Abstract
Meiotic crossover (CO) formation between homologous chromosomes (homologues) entails DNA double strand break (DSB) formation, homology search using DSB ends, and synaptonemal complex (SC) formation coupled with DSB repair. Meiotic progression must be prevented until DSB repair and homologue alignment are completed to avoid forming aneuploid gametes. Here we show that mouse HORMAD1 ensures that sufficient numbers of processed DSBs are available for successful homology search. HORMAD1 is needed for normal SC formation and for the efficient recruitment of ATR checkpoint kinase activity to unsynapsed chromatin. The latter phenomenon was proposed to be important in meiotic prophase checkpoints in both sexes. Consistent with this hypothesis, HORMAD1 is essential for the elimination of SC-defective oocytes. SC formation results in HORMAD1 depletion from chromosome axes. Thus, we propose that SC and HORMAD1 are key components of a negative feedback loop that coordinates meiotic progression with homologue alignment: HORMAD1 promotes homologue alignment and SC formation, and SCs down-regulate HORMAD1 function, thereby permitting progression past meiotic prophase checkpoints.
INTRODUCTION
Physical linkages between homologues ensure correct chromosome segregation during the first meiotic division in mammals. These physical linkages, called chiasmata, depend on the formation of at least one reciprocal recombination event, or CO, between each homologue pair and on cohesion between pairs of sister chromatids (Supplementary Information, Fig. S1a)1, 2. CO formation begins with the introduction of DSBs into the genome by the SPO11 enzyme (Supplementary Information, Fig. S1)3-5. DSBs are processed to produce single-stranded DNA ends that can be used to probe for homology through strand invasion6. Several DSB ends work together on each homologue pair to ensure successful homologue alignment. After successful homology search, SCs form and connect the axes of aligned homologues. SC components promote post-homology search steps in DSB repair and are required for efficient CO formation1, 2. After SCs formation, homology search is no longer needed, most DSBs become repaired from homologues as non-crossovers, and at least one DSB per chromosome pair is turned into a CO1, 2. In mammals, meiotic checkpoint mechanisms eliminate meiocytes with defects in homologue alignment and DSB repair during the first meiotic prophase, thereby ensuring that it is rare for gametes to form with an abnormal chromosome set or with unrepaired DNA7-14. Despite the importance of these meiotic prophase checkpoint mechanisms, they are poorly understood.
In various non-mammalian taxa, meiotic HORMA (Hop1, Rev7 and Mad2)-domain proteins have been implicated in diverse processes linked to CO formation2, 15-38. These include DSB formation, homology search, preferred use of homologous DNA over sister DNA for repair of DSBs, SC formation and the meiotic prophase checkpoint. Here we address the functions of HORMAD1, one of two meiosis-specific mouse HORMA-domain proteins (HORMAD1 and HORMAD2) that were shown to preferentially associate with unsynapsed chromosome axes during first meiotic prophase in mice39-41.
RESULTS
HORMAD1 is required for fertility
Reasoning that functional analysis of HORMADs might provide novel insights into meiotic chromosome behaviour and CO formation in mammals, we disrupted Hormad1 in mouse (Supplementary Information, Fig. S2). While no obvious somatic defects were observed in Hormad1−/− mice, both sexes are sterile, as reported by others as well41. Although spermatocytes in Hormad1−/− mice are present in testis tubules at epithelial cycle stage III-IV, which we identified by the presence of intermediate spermatogonia42, they undergo apoptosis by the end of stage IV, and post-meiotic cells are not found in Hormad1−/− testes (Supplementary Information, Fig. S3). In wild type (WT), stage IV tubules contain mid-pachytene spermatocytes42; thus Hormad1−/− spermatocytes are eliminated at a stage equivalent to mid-pachytene. Since spermatocytes with defects in SC formation and DSB repair are eliminated by the mid-pachytene checkpoint7-14 we examined SC formation on nuclear surface spreads of Hormad1−/− spermatocytes.
HORMAD1 promotes SC formation
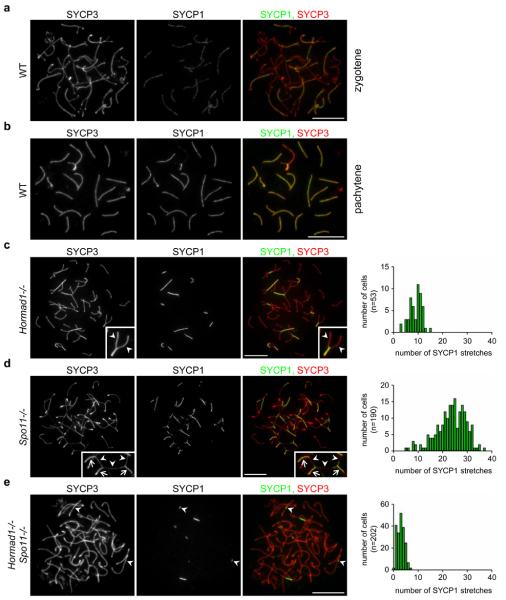
In WT spermatocytes, chromosome axes are fully formed by late-zygotene and SC formation on autosomes is completed by pachytene (Fig. 1a, b). While chromosome axis-cohesion core development and the timing of SC formation are similar in WT and Hormad1−/− spermatocytes, the efficiency of stable SC initiation and SC elongation is reduced in the mutant (Fig. 1c, Supplementary Information, Fig. S4). Autosomal SC formation is never completed in Hormad1−/− cells with fully formed chromosome axes (n=1000); most chromosomes that start SC formation do not complete it, and many chromosomes do not even partially synapse (Fig. 1c). Due to these defects we cannot distinguish between late-zygotene and pachytene in mutant spermatocytes and we refer to these stages as zygotene-pachytene. Unlike in SC transverse filament mutant meiocytes, where unsynapsed chromosomes align along their length8-11, unsynapsed chromosomes do not align in Hormad1−/− spermatocytes (Fig. 1c). Nevertheless, based on the similar axis lengths of synapsed chromosomes, the relatively long stretches of SCs that frequently form in zygotene-pachytene Hormad1−/− spermatocytes appear to connect homologues (Fig. 1c). SC formation between non-homologous chromosomes is unambiguously identifiable only in a small fraction of Hormad1−/− spermatocytes (2.3% n=174) (data not shown). Similar homologue alignment and SC formation defects are found in Hormad1−/− oocytes (Supplementary Information, Fig. S5). Others reported complete lack of SCs in Hormad1−/− spermatocytes based on the lack of tripartite SC structures observable by electron microscopy41. The different conclusion reached by us may reflect differences in strain backgrounds and/or SC detection methods in the two studies.
Figure 1. HORMAD1 promotes SC formation independently of DSB-dependent processes.
Images of SYCP3 (chromosome axis) and SYCP1 (SC transverse filament), detected by immunofluorescence (IF) on nuclear spreads of WT zygotene (a), WT pachytene (b) and mutant zygotene-pachytene spermatocytes (c-e left) collected from 14-weeks-old mice. The frequency distribution of SC stretches in mutant zygotene-pachytene spermatocytes is shown (c-e right). c, SC formation is never completed on all autosomes in Hormad1−/− cells, and unlike in SC transverse filament mutant meiocytes, where unsynapsed chromosomes align along their length8-11, unsynapsed chromosomes do not align in Hormad1−/− spermatocytes. Nevertheless, robust stretches of SC frequently form between chromosomes that appear homologous based on their similar axis length. See enlarged view of boxed partially synapsed autosome: unsynapsed axes (arrowheads) are of similar lengths. d, In contrast, SCs connect multiple non-homologous axes, thereby creating a meshwork of interconnected axes in Spo11−/− mutant, in which strand invasion and homology search is not possible due to lack of DSBs. See enlarged view of boxed chromosome axes: arrowheads mark unsynapsed, arrows mark synapsed axes. e. Both the number and the length of SYCP1 stretches are reduced in Hormad1−/− Spo11−/− spermatocytes relative to the single mutants. The majority (61% in n=144 cells) of the remaining SYCP1 stretches is unambiguously linked to a single chromosome axis (arrowheads) indicating that SYCP1 stretches do not necessarily mark inter-chromosomal SCs. Bars, 10 μm.
Homologue alignment defects could explain the SC defects in the Hormad1−/− mutant. Additionally, HORMAD1 may also have SC promoting functions independent of its role in homologue alignment. To test this possibility, we examined the effect of the Hormad1 mutation in the DSB-deficient Spo11−/− background, where homology search fails and SCs form extensively between non-homologous partners (Fig. 1)4, 5. If HORMAD1 promotes SC formation only through a function in DSB repair and homologue alignment, Spo11−/− Hormad1−/− double mutant meiocytes should be as proficient in non-homologous SC formation as Spo11−/− single mutant meiocytes. Notably, the double mutant has a much more severe SC defect than either single mutant in both sexes (Fig. 1, Supplementary Information, Fig. S5). It is unlikely that the double mutant SC-phenotype is caused by an early block in meiotic progression, because single and double mutant spermatocytes are eliminated at a comparable stage (Supplementary Information, Fig. S6). Hence, our findings indicate that HORMAD1 promotes SC formation independent of homology search and raise the possibility that HORMAD1 has a direct role in SC formation.
