Abstract
CryptoDB (http://CryptoDB.org) represents a collaborative effort to locate all genome data for the apicomplexan parasite Cryptosporidium parvum in a single user-friendly database. CryptoDB currently houses the genomic sequence data for both the human type 1 H strain and the bovine type 2 IOWA strain in addition to all other available EST and GSS sequences obtained from public repositories. All data are available for data mining via BLAST, keyword searches of pre-computed BLASTX results and user-defined or PROSITE motif pattern searches. Release 1.0 of CryptoDB contains ∼19 million bases of genome sequence for the H and IOWA strains and an additional ∼24 million bases of GSS and EST sequence obtained from other sources. Open reading frames greater than 50 and 100 amino acids have been generated for all sequences and all data are available for bulk download. This database, like other apicomplexan parasite databases, has been built utilizing the PlasmoDB model.
INTRODUCTION
Cryptosporidium parvum, an apicomplexan coccidian parasite, is the causative agent of cryptosporidiosis and an important AIDS pathogen. Cryptosporidium is capable of infecting a large number of mammalian hosts and causes an acute gastrointestinal disease that can lead to death in immunosuppressed individuals (1,2). The oocysts produced by Cryptosporidium are extremely hardy, easily spread via water, and difficult to inactivate or remove from water intended for consumption without the use of filtration (3). For these reasons, C.parvum is categorized as an NIH category B biodefense agent (http://www.niaid.nih.gov/biodefense/bandc_priority.htm).
Cryptosoridium parasites are notoriously difficult to work with in the laboratory and long-term culture is not available. To facilitate Cryptosporidium research, two genome projects were undertaken: one for the human type 1 strain H (http://www.parvum.mic.vcu.edu/) and one for the bovine type strain IOWA (http://www.cbc.umn.edu/ResearchProjects/AGAC/Cp/index.htm). The C.parvum genome is quite accessible at a size of ∼9 million bp and eight chromosomes. We are currently several years past physical mapping of the genome (4) and the initiation of EST, GSS and two genome projects for C.parvum (5–8), and the research community is in need of better tools to access these and future genomic data. CryptoDB provides a single, unified, searchable database to transform this large and still growing quantity of data into a tool to facilitate the various lines of research conducted with this and other apicomplexan parasites.
DATA INVENTORY
Release 1.0 of CryptoDB contains the May 15, 2003 ‘locked down’ genomic sequence for two C.parvum strains, the human type 1 strain H [recently renamed Cryptosporidium hominus (9)] and a bovine type 2 strain IOWA. These genome sequences are not yet 100% complete, but they contain sufficient coverage to permit a sequence freeze for draft annotation purposes. The published and annotated sequence of chromosome VI of the IOWA strain (10) is also represented. The database does not currently contain raw trace reads or individual shotgun reads. The database contains assembled genomic contigs provided by the sequence generators ranging in size from hundreds of base pairs to 1.2 million bp in length. In addition to the data provided by the genome sequencing efforts, ∼24 Mbp of genomic survey sequence (GSS) and expressed sequence tag (EST) data are incorporated. Open reading frames (ORFs) greater than 50 and 100 amino acids in length have been calculated for all nucleic acid sequences in all six reading frames. Given the paucity of introns in Cryptosporidium (7), the ORF sets are particularly useful for gene identification. All data sets are available for bulk download or analysis using a variety of tools.
ANALYSIS RESOURCES
CryptoDB is designed to be a resource where users actively search the available data to find their gene(s) of interest. As the data are unannotated, three tools are provided to facilitate gene discovery. The first and most familiar tool is BLAST analysis. All data sets, including ORFs are available for searching individually or in combined ‘all available data’ sets via BLAST. Results from BLAST searches are linked to a sequence retrieval tool (SRT), which permits the user to immediately download all or any portion of a sequence discovered in the search. Optionally, BLAST results can be viewed as a multiple sequence alignment using MView (11).
The second tool is a complement to BLAST searching. In a typical BLAST analysis the user needs to input a sequence that is then used to search a database to obtain other similar sequences. To alleviate the task of fetching a sequence to use for BLAST searching, all nucleotide sequences for Cryptosporidium have been used to perform a BLASTX search of the GenBank non-redundant protein database. All words and EC numbers in the definition line of hits with an E-value ≤ 0.001 were collected and indexed with a pointer to the sequence(s) that gave rise to them. Users search the index by entering keywords, like ‘actin’ or ‘ribo*’ and a list of all sequences that produced BLAST hits containing the keywords along with their BLAST alignments are returned to the user for inspection (Fig. 1).
