-148-9-2735. [DOI] [PubMed] [Google Scholar]
- 20.Sperandio V, et al. Activation of enteropathogenic Escherichia coli (EPEC) LEE2 and LEE3 operons by Ler. Mol Microbiol. 2000;38:781–793. doi: 10.1046/j.1365-2958.2000.02168.x. [DOI] [PubMed] [Google Scholar]
- 21.Bustamante VH, et al. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol Microbiol. 2001;39:664–678. doi: 10.1046/j.1365-2958.2001.02209.x. [DOI] [PubMed] [Google Scholar]
- 22.Huang LH, Syu WJ. GrlA of enterohemorrhagic Escherichia coli O157:H7 activates LEE1 by binding to the promoter region. J Microbiol Immunol Infect. 2008;41:9–16. [PubMed] [Google Scholar]
- 23.Jobichen C, et al. Structure of GrlR and the implication of its EDED motif in mediating the regulation of type III secretion system in EHEC. PLoS Pathog. 2007;3:e69. doi: 10.1371/journal.ppat.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhatt S, et al. The RNA binding protein CsrA is a pleiotropic regulator of the locus of enterocyte effacement pathogenicity island of enteropathogenic Escherichia coli. Infect Immun. 2009;77:3552–3568. doi: 10.1128/IAI.00418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen AM, Kaper JB. Hfq affects the expression of the LEE pathogenicity island in enterohaemorrhagic Escherichia coli. Mol Microbiol. 2009;73:446–465. doi: 10.1111/j.1365-2958.2009.06781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shakhnovich EA, et al. Hfq negatively regulates type III secretion in EHEC and several other pathogens. Mol Microbiol. 2009;74:347–363. doi: 10.1111/j.1365-2958.2009.06856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laaberki MH, et al. Concert of regulators to switch on LEE expression in enterohemorrhagic Escherichia coli O157:H7: interplay between Ler, GrlA, HNS and RpoS. Int J Med Microbiol. 2006;296:197–210. doi: 10.1016/j.ijmm.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 28.Iyoda S, Watanabe H. ClpXP protease controls expression of the type III protein secretion system through regulation of RpoS and GrlR levels in enterohemorrhagic Escherichia coli. J Bacteriol. 2005;187:4086–4094. doi: 10.1128/JB.187.12.4086-4094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lodato PB, Kaper JB. Post-transcriptional processing of the LEE4 operon in enterohaemorrhagic Escherichia coli. Mol Microbiol. 2009;71:273–290. doi: 10.1111/j.1365-2958.2008.06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomoyasu T, et al. ClpXP controls the expression of LEE genes in enterohaemorrhagic Escherichia coli. FEMS Microbiol Lett. 2005;253:59–66. doi: 10.1016/j.femsle.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 31.Roe AJ, et al. Heterogeneous surface expression of EspA translocon filaments by Escherichia coli O157:H7 is controlled at the posttranscriptional level. Infect Immun. 2003;71:5900–5909. doi: 10.1128/IAI.71.10.5900-5909.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grant AJ, et al. Co-ordination of pathogenicity island expression by the BipA GTPase in enteropathogenic Escherichia coli (EPEC) Mol Microbiol. 2003;48:507–521. doi: 10.1046/j.1365-2958.2003.t01-1-03447.x. [DOI] [PubMed] [Google Scholar]
- 33.Campellone KG, et al. Increased adherence and actin pedestal formation by dam-deficient enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 2007;63:1468–1481. doi: 10.1111/j.1365-2958.2007.05602.x. [DOI] [PubMed] [Google Scholar]
- 34.Romeo T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol. 1998;29:1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x. [DOI] [PubMed] [Google Scholar]
- 35.Timmermans J, Van Melderen L. Post-transcriptional global regulation by CsrA in bacteria. Cell Mol Life Sci. 2010;67:2897–2908. doi: 10.1007/s00018-010-0381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jousselin A, et al. On the facultative requirement of the bacterial RNA chaperone, Hfq. Trends Microbiol. 2009;17:399–405. doi: 10.1016/j.tim.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 37.Carpousis AJ. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- 38.Gottesman S, et al. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli. Sequence and in vivo activities. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- 39.Ingmer H, Brondsted L. Proteases in bacterial pathogenesis. Res Microbiol. 2009;160:704–710. doi: 10.1016/j.resmic.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 40.Elliott SJ, Sperandio Vanessa, Giron Jorge A, Shin Sooan, Mellies Jay L, Wainwright Leslie, Hutcheson Steven W, McDaniel Timothy K, Kaper James B. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect and Immun. 2000;68:6115–6126. doi: 10.1128/iai.68.11.6115-6126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, et al. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol. 2005;56:1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- 43.Baker CS, et al. CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J Bacteriol. 2007;189:5472–5481. doi: 10.1128/JB.00529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hengge R. Proteolysis of sigmaS (RpoS) and the general stress response in Escherichia coli. Res Microbiol. 2009;160:667–676. doi: 10.1016/j.resmic.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 45.Nogueira T, Springer M. Post-transcriptional control by global regulators of gene expression in bacteria. Curr Opin Microbiol. 2000;3:154–158. doi: 10.1016/s1369-5274(00)00068-0. [DOI] [PubMed] [Google Scholar]