HORMAD1, DSB formation and DSB repair
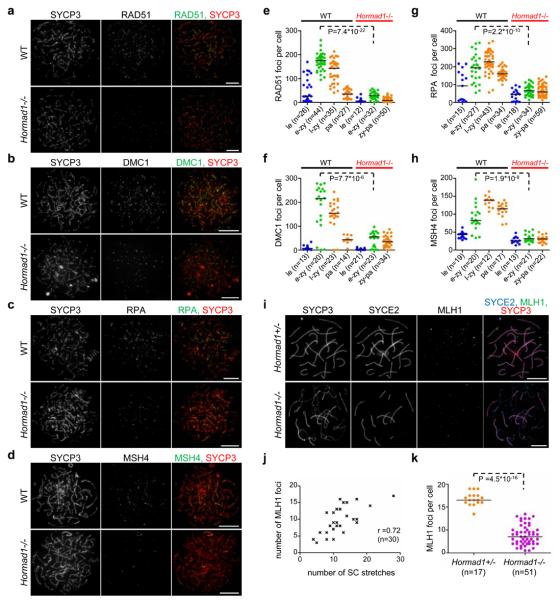
Early DSB repair steps are important for homology search. Therefore, we examined if chromosome alignment-SC defects in Hormad1−/− meiocytes reflect DSB repair defects. The first step in DSB repair is DNA end resection, which creates single-stranded 3′ overhangs bound by two recombinases, RAD51 and DMC1, that facilitate homology search1, 2, 6, 43-45. RAD51- and DMC1-marked meiotic DSB sites are chromosome axes-associated, and RAD51-DMC1 focus counts provide an estimate of the number of DSBs available for homology search1. Upon SC formation, RAD51 and DMC1 are replaced at most DSB sites by markers of intermediate stages of DSB repair, including the single-stranded DNA binding protein RPA and MSH41, 46. RAD51, DMC1, RPA and MSH4 foci numbers are significantly lower in Hormad1−/− spermatocytes than in WT cells during observable prophase stages (Fig. 2a-h, Supplementary Information, Fig. S7a-d)41. During the comparable early-mid-zygotene stage median foci numbers are three- to six-fold reduced in Hormad1−/− spermatocytes. (Fig. 2e-h). RAD51 and RPA focus numbers are similarly reduced in Hormad1−/− oocytes (Supplementary Information, Fig. S7e, f)41. Thus the steady state numbers of single-stranded DSB ends appear strongly reduced in Hormad1−/− meiocytes, which could explain the inefficient homologue alignment and contribute to defective SC formation.
Figure 2. Numbers of early, intermediate and late recombination protein foci are reduced in the absence of HORMAD1 in prophase meiocytes.
a-d, SYCP3 and either RAD51 (a) DMC1 (b), RPA (c) or MSH4 (d) are detected on nuclear spreads of typical early-mid-zygotene WT and Hormad1−/− spermatocytes from 16-days-old mice. Bars, 10 μm. e-h, Foci numbers of early (RAD51-e, DMC1-f) and intermediate (RPA-g, MSH4-h) recombination proteins during leptotene (le) and early-mid-zygotene (e-zy) in WT and Hormad1−/−; late-zygotene (l-zy) and pachytene (pa) in WT and zygotene-pachytene (zy-pa) in Hormad1−/− spermatocytes. Median foci numbers are marked. During the comparable early-mid-zygotene stage, a three- to six-fold reduction (highly significant by Mann–Whitney test) is observed in recombination protein foci numbers in the mutant relative to WT. i-k, Focus numbers of the CO marker MLH1 are reduced in the absence of HORMAD1 in oocytes. i, SYCP3 (chromosome axis), SYCE2 (SC central element) and MLH1 were detected by IF in nuclear spreads of Hormad1+/− and Hormad1−/− oocytes from 19.5 dpc foetuses (a stage when most oocytes are in the late-pachytene or diplotene stage in WT). Fewer chromosome axis-associated MLH1 foci are detected in Hormad1−/− oocytes than in Hormad1+/− oocytes. Note that the majority of MLH1 foci (78%, n=51 cells) are observed on synapsed axes in the mutant. Bars, 10 μm. j, Scatter plot shows positive correlation (Spearman's r=0.72, n=30) between MLH1 foci numbers and the number of SC stretches (immunostaining for SYCE1 or SYCE2) in Hormad1−/− oocytes, indicating that DSBs might be repaired as COs preferentially in chromosome regions that align and synapse, or that synapsis occurs preferentially where COs are successfully designated. k, Chromosome axis-associated MLH1 focus numbers are reduced approximately three-fold in Hormad1−/− oocytes from 18.5-19.5 dpc foetuses relative to Hormad1+/− oocytes. Median focus numbers are marked by horizontal lines.
Two scenarios could explain the reduced numbers of processed DSB ends in Hormad1−/− meiocytes: either fewer processed DSBs are produced, or DSBs are repaired faster. Recombination protein foci first appeared at similar times (but fewer in number) and did not disappear prematurely in Hormad1−/− spermatocytes (Fig. 2e-h). In particular, we did not see dramatic early decline in RPA focus numbers, which would indicate premature repair. We also examined the behaviour of the late recombination marker MLH1, which appears at destined CO sites from mid-pachytene onwards in WT (Fig. 2i-k)46-48. We never observed MLH1 foci in Hormad1−/− spermatocytes (n=378). Defective MLH1 foci formation in Hormad1−/− spermatocytes41 is unlikely to be the result of a direct block in DSB repair, because MLH1 foci are detected (preferentially associated with synapsed axes) in Hormad1−/− oocytes, albeit in reduced numbers (~30% of WT) (Fig. 2i-k). This difference between oocytes and spermatocytes is likely due to different timing of elimination of defective meiocytes, with oocytes eliminated later, allowing stages corresponding to late-pachytene to be examined. Although we cannot exclude that some DSB repair steps are accelerated in Hormad1−/− meiocytes, the behaviour of recombination proteins does not indicate significant acceleration.
We next asked how efficiently DSBs and/or single-stranded DSB ends are produced in the Hormad1−/− mutant. DSBs trigger accumulation of phospho-histone H2AX (γH2AX) during leptotene-early-zygotene through activation of ATM kinase49, 50. Levels of chromatin-bound γH2AX are significantly reduced in Hormad1−/− leptotene-early-zygotene spermatocytes (Supplementary Information, Fig. S8). Consistent with this, others reported reduced γH2AX levels and reduced ATM autophosphorylation, implying reduced ATM activation, in Hormad1−/− spermatocytes41.
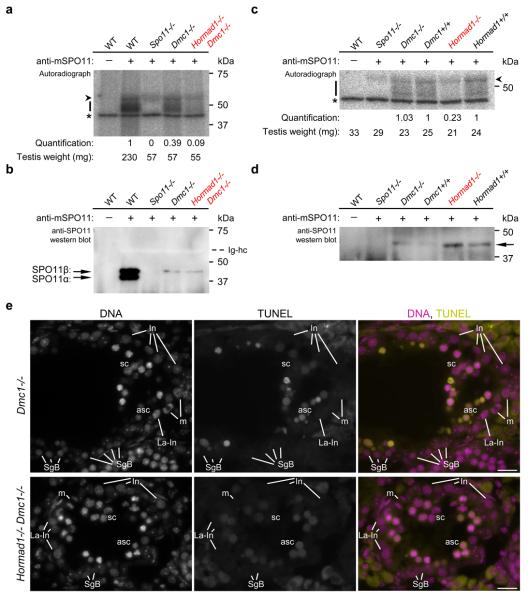
Since this result may indicate reduced DSB formation, we measured the effect of Hormad1−/− mutation on amounts of SPO11-oligonucleotide complexes that are produced when DSB ends are resected45 (Fig. 3a-d). To control for the fact that the Hormad1−/− mutant displays a mid-pachytene spermatogenic block, we employed the DSB repair-defective Dmc1−/− background: DMC1-deficient spermatocytes arrest in mid-pachytene irrespective of Hormad1 genotype (Fig. 3e)12, 43, 44. In three independent experiments with adult mice we observed 2-, 3.1- and 4.2-fold reduction in testis-weight-normalised SPO11-oligonucleotide levels in Hormad1−/− Dmc1−/− testes relative to Hormad1+/+ Dmc1−/− controls (Fig. 3a). We also examined SPO11-oligonucleotide levels in testes of juvenile 14 days postpartum-dpp mice (Fig. 3c), in which the sizes and cellularities of WT and mutant testes are comparable because the vast majority of spermatocytes have not yet progressed far enough in meiosis to be affected by the mid-pachytene checkpoint. In two independent experiments testis-weight-normalised SPO11-oligonucleotide levels were reduced 3.8- and 4.8-fold in Hormad1−/− testes relative to WT controls, whereas WT and Dmc1−/− testes were similar. Although we cannot exclude that turnover of both SPO11-oligonucleotide complexes and single-stranded DSB ends are faster in the Hormad1−/− mutant, the simplest interpretation of our observations is that HORMAD1 is needed for efficient formation of DSBs and/or single-stranded DSB ends.
