Abstract
Inteins self-splice from precursor polypeptides to reconstitute functional proteins. Here we describe inteins as redox-responsive switches in bacteria. Regulation was achieved by engineering a disulfide bond between the intein’s catalytic cysteine and the flanking polypeptide. This interaction was validated by an X-ray structure, which includes a transient splice junction. A natural analogue of the designed system was identified in Pyrococcus abysii, suggesting an unprecedented form of adaptive, post-translational regulation.
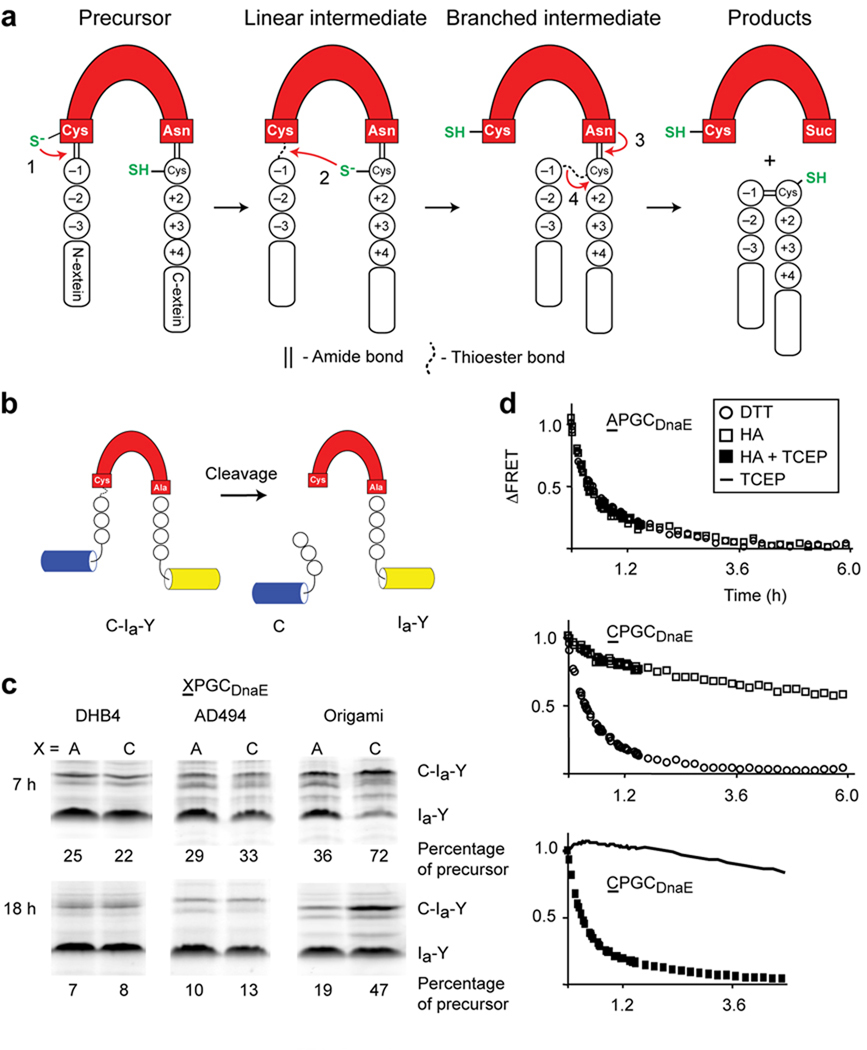
Inteins are excised from internal positions of precursor polypeptides through an autocatalytic protein splicing reaction (Fig. 1a)1–3. Inteins share distinctive sequence features and occur in all domains of life4, where they exist ostensibly as parasitic elements5. Proteins that harbor inteins, by contrast, are diverse and carry out vital metabolic functions once spliced. While there is general agreement on the overall pathway leading from an intein precursor to spliced products6, many fundamental questions remain. These include how the intein rearranges a stable peptide bond into a reactive thioester to initiate splicing, and the precise relationship between the intein and the flanking amino- and carboxyl-terminal sequences, called N- and C-exteins, respectively7–9. Experiments to interrogate these issues are stymied by the difficulties of isolating unspliced precursor and by the instability of protein splicing intermediates10.
Figure 1.
Engineered redox-responsive intein precursor in vivo and in vitro. (a) Scheme for intein splicing. Conserved terminal intein residues are indicated as is the extein C+1 residue. N-extein and C-extein residues are designated by minus and plus signs, respectively. (b). FRET-based intein reporter with the DnaE intein (red) inserted between cyan and yellow fluorescent proteins. (c). Biomimetic Cys-3 variant, CPGCDnaE, displays enhanced redox sensitivity in vivo. Activities of CPGCDnaE and APGCDnaE inteins were quantified as the percentage of precursor (C-Ia-Y) remaining, 7 h and 18 h after induction of protein expression in E. coli DHB4, AD494 and origami. Data are representative of ≥2 independent experiments. Additional bands, observed previously with unboiled samples, were excluded from the quantitation9 (d). Effect of redox conditions on the in vitro cleavage kinetics of APGCDnaE and CPGCDnaE. N-extein cleavage was monitored by FRET loss that occurs upon cleavage by DTT or by hydroxylamine (HA). Each data point is an average of three measurements.