- 46.Hengge R, Turgay K. Proteolysis in prokaryotes--from molecular machines to a systems perspective. Res Microbiol. 2009;160:615–617. doi: 10.1016/j.resmic.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 47.Deng W, et al. Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect Immun. 2005;73:2135–2146. doi: 10.1128/IAI.73.4.2135-2146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang D, et al. Hierarchal type III secretion of translocators and effectors from Escherichia coli O157:H7 requires the carboxy terminus of SepL that binds to Tir. Mol Microbiol. 2008;69:1499–1512. doi: 10.1111/j.1365-2958.2008.06377.x. [DOI] [PubMed] [Google Scholar]
- 49.Romeo T, et al. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J Bacteriol. 1993;175:4744–4755. doi: 10.1128/jb.175.15.4744-4755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schubert M, et al. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat Struct Mol Biol. 2007;14:807–813. doi: 10.1038/nsmb1285. [DOI] [PubMed] [Google Scholar]
- 51.Dubey AK, et al. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA. 2005;11:1579–1587. doi: 10.1261/rna.2990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu MY, et al. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- 53.Weilbacher T, et al. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol Microbiol. 2003;48:657–670. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- 54.Tsui HC, et al. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 55.Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr Opin Microbiol. 2010;13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 56.Link TM, et al. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci U S A. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valentin-Hansen P, et al. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 58.Soper T, et al. Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci U S A. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toledo-Arana A, et al. Small noncoding RNAs controlling pathogenesis. Curr Opin Microbiol. 2007;10:182–188. doi: 10.1016/j.mib.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 60.Sledjeski DD, et al. Hfq is necessary for regulation by the untranslated RNA DsrA. J Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lease RA, Belfort M. Riboregulation by DsrA RNA: trans-actions for global economy. Mol Microbiol. 2000;38:667–672. doi: 10.1046/j.1365-2958.2000.02162.x. [DOI] [PubMed] [Google Scholar]
- 62.Majdalani N, et al. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci U S A. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lease RA, et al. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc Natl Acad Sci U S A. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010;29:3094–3107. doi: 10.1038/emboj.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grimaud R, et al. Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J Biol Chem. 1998;273:12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- 66.Perez JC, Groisman EA. Evolution of transcriptional regulatory circuits in bacteria. Cell. 2009;138:233–244. doi: 10.1016/j.cell.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heusipp G, et al. DNA adenine methylation and bacterial pathogenesis. Int J Med Microbiol. 2007;297:1–7. doi: 10.1016/j.ijmm.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 68.Geier GE, Modrich P. Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease. J Biol Chem. 1979;254:1408–1413. [PubMed] [Google Scholar]
- 69.Iyoda S, et al. The GrlR-GrlA regulatory system coordinately controls the expression of flagellar and LEE-encoded type III protein secretion systems in enterohemorrhagic Escherichia coli. J Bacteriol. 2006;188:5682–5692. doi: 10.1128/JB.00352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Erdem AL, et al. Host protein binding and adhesive properties of H6 and H7 flagella of attaching and effacing Escherichia coli. J Bacteriol. 2007;189:7426–7435. doi: 10.1128/JB.00464-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miranda RL, et al. Glycolytic and gluconeogenic growth of Escherichia coli O157:H7 (EDL933) and E. coli K-12 (MG1655) in the mouse intestine. Infect Immun. 2004;72:1666–1676. doi: 10.1128/IAI.72.3.1666-1676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Flynn JM, et al. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 73.Perez JC, et al. Evolution of a bacterial regulon controlling virulence and Mg(2+) homeostasis. PLoS Genet. 2009;5:e1000428. doi: 10.1371/journal.pgen.1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sonnleitner E, et al. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb Pathog. 2003;35:217–228. doi: 10.1016/s0882-4010(03)00149-9. [DOI] [PubMed] [Google Scholar]
- 75.Mitobe J, et al. Involvement of RNA-binding protein Hfq in the osmotic-response regulation of invE gene expression in Shigella sonnei. BMC Microbiol. 2009;9:110. doi: 10.1186/1471-2180-9-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mitobe J, et al. Involvement of RNA-binding protein Hfq in the post-transcriptional regulation of invE gene expression in Shigella sonnei. J Biol Chem. 2008;283:5738–5747. doi: 10.1074/jbc.M710108200. [DOI] [PubMed] [Google Scholar]
- 77.Ding Y, et al. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol Microbiol. 2004;53:345–354. doi: 10.1111/j.1365-2958.2004.04142.x. [DOI] [PubMed] [Google Scholar]