Figure 3. Amounts of SPO11-oligonucleotide complexes in testes are reduced in the absence of HORMAD1.
a, c, Measurement of SPO11-oligonucleotide complexes in testes of adult 14-weeks-old (a) and juvenile 14 dpp (c) mice. SPO11-oligonucleotide complexes were immunoprecipitated with or without anti-SPO11 antibodies, and covalently-linked oligonucleotides were radioactively labelled. Each sample represents one testis-equivalent SPO11-oligonucleotide complexes. Bars mark SPO11-specific signals and asterisks indicate non-specific labelling of a contaminant in the terminal deoxytransferase preparations. Arrowhead marks an artifactual radioactive signal attributable to presence of immunoglobulin heavy chain (Ig-hc in b). Quantified radioactive signals in the mutants have been background-corrected and normalised: in a signals are normalised to the adult WT control; in c Dmc1−/− and Hormad1−/− signals are normalised to their litter mate Dmc1+/+ and Hormad1+/+ controls, respectively (see Methods). Blots of immunoprecipitates from a and c were probed with anti-SPO11 antibodies in b and d, respectively. b, In WT adults, two alternative forms of SPO11 (α and β) are present5. Total SPO11 amounts are similar in Dmc1−/− and Hormad1−/− Dmc1−/− mutants, and are lower in the mutants than in WT. Only SPO11β, the form that appears early in meiosis, is detected in the mutants. Arrowhead marks the immunoglobulin heavy chain (bleached out signal). Mid-pachytene spermatogenic block in the mutants (e) is the likely cause of reduced SPO11 amounts in Dmc1−/− and Hormad1−/− Dmc1−/− testes, and of reduced SPO11-oligonucleotide amounts in Dmc1−/− testes12, 43, 44. d, In juveniles, total SPO11 protein levels are low and only the long β form of SPO11 (arrow) is detectable5. For full scan gel-images of a-d see Supplementary Information, Fig, S10. e, DNA was detected by DAPI, and apoptosis was detected by IF-TUNEL assay on cryosections of testes of 15-weeks-old mice. Dmc1−/− and Hormad1−/− Dmc1−/− spermatocytes undergo apoptosis in stage IV tubules as identified by the concomitant presence of intermediate spermatogonia (In), late-prometaphase intermediate spermatogonia (La-In), mitotic intermediate spermatogonia (m) and spermatogonia B (SgB). Both non-apoptotic (sc) and apoptotic (asc) spermatocytes are present in the stage IV tubules shown. Spermatocytes are fully eliminated upon progression to stage V (data not shown). Bars, 20 μm.
HORMAD1 and meiotic prophase checkpoints
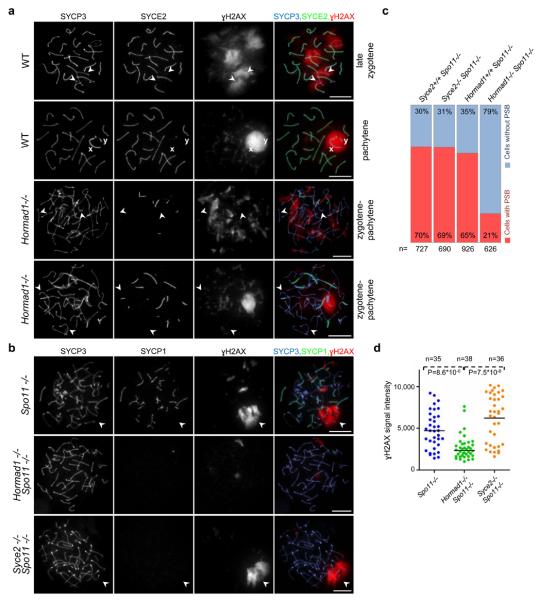
Collaboration between Hop1 and Mec1, the yeast orthologues of HORMAD1 and ATR respectively, is required for the meiotic prophase checkpoint in budding yeast35. Interestingly, HORMAD1 preferentially associates with unsynapsed chromosome axes, resembling the behaviour of ATR and two ATR activators, BRCA1 and TOPBP1 in WT meiocytes51-57. Active ATR phosphorylates H2AX on unsynapsed chromatin from zygotene onwards, thereby promoting meiotic silencing of unsynapsed chromosomes (MSUC), a phenomenon that is crucial for the mid-pachytene checkpoint13, 14, 57. Therefore, we asked whether HORMAD1 also has a role in these processes. In spermatocytes, sex chromosomes only synapse and recombine in their short pseudoautosomal regions. Consequently, most sex chromosome regions remain unsynapsed and silenced in a γH2AX-rich structure termed the sex body57-59. Efficient sex chromosome silencing is essential for progression beyond mid-pachytene60, 61. In DSB repair-SC defective spermatocytes, ATR activity (γH2AX) persists on unsynapsed autosomes, and fails to accumulate strongly on sex chromosomes8-12, 60. Consequently, sex body formation and sex chromosome silencing fail, resulting in mid-pachytene apoptosis60, 61. Sex body formation is defective in Hormad1−/− spermatocytes (Fig. 4a)41. Nevertheless, γH2AX was observed accumulating in chromatin regions that are not associated strongly with unsynapsed chromosome axes in Hormad1−/− spermatocytes. This abnormal localisation of γH2AX could result either from reduced DSB numbers and incomplete SC formation or from a more direct involvement of HORMAD1 in the recruitment of ATR activity to unsynapsed chromatin.
Figure 4. HORMAD1 is required for sex body and pseudo-sex body formation in the Spo11+/+ and Spo11−/− backgrounds, respectively.
a, SYCP3 (chromosome axis), SYCE2 (SC central element) and γH2AX were detected in nuclear spreads of WT late-zygotene and pachytene, and Hormad1−/− zygotene-pachytene spermatocytes collected from 16-days-old mice. Matched exposure images of γH2AX are shown. In WT late-zygotene spermatocytes, γH2AX chromatin domains associate with unsynapsed chromosome axes. In WT pachytene cells, unsynapsed regions of X and Y sex chromosome axes (marked by x and y) are surrounded by one large γH2AX-rich chromatin domain, the sex body. Anti-γH2AX staining is patchy in the majority (59%) of Hormad1−/− spermatocytes, with no clear correlation between lack of synapsis and γH2AX localisation (third row). A few large γH2AX-rich chromatin domains form in a minority of Hormad1−/− spermatocytes (41%, n=512), but only a subset of unsynapsed axes overlap with γH2AX-rich chromatin, and synapsed axes overlapping with γH2AX-rich chromatin domains are also observed regularly (bottom row). Arrowheads mark two unsynapsed axes in WT late-zygotene and in each mutant cell. Bars, 10 μm. b, Matched exposure images of SYCP3 (chromosome axis), SYCP1 (SC transverse filament) and γH2AX in nuclear spreads of Spo11−/− , Hormad1−/− Spo11−/− and Syce2−/− Spo11−/− spermatocytes of adult (9-weeks-old) mice. Large γH2AX-rich chromatin domains-pseudo-sex bodies (marked by arrowheads) frequently form in Spo11−/− and Syce2−/− Spo11−/− spermatocytes. Bars, 10 μm. c, Quantification of pseudo-sex body formation in spermatocytes with full-length chromosome axes (collected from 24-days-old mice). The percentage of cells with no pseudo-sex body (cells without PSB) or with one to three clear pseudo-sex bodies (cells with PSB) is shown. d, Quantification of γH2AX signal in spermatocytes with full-length chromosome axes (collected from 24-days-old mice) shows a significant reduction in total nuclear γH2AX amounts in Hormad1−/− Spo11−/− relative to Spo11−/− and Syce2−/− Spo11−/− (Mann–Whitney test).