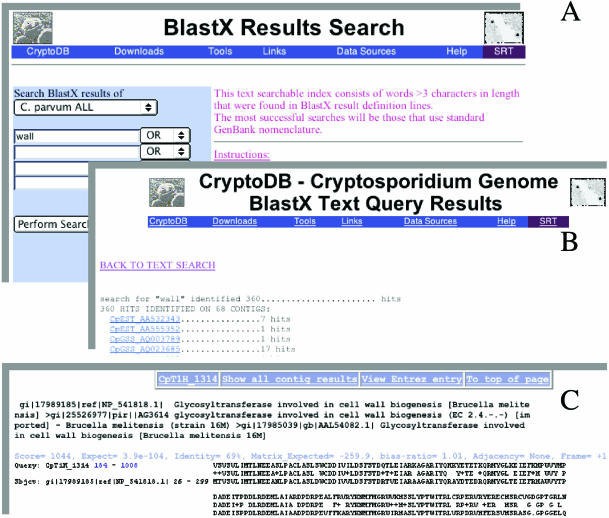
Figure 1.
Composite of screen displays demonstrating the text search tool. (A) The BLASTX results for ALL C.parvum nucleotide sequences, including ESTs, were searched for the keyword ‘wall’. (B) 360 BLASTX definition lines, generated by 68 sequences within the database, were found. (C) Examination of one of the 360 hits reveals the definition line and alignment. We see that the unannotated genomic sequence contig CpT1H_1314 (a fragment from type 1, strain H) contains a region with high similarity to glucosyltransferase, a protein described as being involved in cell wall biogenesis. We discovered this region because the GenBank definition line for this record contained our keyword, ‘wall’. A more specific search for ‘Cryptosporidium’ AND ‘oocyst’ AND ‘wall’ AND ‘protein’, would have yielded different results.
Another useful way to search for genes of interest is to search for protein motifs. In the absence of predicted genes, the motif search tool permits searches of any of the six-frame ORF data sets with defined PROSITE motifs (12), or a user-defined motif. Instructions are provided on how to generate a PERL regular expression describing your pattern including ambiguous characters (e.g. two cysteine residues followed by anything followed by a basic residue).
Pub-Crawler searches (13) of the NCBI PubMed and sequence databases are performed nightly and users are alerted to new research articles or sequence depositions for Cryptosporidium. Additional features include a ‘Links’ page with links to numerous other Cryptosporidium resources located on the internet.
SYSTEM DESIGN AND IMPLEMENTATION
Release 1.0 of CryptoDB is a flat file database which combines bioinformatics applications such as WUBLAST [W. Gish (1996–2003) http://blast.wustl.edu] and PubCrawler (13) with custom software developed and kindly provided by the PlasmoDB and GUS database projects (14–16) and subsequently modified to suit the needs of this project. The appearance of the system has intentionally been maintained in a form similar to PlasmoDB (15,16) and ToxoDB (17) to facilitate usage by researchers in the apicomplexan parasite community.
The website software is constructed with CGI scripts written in PERL and pages are currently served from an Apache web server at the University of Georgia. The SRT employs a daemon which continually listens for, and processes, sequence retrieval requests. Any sequence can be retrieved in part or whole. The data download site permits download of bulk sequence data sets in FASTA format. Live and development versions of the database are maintained at all times to ensure smooth operation of the site.
FUTURE DIRECTIONS
The current implementation of CryptoDB is designed to facilitate data mining of unannotated draft sequence. As annotation, expression and proteomic data become available, the database will migrate into the relational GUS framework to facilitate data integration and queries across data types (e.g. ‘list all genes on chromosome III that contain transmembrane domains and are up-regulated at time-point X’ or ‘list all genes indicated to be involved in glycolysis’).
Cryptosporidium is the first apicomplexan parasite to have two such closely related isolates sequenced. Since these isolates differ in aspects of their biology and normal host range, detailed comparative and SNP analyses will be of particular interest. Genome-wide synteny and SNP determinations will be available and searchable. A Cryptosporidium strain registry containing multiple sequence alignments of sequences used for classification will be created to facilitate identification of strains. Users will be able to register and deposit new strain-specific sequences in the database.
Collaborations are under way with groups at the University of Pennsylvania to create an apicomplexan comparative genome resource ‘ApiDB’, by linking CryptoDB to PlasmoDB and ToxoDB. This linkage will not disturb the individual databases and will enhance comparative studies by allowing cross-genus searches and queries.
CITING CryptoDB AND ORIGINAL DATA SOURCES
Publications and presentations benefiting from the use of this database should cite the original data source(s), the data release date(s) and the database. A table of data sources, release dates, source contact information and publications related to data contained within the database are linked to the front page of the website. CryptoDB should be cited via the URL (http://CryptoDB.org) and this publication.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank the GUS and PlasmoDB development teams for their generosity and software support.