We are interested in imparting control over intein activity without mutating catalytic residues. We also wish to use natural triggers to probe structure/function and to investigate biologically relevant regulatory functions. Here we explore the possibility of coupling intein catalysis to redox conditions. Perturbations in redox status occur naturally, reversibly, and have the potential to influence inteins, as protein splicing commonly depends on reduced sulfhydryl groups11, 12. The fused Synechocystis sp. strain PCC6803 DnaE intein studied here utilizes two nucleophilic cysteine residues for splicing: intein Cys1 and C-extein Cys+1 (Fig. 1a, steps 1 and 2)13, 14. Cys1 initiates splicing by attacking the backbone amide bond of the preceding N-extein residue at −1. Transfer of the resulting thioester to Cys+1 yields a branched intermediate (Fig. 1a)15. To implement redox control over this autoprocessing activity we mutated the N- and C-exteins exclusively, leaving the intein unchanged.
We employed cysteine scanning mutagenesis and in vivo screening to find disulfide-bond partners for Cys1 and Cys+1. Six potential redox-active cysteine substitutions were prepared at positions −3, −2, and −1 of the N-extein and at +2, +3, or +4 of the C-extein, to block the first or second steps of splicing, respectively. The six constructs were inserted between cyan and yellow fluorescent proteins in a FRET-active intein reporter we described previously9 (Fig. 1b). For screening, the intein contained an Ala substitution of its C-terminal Asn to block processing past step 2. Using this construct, activity can be measured in solution, in gels, and in live cells9.
To test the cysteine variants for redox-regulated processing in living cells, we utilized an E. coli mutant, origami, with an oxidizing cellular environment. This strain, deficient in two oxidoreductases (ΔtrxB ΔgorA), allows disulfide bonds to form more efficiently than in isogenic parental strains, DHB4 and AD494 (ΔtrxB)16. Of the six mutants screened, only the Cys-3 mutant processed more slowly than the wild-type when expressed in origami (Supplementary Fig. 1a, b). Interestingly, Cys mutants +2, +3 and +4 showed enhanced processing, as apparent from the precursor-to-product ratios. The activating influence of the Cys +2 and Cys +3 mutations agrees with previous work9. We conclude that the cysteine substitution at N-extein −3 provides the single route to oxidative intein inhibition in vivo.
We adopted a biomimetic approach to stimulate the redox phenotype of the Cys-3 mutant. The N-3 substitution creates the oxidoreductase “CXXC” motif with Cys1. In thioredoxin, the most reducing of E. coli’s oxidoreductases17, proline and glycine, corresponding to XX in CXXC, create a tight turn in the protein backbone, favoring disulfide formation18. We therefore modified the Cys-3 mutant by replacing Glu and Tyr at −2 and −1 with Pro and Gly, respectively. We refer to this construct as CPGCDnaE. As a control, we prepared APGCDnaE with Ala at −3. The two variants were expressed in origami, AD494, and DHB4 to evaluate redox-regulated activity. Yields of CPGCDnaE and APGCDnaE precursor were nearly identical in AD494 and DHB4, indicating similar intein activity (Fig. 1c). However, the yield of intact CPGCDnaE precursor is >2-fold that of APGCDnaE precursor in the origami strain, at 7 h or 16 h post-induction (Fig. 1c). Also, the yield of CPGCDnaE precursor was 3-fold greater in origami compared with DHB4 at 7 h, and at 18 h the ratio approaches 6.
Using an in vitro intein activity assay, we determined that CPGCDnaE and APGCDnaE precursors can generate thioester for splicing at levels comparable to the wild-type intein. However, with CPGCDnaE precursor, that activity depends on the presence of a reducing agent. In the first step of splicing, the N-extein residue at −1 is transferred to the side chain of Cys1, generating a reactive thioester (Fig. 1a). The presence of this intermediate was probed by incubating bacterially-expressed FRET-active CPGCDnaE and APGCDnaE precursors in the presence of thioester-cleavage reagents under reducing and nonreducing conditions. Thioester cleavage was followed continuously by FRET loss that occurs once the CFP-fused N-extein separates from the precursor (Fig. 1b)9. With dithiothreitol (DTT) as the thioester cleavage reagent, we observed rapid conversion of FRET-active precursor into products for both APGCDnaE and CPGCDnaE (Fig. 1d, top and middle panels), at rates within 2-fold of wild-type (t½, 20 min)9. This consistency in cleavage rates suggests that the intein activity is relatively unchanged despite mutated N-extein residues.
Because DTT functions both as a nucleophile and as a reducing agent, we repeated the experiment using the nonreducing nucleophile, hydroxylamine. The CPGCDnaE precursor cleavage rate appeared markedly suppressed with hydroxylamine (t½ > 6 h; Fig. 1d, middle). In contrast, the hydroxylamine cleavage rate of the APGCDnaE precursor was similar as with DTT (Fig. 1d, top). To address whether the suppressed hydroxyaminolysis of the CPGCDnaE precursor was attributable to oxidative inhibition, we conducted the experiment with hydroxylamine plus the reducing agent Tris(2-carboxyethyl) phosphine (TCEP). Under these conditions, cleavage susceptibility of the CPGCDnaE precursor was restored to that observed with DTT (Fig.1d, bottom), further supporting the Cys-3-dependent redox-sensitivity of the precursor.