To test these two possibilities, we examined the effect of Hormad1−/− mutation on the MSUC pathway in a Spo11−/− background4, 5. Although SC formation is abnormal in Spo11−/− spermatocytes, MSUC is active and a random subset of unsynapsed chromosome axes is frequently associated with one or a few γH2AX-rich silenced chromatin domains termed pseudo-sex bodies (Fig. 4b)12, 50. Since SC formation is severely compromised in Hormad1−/− Spo11−/− meiocytes relative to Spo11−/− mutants (Fig. 1), we controlled for loss of SCs by comparing pseudo-sex body formation and the behaviour of MSUC proteins in the Hormad1−/− Spo11−/− double mutant with the Syce2−/− Spo11−/− double mutant, which lacks an SC central element component and does not form SCs9. Formation of pseudo-sex body-like chromatin domains is strongly reduced in Hormad1−/− Spo11−/− double mutant spermatocytes relative to both Spo11−/− and Syce2−/− Spo11−/− controls (Fig. 4b, c). Lack of HORMAD1 strongly reduces total nuclear γH2AX immunofluorescence staining in the Spo11−/− background, whereas loss of SC formation does not (Fig. 4d). Both ATR and TOPBP1 accumulate within pseudo-sex bodies, and BRCA1 accumulates on chromosome axes associated with pseudo-sex bodies (Fig. 5)12, 50, 60. We observed reduced localisation of TOPBP1, ATR and BRCA1 to unsynapsed chromatin in the Hormad1−/− Spo11−/− double mutant in comparison with both control strains (Fig. 5). This phenotype is probably not caused by an early block in meiotic progression, since Spo11−/−, Syce2−/− Spo11−/− and Hormad1−/− Spo11−/− spermatocytes are eliminated in stage IV tubules (Supplementary Information, Fig. S6). The most straightforward interpretation is that HORMAD1 promotes recruitment of MSUC proteins to unsynapsed chromatin regions independently of its role in the formation of processed DSBs and SCs.
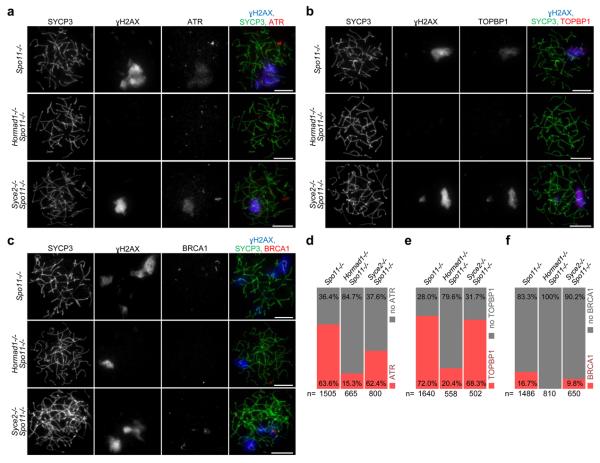
Figure 5. I HORMAD1 is required for efficient accumulation of ATR, TOPBP1 and BRCA1 on chromatin in the absence of programmed DSBs.
Matched exposure images of SYCP3 (chromosome axis), γH2AX and either ATR (a), TOPBP1 (b) or BRCA1 (c) in nuclear spreads of spermatocytes collected from 24-days-old mice. Spo11−/− and Syce2−/− Spo11−/− spermatocytes are shown with cloud-like ATR (a) or TOPBP1 (b) accumulation or with BRCA1 localised to axes (c) in γH2AX-marked pseudo-sex bodies. In a and b, Hormad1−/− Spo11−/− cells are shown without a pseudo-sex body and without ATR or TOPBP1 accumulation on chromatin, respectively. In c, Hormad1−/− Spo11−/− cell is shown with a pseudo-sex body, within which no BRCA1 association with the axis was detected. Note that only a small minority of Hormad1−/− Spo11−/− spermatocytes shows pseudo-sex body-like accumulation of γH2AX (Fig. 4c). Bars, 10 μm. d-f, Frequency of cloud-like ATR (d) and TOPBP1 (e) accumulation and frequency of BRCA1 association with axes (f) in spermatocytes with fully formed chromosome axes. ATR- and TOPBP1-rich chromatin domains are frequently observed in Spo11−/− and Syce2−/− Spo11−/− spermatocytes, and BRCA1 association with axes is also observed in these mutants. ATR and TOPBP1 are virtually absent from chromatin in the large majority of Hormad1−/− Spo11−/− cells, and BRCA1 localisation to axes was never observed in Hormad1−/− Spo11−/− spermatocytes.
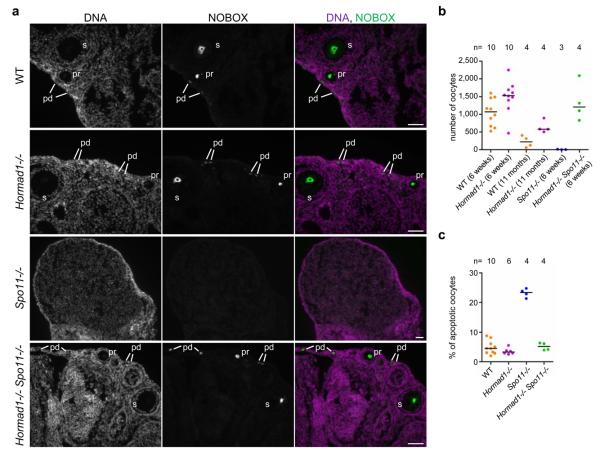
The prophase checkpoint is less well understood in oocytes than in spermatocytes, although ATR activation and MSUC has been implicated in the female checkpoint13, 57. In the absence of heterologous sex chromosomes, oocytes do not form sex bodies. Nevertheless Spo11−/− oocytes frequently contain male pseudo-sex body-like γH2AX-rich chromatin domains that also contain TOPBP1 and BRCA1 (Supplementary Information, Fig. S9)13. Formation of such female “pseudo-sex bodies” is strongly reduced in Hormad1−/− Spo11−/− oocytes (Supplementary Information, Fig. S9), indicating that HORMAD1 may have a role in MSUC and the prophase checkpoint in females. Therefore, we compared the number of oocytes in ovaries from 6-weeks-old WT, Hormad1−/−, Spo11−/− and Hormad1−/− Spo11−/− animals and in ovaries from 11-month-old WT and Hormad1−/− animals (Fig. 6a, b). In the SC-defective Spo11−/− mutant, very few oocytes survive until 6 weeks. Despite defective SC formation, oocyte numbers are not reduced in Hormad1−/−41 or Hormad1−/− Spo11−/− animals relative to WT. Notably, oocyte apoptosis rates are similar in WT, Hormad1−/− and Hormad1−/− Spo11−/− mice, and are greatly elevated in the Spo11−/− mutant one day after birth, when many of the SC-defective oocytes are eliminated (Fig. 6c). Thus, Hormad1−/− suppresses oocyte loss in the Spo11−/− background. These observations demonstrate that HORMAD1 is essential for the female prophase checkpoint that eliminates oocytes with abnormal SC formation and homologue alignment.
Figure 6. I Lack of HORMAD1 allows survival of oocytes in the SC-defective Spo11−/− mutant.
a, NOBOX (postnatal oocyte marker) was detected by IF on cryosections of ovaries from 6-weeks-old mice. DNA was detected by DAPI. Oocytes in primordial (pd), primary (pr) and secondary (s) follicular stages are shown in WT, Hormad1−/− and Hormad1−/− Spo11−/− ovaries. In the SC-defective Spo11−/− mutant, oocyte numbers are strongly reduced. A lower magnification section of a Spo11−/− ovary is shown to better illustrate the absence of oocytes. Bars, 50 μm. b, Sum of oocyte numbers on every tenth section of sectioned-through ovary pairs at the indicated ages. Each data point represents a mouse. c, Fraction of apoptotic oocytes in the ovaries of 1-day-old (1 dpp) mice of indicated genotypes. Horizontal lines show medians in b and c.
Chromosome segregation in Hormad1−/− oocytes
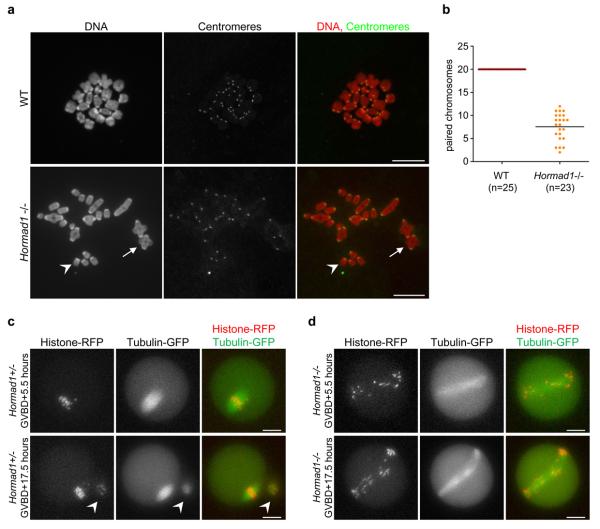
Follicular oocyte development does not require HORMAD141 since we found oocytes at all stages of follicular development in Hormad1−/− and Hormad1−/− Spo11−/− animals (Fig. 6a). This allowed us to test whether chiasmata form during the first meiotic metaphase in the absence of HORMAD1. On average, roughly one third of chromosomes were connected by chiasmata in in vitro-matured metaphase Hormad1−/− oocytes, which corresponds well to the number of MLH1 foci observed in pachytene oocytes (Fig. 2k, 7). Chiasmata formation depends both on sister chromatid cohesion along chromosome arms and on CO formation. Thus, our observations suggest that sister chromatid cohesion is intact and that MLH1-marked DSB sites are efficiently resolved as COs in the Hormad1−/− mutant.