REFERENCES
- 1.Dubey J.P., Speer,C.A. and Fayer,R. (1990) Cryptosporidiosis of Man and Animals. CRC Press, Inc., Boca Raton, FL. [Google Scholar]
- 2.Peterson C. (1992) Cryptosporidiosis in patients infected with the human immunodeficiency virus. Clin. Infect. Dis., 15, 903–909. [DOI] [PubMed] [Google Scholar]
- 3.Fayer R. (1997) Cryptosporidium and Cryptosporidiosis. CRC Press, Inc., Boca Raton, FL. [Google Scholar]
- 4.Piper M.B., Bankier,A.T. and Dear,P.H. (1998) A HAPPY map of Cryptosporidium parvum. Genome Res., 8, 1299–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strong W. and Nelson,R. (2000) Gene discovery in Cryptosporidium parvum: expressed sequence tags and genome survey sequences. Contrib. Microbiol., 6, 92–115. [DOI] [PubMed] [Google Scholar]
- 6.Strong W.B. and Nelson,R.G. (2000) Preliminary profile of the Cryptosporidium parvum genome: an expressed sequence tag and genome survey sequence analysis. Mol. Biochem. Parasitol., 107, 1–32. [DOI] [PubMed] [Google Scholar]
- 7.Liu C., Vigdorovich,V., Kapur,V. and Abrahamsen,M.S. (1999) A random survey of the Cryptosporidium parvum genome. Infect. Immun., 67, 3960–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Widmer G., Akiyoshi,D., Buckholt,M.A., Feng,X., Rich,S.M., Deary,K.M., Bowman,C.A., Xu,P., Wang,Y., Wang,X. et al. (2000) Animal propagation and genomic survey of a genotype 1 isolate of Cryptosporidium parvum. Mol. Biochem. Parasitol., 108, 187–197. [DOI] [PubMed] [Google Scholar]
- 9.Morgan-Ryan U.M., Fall,A., Ward,L.A., Hijjawi,N., Sulaiman,I., Fayer,R., Thompson,R.C., Olson,M., Lal,A. and Xiao,L. (2002) Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J. Eukaryot. Microbiol., 49, 433–440. [DOI] [PubMed] [Google Scholar]
- 10.Bankier A.T., Spriggs,H.F., Fartmann,B., Konfortov,B.A., Madera,M., Vogel,C., Teichmann,S.A., Ivens,A. and Dear,P.H. (2003) Integrated mapping, chromosomal sequencing and sequence analysis of Cryptosporidium parvum. Genome Res., 13, 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown N.P., Leroy,C. and Sander,C. (1998) MView: a web-compatible database search or multiple alignment viewer. Bioinformatics, 14, 380–381. [DOI] [PubMed] [Google Scholar]
- 12.Sigrist C.J., Cerutti,L., Hulo,N., Gattiker,A., Falquet,L., Pagni,M., Bairoch,A. and Bucher,P. (2002) PROSITE: a documented database using patterns and profiles as motif descriptors. Brief. Bioinform., 3, 265–274. [DOI] [PubMed] [Google Scholar]
- 13.Hokamp K. and Wolfe,K. (1999) What’s new in the library? What’s new in GenBank? Let PubCrawler tell you. Trends Genet., 15, 471–472. [DOI] [PubMed] [Google Scholar]
- 14.Davidson S.B., Crabtree,J., Brunk,B., Schug,J., Tannen,V., Overton,G.C. and Stoeckert,J.C.J. (2001) K2/Kleisli and GUS: Experiments in integrated access to genomic data sources. IBM Systems J., 40, 512–531. [Google Scholar]
- 15.Kissinger J.C., Brunk,B.P., Crabtree,J., Fraunholz,M.J., Gajria,B., Milgram,A.J., Pearson,D.S., Schug,J., Bahl,A., Diskin,S.J. et al. (2002) The Plasmodium genome database. Nature, 419, 490–492. [DOI] [PubMed] [Google Scholar]
- 16.Bahl A., Brunk,B., Crabtree,J., Fraunholz,M.J., Gajria,B., Grant,G.R., Ginsburg,H., Gupta,D., Kissinger,J.C., Labo,P. et al. (2003) PlasmoDB: the Plasmodium genome resource. A database integrating experimental and computational data. Nucleic Acids Res., 31, 212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kissinger J.C., Gajria,B., Li,L., Paulsen,I.T. and Roos,D.S. (2003) ToxoDB: accessing the Toxoplasma gondii genome. Nucleic Acids Res., 31, 234–236. [DOI] [PMC free article] [PubMed] [Google Scholar]