We evaluated the consequences of reduced CO numbers on the first meiotic division in in vitro-matured oocytes. Although nuclear envelope breakdown efficiency was similar in WT (142 of 149) and Hormad1−/− (73 of 77) oocytes, polar body extrusion was greatly reduced in the mutant (5 of 60) relative to WT (92 of 95). Video microscopy shows that Hormad1−/− oocytes have abnormally long meiosis I spindles, on which chromosomes fail to align (Fig. 7c, d, Supplementary Information, Movies 1, 2). This defect likely causes anaphase failure and the polar body extrusion defect. Despite the abnormal first meiotic division, some of the Hormad1−/− oocytes are fertilised in vivo since in a few cases (3 of 11 breeding pairs) we found one to three reabsorbing embryos five to eight days after breeding began. Nevertheless, embryos never developed to full term in Hormad1−/− females (19 females, 125 breeding weeks). Infertility and loss of embryos are consistent with the meiotic chromosome segregation defects (Fig. 7c, d, Supplementary Information, Movies 1, 2) and with the reported widespread aneuloidy in early embryos in Hormad1−/− females41.
Figure 7. Reduced numbers of chiasmata form in Hormad1−/− oocytes.
a, Centromeres were detected by IF and DNA was detected by propidium iodide on nuclear spreads of in vitro-matured metaphase stage oocytes. In WT cells, 20 pairs of chromosomes are connected by chiasmata. In the Hormad1−/− mutant, a large fraction of chromosomes does not have chiasmata (one such chromosome is marked by arrowhead). Arrow marks a pair of chromosomes connected via a chiasma in the mutant oocyte. Note that bivalents are symmetrical and chiasmata form between chromosomes of identical length in the mutant, indicating that CO formation took place between homologous chromosomes. Chromosomes scatter over a larger area during nuclear spreading in the mutant, therefore it was not possible to include all 40 chromosomes of a meiosis I oocyte in the image. Bars, 10 μm. b, The average numbers of paired chromosomes connected by chiasmata (marked by line) are reduced nearly three-fold in in vitro-matured Hormad1−/− oocytes relative to WT oocytes. c and d, mRNAs encoding β-tubulin-GFP and histone H2B-RFP were injected into WT (c) and Hormad1−/− (d) oocytes during the germinal vesicle stage (prophase), and the oocytes were matured in vitro. The fluorescent proteins in the oocytes were imaged 5.5 hours after germinal vesicle break down (GVBD), at a time when WT oocytes are in the first meiotic metaphase, and 17.5 hours after GVBD, at a time when WT oocytes are arrested in the second meiotic metaphase. A polar body (marked by arrowhead in c), which was extruded 7 hours after GVBD (Supplementary Information, Movie 1) is observed next to the metaphase II stage WT oocyte at the 17.5 hour time point. In the Hormad1−/− oocyte (d), the meiotic spindle is abnormally long and chromosomes fail to align at both time points. Meiotic anaphase did not take place in the displayed Hormad1−/− oocyte (Supplementary Information, Movie 2), and no polar body can be observed at either time points (d). Bars, 20 μm.
DISCUSSION
We analysed the functions of HORMAD1 using mouse genetics. We propose three distinct biological functions for mouse HORMAD1. First, HORMAD1 increases the steady state numbers of single-stranded DSB ends, thereby facilitating homology search. Our data suggest that HORMAD1 promotes either efficient DSB formation or resection of DSB ends or both. Because budding yeast Hop1 is required for efficient DSB formation29, 35 we favour the first possibility. In budding yeast, inter-homologue CO numbers remain relatively stable when DSB numbers are reduced, a phenomenon called CO homeostasis62. Comparable reduction in the numbers of early-intermediate recombination protein foci (particularily RPA) and of chiasmata in the Hormad1−/− mutant might be interpreted as a sign of lack of CO homeostasis in mice (Fig. 2, 7). However, a trivial explanation is that CO homeostasis can function only if homology search succeeds and homologues align. Hence, the roughly three-fold reduction in CO formation may simply reflect the low number of homologues that manage to partially or fully align in Hormad1−/− meiocytes. The observed correlation between numbers of SYCP1 stretches and MLH1 foci in oocytes is consistent with this latter possibility (Fig. 2j). Our data suggest that Hormad1−/− meiocytes are proficient in DSB repair steps that are important for homology search and inter-homologue CO formation, and reduced CO formation is caused by reduced levels of single-stranded DSB ends. Consistently with this hypothesis some homologues appear to synapse at least partially in Hormad1−/− meiocytes, a conclusion based on the observations that incomplete SCs form between chomosomes of similar axis length, and that SC formation largely depends on SPO11-DSBs and presumably on the downstream process of homology search in Hormad1−/− cells (Fig. 1, Supplementary Information, Fig. S5). Furthermore, chiasmata form between chromosomes of identical length indicating that inter-homologue chiasmata, which are consequences of inter-homologue COs, form in Hormad1−/− oocytes (Fig. 7a).
Second, HORMAD1 promotes SC formation independently of its role in homology search. This conclusion is based on the observation that HORMAD1 is needed for efficient non-homologous SC formation (Fig. 1, Supplementary Information, Fig. S5). It is formally possible that HORMAD1 is required only for pathological non-homologous synapsis. However, we consider it unlikely that as a prelude to homology search unsynapsed chromosome axes are loaded with a protein (HORMAD1) that specifically promotes non-homologous synapsis. It seems more likely that HORMAD1 promotes SC formation between axes that come into close proximity, irrespective of underlying homology.
Finally, HORMAD1 plays a key role in the male mid-pachytene checkpoint and the female meiotic prophase checkpoint, both of which eliminate meiocytes with defects in DSB repair and/or in homologue alignment-SC formation. HORMAD1 is required for efficient build up of ATR activity on unsynapsed chromosome regions, a process that is believed to form the basis of MSUC and meiotic prophase quality control in both sexes13, 14, 57 (Fig. 4, 5, Supplementary Information, Fig. S9). In males, mid-pachytene apoptosis of DSB repair-SC defective spermatocytes is triggered by failure of MSUC on sex chromosomes, allowing transcription of genes whose expression is incompatible with survival beyond mid-pachytene13, 60, 61. In females, elimination of defective oocytes might be triggered by inappropriate MSUC or persistent ATR activity during prophase13. Therefore, the apparently opposite effect of Hormad1 mutation in the two sexes (i.e., apoptosis of spermatocytes and survival of SC-defective oocytes) is likely caused by a common defect in recruitment of ATR activity to unsynapsed chromosomes. ATR is recruited to DSB sites in somatic cells, where it becomes active as part of the G2/M DSB repair checkpoint14. In meiosis, ATR and its activators are recruited to both DSB sites and unsynapsed chromatin13, 14, 51-57, 60. Our data raise the possibility that HORMAD1 fulfils its role in meiotic progression control by recruiting ATR and/or other components of the somatic DSB checkpoint machinery to unsynapsed chromatin, thereby adapting their functions for the detection of unsynapsed chromosome axes.
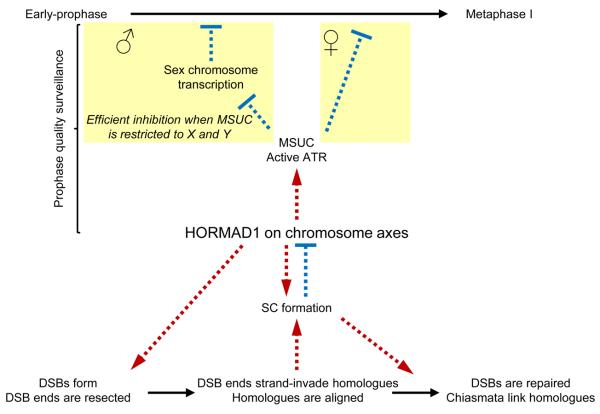
A comprehensive model for meiotic progression control emerges from this study and previous work showing that HORMAD1 is depleted from chromosome axes in response to SC formation (Fig. 8)40. We propose that the SC and HORMAD1 interact in a negative feedback-loop, to coordinate meiotic progression with successful homologue alignment. HORMAD1 promotes homologue alignment and SC formation. In turn, SC formation down-regulates HORMAD1 function by promoting HORMAD1 depletion from aligned homologues, which is a prerequisite for progression beyond the first meiotic prophase.
Figure 8. Model for meiotic progression: negative feedback-loop of HORMAD1 and SC coordinates homology search and meiotic progression.
Processes, activation-promotion and inhibition are marked by continuous black arrows, red dashed arrows and blue flat-ended dashed arrows, respectively. HORMAD1 associates with forming chromosome axes at the beginning of meiosis where it promotes DSB formation and/or processing of DSBs. It thereby ensures that adequate numbers of single-stranded DSBs are available for homology search. As part of the homology search process DSB ends strand-invade into homologues. Multiple strand invasion events along the length of chromosomes lead to full alignment of pairs of homologues, which is a prerequisite for the completion of SC formation. HORMAD1 also promotes SC formation through a mechanism that is independent of homology search. SC formation leads to depletion of HORMAD1 from axes and down-regulation of HORMAD1 function40. Hence, in spermatocytes, full autosomal SC formation leads to a restriction of HORMAD1 and ATR activity to sex chromosomes, thereby promoting efficient silencing of sex chromosomes, which is a prerequisite for progression beyond mid-pachytene13, 60, 61. In oocytes, completion of SC formation on all chromosomes leads to complete inactivation of HORMAD1, which in turn leads to down-regulation of ATR and MSUC. Since sustained ATR activity and/or sustained MSUC is believed to block progression beyond meiotic prophase13, full SC formation and HORMAD1 inactivation link successful homologue alignment with progression beyond meiotic prophase. Note that successful DSB repair is probably also required for full down-regulation of ATR activity and for meiotic progression in oocytes (for the sake of simplicity, this branch of the prophase checkpoint is not displayed in the model).
Although our observations and consistent observations of others41 clearly show the importance of HORMAD1 during mice meiosis, Hormad1−/− phenotypes appear partial. Conceivably, HORMAD1's paralogue, HORMAD2, might partially substitute for HORMAD1 during meiosis. It will be possible to resolve this question through analysis of Hormad1−/− Hormad2−/− mice when such mutant becomes available.
Methods
Hormad1 targeting
The Hormad1 targeting construct was designed according to the multi-purpose allele strategy63. The Splice-Acceptor (SA)-IRES-LacZneo-pA and PGK-Blasticidin-pA cassettes flanked by FRT sites were inserted in intron 3 (Supplementary Information, Fig. S2). The frame-shifting exon 4 was flanked by loxP sites. Plasmids were constructed using recombineering methods available on request64. To modify the Hormad1 locus, mouse R1 embryonic stem (ES) cells were cultured (using Mitomycin-C inactivated mouse embryonic fibroblasts as feeders) and electroporated with the linearised targeting constructs using standard protocols. Southern blot was used to identify correctly targeted ES clones: DNA was digested overnight with BclI, Bsu36I or HindIII for Southern blots with internal-, 5′- or 3′-probe, respectively, and DNA fragments were separated on 0.8% agarose gels (data not shown).
Generation of knockouts and genotyping
We generated mice that carried either of two different Hormad1 null alleles (Hormad1insertion and Hormad1deletion) or an allele with restored functionality (Hormad1restored) (Supplementary Information, Fig. S2). Two independent ES cell clones with a single integration of the targeting construct into the Hormad1 locus were used to generate chimeric animals. Progeny of the chimeric animals crossed to C57BL/6JOlaHsd females were genotyped by Southern blot and PCR (Supplementary Information, Fig. S2). In subsequent crosses, only PCR was used for genotyping the various alleles of Hormad1 from tail tip DNA. Genotyping primers: LacZfor 5′-TGGCTTTCGCTACCTGGAGAGAC, LacZrev 5′-AATCACCGCCGTAAGCCGACCAC, 5loxD 5′-TCCCTTCTGTCCTCCCATCTCC, loxP1 5′-GCTATACGAAGTTATAGCGGCAC, 3loxD 5′-TGGGTGCAAGCCTTTAATCCCC. PCR product sizes with LacZfor/LacZrev primers: Hormad1insertion template-208 bp, other alleles-no specific product; with LoxP1/3loxD primers: Hormad1insertion, Hormad1restored and Hormad1deletion templates-267 bp, WT allele-no specific product; with 5loxD/3loxD primers: Hormad1insertion and Hormad1restored templates-487 bp, WT allele-377 bp, Hormad1deletion allele-no specific product. To generate Hormad1restored/+ mice, we crossed ES26 derived Hormad1insertion/+ mice with hACTB:FLPe transgenic mice65. To generate Hormad1deletion/+ mice, we crossed Hormad1restored/+ with PGK-Cre transgenic mice66.
Animal experiments
Dmc1, Spo11 or Syce2 knockout mice were reported earlier4, 9, 43. Experiments were performed in a mixed background (with 75-87.5% contribution from the C57BL/6JOlaHsd inbred line). Whenever possible, experimental animals were compared with litter mate controls or with age-matched non-litter mate controls from the same colony. Although some of the figures show data from juveniles, all of our conclusions were reconfirmed from adult mice (more than 8-weeks-old). In Fig. 2 e-h data points from different ages (16 days and 8 weeks) were pooled when WT and Hormad1−/− litter mates were compared, since data at different ages were similar in respect of averages and ranges. HORMAD1 is not detected by western blot in Hormad1insertion/insertion testis extract (Supplementary Information, Fig. S2) and HORMAD1 is not detected by immunofluorescence on chromosome axes of Hormad1insertion/insertion meiocytes (data not shown), indicating that Hormad1insertion is a null allele. All of our conclusions are based on experiments that were performed at least once in the ES26 Hormad1insertion lineage. Apart from the kinetic analysis of axis and SC formation all of our conclusions involving experiments with single knockout mice were reconfirmed in the ES21 Hormad1insertion and ES26 Hormad1deletion lineages. The phenotypes of ES26 Hormad1insertion/insertion, ES21 Hormad1insertion/insertion and ES26 Hormad1deletion/deletion are indistinguishable. Therefore, we do not distinguish between Hormad1insertion and Hormad1deletion alleles, and we refer to them as the Hormad1- allele. ES26 Hormad1restored/restored mice are fertile. Testis size and SC formation in spermatocytes are indistinguishable in Hormad1restored/restored and WT mice (data not shown), which reconfirms that the phenotypes of Hormad1insertion/insertion and Hormad1deletion/deletion animals are caused by interference with production of WT Hormad1 transcripts. For staging embryonic development, the day of detection of a vaginal plug was marked as 0.5 days post coitum (dpc). All animals were used and maintained according to regulations provided by the animal ethics committee of the Technische Universität Dresden.
Immunofluorescence (IF)
Preparation and immunostaining of testis-ovary cryosections and nuclear surface spreads of meiocytes were performed as described previously with a few modifications40, 67. Ovaries and testes were fixed for 25 and 40 min, respectively, in 3.6% formaldehyde buffered with pH7.4 100 mM Na-phosphate. 8 μm (ovary) or 7 μm (testis) thick sections were cut from frozen specimens embedded in “O.C.T.” (Sakura Finetek Europe). Sections dried onto slides were immunostained following washes in PBS pH7.4 and PBS pH 7.4, 0.1% Triton X-100. IF-TUNEL assay was performed with Millipores′s ApopTag® Plus In Situ Apoptosis Fluorescein Detection Kit (S 7111). Antibodies: as described before40, rabbit anti-SMC3 (1:200) and rabbit anti-STAG3 (1:500) were gifts from Rolf Jessberger), guinea pig anti-SYCE1 (1:500) and guinea pig anti-SYCE2 (1:200) were gifts from Christer Höög, rabbit anti-RAD51 (1:200, Santa Cruz: sc-8349), rabbit anti-DMC1 (1:50, Santa Cruz: sc-22768), rabbit anti-MSH4 (1:150, Abcam: ab58666), mouse anti-MLH1 (1:50, BD Biosciences: 551092), rabbit anti-ATR (1:300, Calbiochem: PC538), goat anti-BRCA1 (1:30, Santa Cruz: sc-1553), rabbit anti-NOBOX (1:500, Abcam: ab41521), rat IgM anti-GCNA168 (1:5, gift from George C. Enders), rabbit anti-cleaved PARP (Asp214) (1:250, Cell Signalling: 9544) and CREST Human centromere auto-antibody HCT-100 (1:50, Cellon SA). For quantification of γH2AX signal in spread spermatocytes (Fig. 4d) matched exposure images were taken as described earlier40. We measured total IF signal intensity of γH2AX with ImageJ in squares that covered spread nuclei identified by DAPI staining. Signal intensities were measured in four regions around the examined nuclei to estimate the background. The signal intensity values in Fig. 4d are background corrected. To compare oocyte numbers in adults, we sectioned both ovaries of each mouse (8 μm thick sections), identified oocytes by anti-NOBOX (oocyte marker69) immunostaining and counted oocytes in every tenth section. To measure the rate of apoptosis in ovaries of 1 dpp animals, we sectioned both ovaries of each animal (8 μm thick sections). The sections were stained for GCNA1 (oocyte marker68) and for cleaved PARP1 (apoptosis marker). The numbers of cleaved PARP-positive and negative oocytes were counted on every eighth section.
Staging meiotic prophase
First, we compared axis development and SC formation in WT nuclear spreads to determine the stages of axis development corresponding to prophase stages. Thereafter, nuclear spreads were staged based on axis development (see details in the legend of Supplementary Information, Fig. S4).
Statistics
Statistical analysis was performed with GraphPad Prism5 and SPSS 11.5 for windows. For the comparison of independent samples, the two-tailed non-parametric Wilcoxon–Mann–Whitney two-sample rank-sum test was used.
Measurement of SPO11-oligonucleotide complexes and testis extracts
Immunoprecipitations of SPO11 and SPO11-oligonucleotide detection were performed as published previously with minor modifications45. Both testes from one juvenile or adult mouse were used for each experiment. Testes were decapsulated, then lysed in 800 μl lysis buffer (1% Triton X-100, 400 mM NaCl, 25 mM HEPES-NaOH at pH 7.4, 5 mM EDTA). Lysates were centrifuged at 100,000 rpm (355,040 g) for 25 min in a TLA100.2 rotor. Supernatants were incubated with anti-mSPO11 antibody 180 (5 μg per pair of testes) at 4°C for 1 h, followed by addition of 30-40 μl protein-A–agarose beads (Roche) and incubation for another 3 h. Beads were washed three times with IP buffer (1% Triton X-100, 150 mM NaCl, 15 mM Tris-HCl at pH 8.0). Immunoprecipitates were eluted with Laemmli sample buffer and diluted six- to seven-fold in IP buffer. Eluates were incubated with anti-mSPO11 antibody 180 at 4°C for 1 h, followed by addition of 30-40μl protein-A–agarose beads (Roche) and incubation overnight. Beads were washed three times with IP buffer and twice with buffer NEB4. SPO11-oligonucleotides were radiolabelled at 37 °C for 1 h using terminal deoxynucleotidyl transferase (Fermentas) and [α-32P] dCTP. Beads were washed three times with IP buffer, boiled in Laemmli sample buffer and fractionated on 8% SDS–PAGE. Immunoprecipitates were transferred to a PVDF membrane by semi-dry transfer (Bio-Rad). Radiolabelled species were detected and quantified with Fuji phosphor screens and ImageGuage software. The quantified signal displayed on the figures is background-corrected i.e. the signal found in the Spo11−/− testes was subtracted (Fig. 3a, c). To calculate the ratios of testes-weight-normalised SPO11-oligonucleotide levels, the background corrected signal was divided by the weight of the testes. For western analysis, membranes were probed with anti-mSPO11 antibody 180 (1:2000 in PBS containing 0.1% Tween 20 and 5% non-fat dry milk), then horseradish-peroxidase-conjugated protein A (Abcam; 1:10,000 in PBS containing 0.1% Tween 20 and 5% non-fat dry milk), and detected using the ECL+ reagent (GE Healthcare). The anti-SPO11 antibody was produced by the hybridoma cell line 180, which was generated by M.P. Thelen at Lawrence Livermore National Laboratory, California.
For anti-HORMAD1 western blot analysis (Supplementary Information, Fig. S2), total testis extracts were prepared by boiling resuspended testes in Laemmli buffer. Standard methods were used for SDS-PAGE and immunoblotting.
In vitro culture and video microscopy of oocytes
Oocytes were collected from antral follicles of 9-16 weeks old mice into M2 medium (38°C) supplemented with 100 μg/ml dibutyryl cyclic AMP (dbcAMP), which arrested oocytes in germinal vesicle stage (GV). Oocytes were induced to undergo meiotic maturation by rinsing and culture in M2 medium without dbcAMP70. Only oocytes where germinal vesicle breakdown (GVBD) was observed within 90 min after release were used for further experiments. Extrusion of the first polar body was determined by visual observation with light microscopy after overnight incubation (approximately 15-19 h after GVBD).
Video microscopy was carried out following injection of GV oocytes with in vitro-transcribed mRNAs (Ambion mMessage mMachine kit) coding for histone-H2B-RFP and β-tubulin-GFP70. Movies were made on a Nikon TE2000E microscope with PrecisExite High Power LED Fluorescence (LAM 1: 400/465, LAM2: 585), equipped with a temperature chamber (Life Imaging Services), Märzhäuser Scanning Stage, CoolSNAP HQ2 camera, and controlled by Metamorph software. Acquisitions (8z sections, every 2 μm, Plan APO 20x/0.75NA objective) were obtained every 30 min, starting from GVBD+5.5 h. GFP was set to 3% intensity, exposure time to 20 ms, RFP to 5% intensity with a 15 ms exposure time.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Hientzsch and K. Duerschke for general lab support and technical assistance; R. Jessberger for sharing ideas, antibodies (anti-SYCP3, anti-STAG3 and anti-SMC3) and support; F. Baudat and B. De Massy for providing us with Spo11+/− mice; J. Chen for the anti-TOPBP1 antibody; E. Marcon for the anti-RPA antibody; C. Höög for anti-SYCE1 and anti-SYCE2 antibodies; G.C. Enders for the anti-GCNA antibody; and M.P. Thelen for the anti-SPO11 antibody. We are grateful to M. Siomos and D. Knapp for discussion, revising and proofreading the manuscript. The Deutsche Forschungsgemeinschaft (DFG; grants: TO421/4-1 SPP1384 and TO421/3-1) and the Sächsisches Staatsministerium für Wissenschaft und Kunst supported K.D. and A.T.; Grants HD53855 and HD40916 from the US National Institutes of Health supported J.L., I.R., M.J. and S.K.; ARC (Subvention fixe 1143) and La Ligue Regionale (RS09/75-39) supported K.H. and K.W.; the Medical Research Council UK supported HJC; the DFG research center and cluster of excellence to the Center for Regenerative Therapies Dresden (CRTD) supported the work of K.A.; a CRTD seed grant supported K.A. and A.T.; and an EFRE grant (Europäischer Fonds für Regionale Entwicklung) supported K.A., J.F. and A.F.S.
Footnotes
AUTHOR CONTRIBUTIONS
K.D. designed, performed and analysed most of the experiments; J.L., I.R., M.J. and S.K. contributed with SPO11-oligo measurements; J.F., K.A. and A.F.S. designed and generated the targeting construct and targeted ES cells; K.H. and K.W. carried out oocyte maturation experiments and oocyte video microscopy; H.J.K. provided the Syce2+/− mouse, A.T. was involved in oocyte maturation experiments and oocyte counts, helped K.D. in experimental design and wrote the paper together with K.D. All authors were involved in discussions and commented on the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interest
References
- 1.Baudat F, de Massy B. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res. 2007;15:565–577. doi: 10.1007/s10577-007-1140-3. [DOI] [PubMed] [Google Scholar]
- 2.Hunter N. Meiotic recombination in Molecular genetics of recombination. Springer-Verlag; Berlin Heidelberg: 2007. [Google Scholar]
- 3.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 4.Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell. 2000;6:989–998. doi: 10.1016/s1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- 5.Romanienko PJ, Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 6.Sun H, Treco D, Szostak JW. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991;64:1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- 7.Hamer G, Novak I, Kouznetsova A, Hoog C. Disruption of pairing and synapsis of chromosomes causes stage-specific apoptosis of male meiotic cells. Theriogenology. 2008;69:333–339. doi: 10.1016/j.theriogenology.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 8.Hamer G, et al. Progression of meiotic recombination requires structural maturation of the central element of the synaptonemal complex. J Cell Sci. 2008;121:2445–2451. doi: 10.1242/jcs.033233. [DOI] [PubMed] [Google Scholar]
- 9.Bolcun-Filas E, et al. SYCE2 is required for synaptonemal complex assembly, double strand break repair, and homologous recombination. J Cell Biol. 2007;176:741–747. doi: 10.1083/jcb.200610027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolcun-Filas E, et al. Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genet. 2009;5:e1000393. doi: 10.1371/journal.pgen.1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Vries FA, et al. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 2005;19:1376–1389. doi: 10.1101/gad.329705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barchi M, et al. Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Mol Cell Biol. 2005;25:7203–7215. doi: 10.1128/MCB.25.16.7203-7215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burgoyne PS, Mahadevaiah SK, Turner JM. The consequences of asynapsis for mammalian meiosis. Nat Rev Genet. 2009;10:207–216. doi: 10.1038/nrg2505. [DOI] [PubMed] [Google Scholar]
- 14.Burgoyne PS, Mahadevaiah SK, Turner JM. The management of DNA double-strand breaks in mitotic G2, and in mammalian meiosis viewed from a mitotic G2 perspective. Bioessays. 2007;29:974–986. doi: 10.1002/bies.20639. [DOI] [PubMed] [Google Scholar]
- 15.Aravind L, Koonin EV. The HORMA domain: a common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem Sci. 1998;23:284–286. doi: 10.1016/s0968-0004(98)01257-2. [DOI] [PubMed] [Google Scholar]
- 16.Martinez-Perez E, Villeneuve AM. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev. 2005;19:2727–2743. doi: 10.1101/gad.1338505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nonomura K, Nakano M, Eiguchi M, Suzuki T, Kurata N. PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J Cell Sci. 2006;119:217–225. doi: 10.1242/jcs.02736. [DOI] [PubMed] [Google Scholar]
- 18.Couteau F, Zetka M. HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes Dev. 2005;19:2744–2756. doi: 10.1101/gad.1348205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodyer W, et al. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev Cell. 2008;14:263–274. doi: 10.1016/j.devcel.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Perez E, et al. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev. 2008;22:2886–2901. doi: 10.1101/gad.1694108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Severson AF, Ling L, van Zuylen V, Meyer BJ. The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev. 2009;23:1763–1778. doi: 10.1101/gad.1808809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zetka MC, Kawasaki I, Strome S, Muller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 1999;13:2258–2270. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Couteau F, Nabeshima K, Villeneuve A, Zetka M. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr Biol. 2004;14:585–592. doi: 10.1016/j.cub.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 24.MacQueen AJ, Colaiacovo MP, McDonald K, Villeneuve AM. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 2002;16:2428–2442. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nabeshima K, Villeneuve AM, Hillers KJ. Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics. 2004;168:1275–1292. doi: 10.1534/genetics.104.030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez-Moran E, Santos JL, Jones GH, Franklin FC. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 2007;21:2220–2233. doi: 10.1101/gad.439007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hollingsworth NM, Byers B. HOP1: a yeast meiotic pairing gene. Genetics. 1989;121:445–462. doi: 10.1093/genetics/121.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loidl J, Klein F, Scherthan H. Homologous pairing is reduced but not abolished in asynaptic mutants of yeast. J Cell Biol. 1994;125:1191–1200. doi: 10.1083/jcb.125.6.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwacha A, Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 30.Hollingsworth NM, Ponte L. Genetic interactions between HOP1, RED1 and MEK1 suggest that MEK1 regulates assembly of axial element components during meiosis in the yeast Saccharomyces cerevisiae. Genetics. 1997;147:33–42. doi: 10.1093/genetics/147.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bailis JM, Smith AV, Roeder GS. Bypass of a meiotic checkpoint by overproduction of meiotic chromosomal proteins. Mol Cell Biol. 2000;20:4838–4848. doi: 10.1128/mcb.20.13.4838-4848.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woltering D, et al. Meiotic segregation, synapsis, and recombination checkpoint functions require physical interaction between the chromosomal proteins Red1p and Hop1p. Mol Cell Biol. 2000;20:6646–6658. doi: 10.1128/mcb.20.18.6646-6658.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niu H, et al. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol Biol Cell. 2005;16:5804–5818. doi: 10.1091/mbc.E05-05-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niu H, et al. Mek1 kinase is regulated to suppress double-strand break repair between sister chromatids during budding yeast meiosis. Mol Cell Biol. 2007;27:5456–5467. doi: 10.1128/MCB.00416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carballo JA, Johnson AL, Sedgwick SG, Cha RS. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell. 2008;132:758–770. doi: 10.1016/j.cell.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 36.Joshi N, Barot A, Jamison C, Borner GV. Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLoS Genet. 2009;5:e1000557. doi: 10.1371/journal.pgen.1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zetka M. Homologue pairing, recombination and segregation in Caenorhabditis elegans. Genome Dyn. 2009;5:43–55. doi: 10.1159/000166618. [DOI] [PubMed] [Google Scholar]
- 38.Hayashi M, Chin GM, Villeneuve AM. C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet. 2007;3:e191. doi: 10.1371/journal.pgen.0030191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukuda T, Daniel K, Wojtasz L, Toth A, Hoog C. A novel mammalian HORMA domain-containing protein, HORMAD1, preferentially associates with unsynapsed meiotic chromosomes. Exp Cell Res. 2010;316:158–171. doi: 10.1016/j.yexcr.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 40.Wojtasz L, et al. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 2009;5:e1000702. doi: 10.1371/journal.pgen.1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin YH, et al. Hormad1 mutation disrupts synaptonemal complex formation, recombination, and chromosome segregation in mammalian meiosis. PLoS Genet. 2010;6:e1001190. doi: 10.1371/journal.pgen.1001190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmed EA, de Rooij DG. Staging of mouse seminiferous tubule cross-sections. Methods Mol Biol. 2009;558:263–277. doi: 10.1007/978-1-60761-103-5_16. [DOI] [PubMed] [Google Scholar]
- 43.Pittman DL, et al. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida K, et al. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol Cell. 1998;1:707–718. doi: 10.1016/s1097-2765(00)80070-2. [DOI] [PubMed] [Google Scholar]
- 45.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moens PB, Marcon E, Shore JS, Kochakpour N, Spyropoulos B. Initiation and resolution of interhomolog connections: crossover and non-crossover sites along mouse synaptonemal complexes. J Cell Sci. 2007;120:1017–1027. doi: 10.1242/jcs.03394. [DOI] [PubMed] [Google Scholar]
- 47.Baker SM, et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 48.Marcon E, Moens P. MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics. 2003;165:2283–2287. doi: 10.1093/genetics/165.4.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernandez-Capetillo O, Liebe B, Scherthan H, Nussenzweig A. H2AX regulates meiotic telomere clustering. J Cell Biol. 2003;163:15–20. doi: 10.1083/jcb.200305124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bellani MA, Romanienko PJ, Cairatti DA, Camerini-Otero RD. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/− spermatocytes. J Cell Sci. 2005;118:3233–3245. doi: 10.1242/jcs.02466. [DOI] [PubMed] [Google Scholar]
- 51.Moens PB, et al. The association of ATR protein with mouse meiotic chromosome cores. Chromosoma. 1999;108:95–102. doi: 10.1007/s004120050356. [DOI] [PubMed] [Google Scholar]
- 52.Plug AW, et al. Changes in protein composition of meiotic nodules during mammalian meiosis. J Cell Sci. 1998;111(Pt 4):413–423. doi: 10.1242/jcs.111.4.413. [DOI] [PubMed] [Google Scholar]
- 53.Plug AW, et al. ATM and RPA in meiotic chromosome synapsis and recombination. Nat Genet. 1997;17:457–461. doi: 10.1038/ng1297-457. [DOI] [PubMed] [Google Scholar]
- 54.Keegan KS, et al. The Atr and Atm protein kinases associate with different sites along meiotically pairing chromosomes. Genes Dev. 1996;10:2423–2437. doi: 10.1101/gad.10.19.2423. [DOI] [PubMed] [Google Scholar]
- 55.Perera D, et al. TopBP1 and ATR colocalization at meiotic chromosomes: role of TopBP1/Cut5 in the meiotic recombination checkpoint. Mol Biol Cell. 2004;15:1568–1579. doi: 10.1091/mbc.E03-06-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turner JM, et al. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr Biol. 2004;14:2135–2142. doi: 10.1016/j.cub.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 57.Turner JM, et al. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet. 2005;37:41–47. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- 58.Baarends WM, et al. Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Mol Cell Biol. 2005;25:1041–1053. doi: 10.1128/MCB.25.3.1041-1053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turner JM, Mahadevaiah SK, Ellis PJ, Mitchell MJ, Burgoyne PS. Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Dev Cell. 2006;10:521–529. doi: 10.1016/j.devcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 60.Mahadevaiah SK, et al. Extensive meiotic asynapsis in mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation. J Cell Biol. 2008;182:263–276. doi: 10.1083/jcb.200710195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Royo H, et al. Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr Biol. 2010;20:2117–2123. doi: 10.1016/j.cub.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 62.Martini E, Diaz RL, Hunter N, Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006;126:285–295. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Testa G, et al. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis. 2004;38:151–158. doi: 10.1002/gene.20012. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 65.Rodriguez CI, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 66.Lallemand Y, Luria V, Haffner-Krausz R, Lonai P. Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic Res. 1998;7:105–112. doi: 10.1023/a:1008868325009. [DOI] [PubMed] [Google Scholar]
- 67.Hodges CA, Hunt PA. Simultaneous analysis of chromosomes and chromosome-associated proteins in mammalian oocytes and embryos. Chromosoma. 2002;111:165–169. doi: 10.1007/s00412-002-0195-3. [DOI] [PubMed] [Google Scholar]
- 68.Enders GC, May JJ., 2nd Developmentally regulated expression of a mouse germ cell nuclear antigen examined from embryonic day 11 to adult in male and female mice. Dev Biol. 1994;163:331–340. doi: 10.1006/dbio.1994.1152. [DOI] [PubMed] [Google Scholar]
- 69.Suzumori N, Yan C, Matzuk MM, Rajkovic A. Nobox is a homeobox-encoding gene preferentially expressed in primordial and growing oocytes. Mech Dev. 2002;111:137–141. doi: 10.1016/s0925-4773(01)00620-7. [DOI] [PubMed] [Google Scholar]
- 70.Niault T, et al. Changing Mad2 levels affects chromosome segregation and spindle assembly checkpoint control in female mouse meiosis I. PLoS One. 2007;2:e1165. doi: 10.1371/journal.pone.0001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.