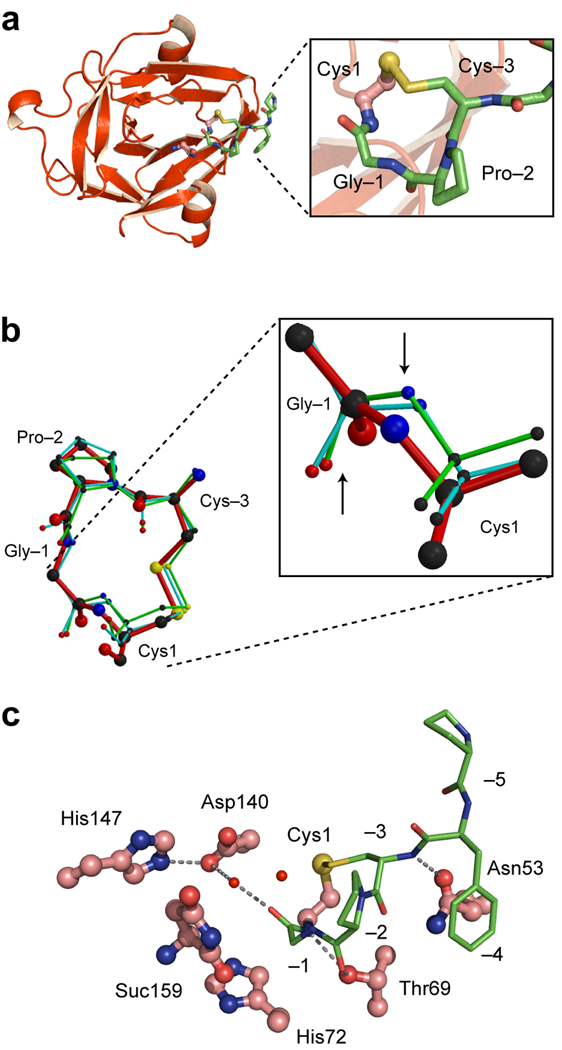
We next determined the X-ray structure of the CPGCDnaE precursor. The protein used for crystallographic studies was a splicing-competent precursor, consisting of wild-type intein, with the native C-terminal Asn, flanked by N- and C-extein sequences KSPDPFCPG and CFNVQ, respectively. Catalytically active intein precursors are ordinarily too reactive for isolation, much less crystallization; however, the N-extein in this CPGCDnaE precursor is stable in phosphate buffer (pH 8) in the absence of reducing agents for ≥2 weeks at 23 °C. Crystals of CPGCDnaE precursor, containing its full complement of active-site residues, appeared after ~4 days and diffracted to 1.55 Å.
The electron density map of the CPGCDnaE precursor showed clear density for the intein and the N-extein. Density for the C-extein was absent owing to intein-catalyzed cleavage at the C-terminal splicing junction, as evidenced by the intein’s C-terminal Asn in cyclized succinimide form (Fig. 1a, step 3). This side reaction has been observed before with inteins harboring a Cys1Ala mutation to block the first step of splicing19, 20. The N-extein is connected to the intein through an amide bond with trans-peptide configuration. Cys-3 and Cys1 are joined by a disulfide bond as expected, explaining the stability of the precursor and the effect of redox changes on CPGCDnaE intein activity (Fig. 2a). However, the Gly-1, which corresponds to the splice junction residue, adopts an unusual conformation, as defined by Molprobity21. Thus, Gly-1 has backbone angles of φ =−122.5° and ψ=86.9°, which fall outside the geometery typical of glycine.
Figure 2.
Crystal structure of the CPGCDnaE intein. (a). Ribbon diagram showing the intein (red) and the N-terminal extein (green). Magnified view of the Cys1-to-Cys-3 disulfide loop is shown in the box (b). Conformation of the CPGC disulfide loop. Comparison of the structure of the disulfide loop in the CPGCDnaE intein (red) with the two other observations of this loop in the Protein Data Bank (1JBQ, green; 1M6Y, cyan). Magnified view shows Gly-Cys peptide bonds, with an upward arrow pointing to the carbonyl oxygen atoms and a downward arrow pointing toward the amide nitrogen atoms. (c). Active-site interactions in the CPGCDnaE intein (intein, red; extein, green). Thr69 of the conserved TXXH motif contacts the amide nitrogen of Cys1 and the carbonyl oxygen of Pro-2 while His72, also of the TXXH motif, does not interact directly with the N-extein. Asp140 makes a water-mediated contact with the −1 carbonyl and a hydrogen bond with His147, also a conserved residue. Suc 159 is the C-terminal succinimide.
A reasonable concern is that these unusual φ and ψ angles of the splice junction Gly-1 residue result from the engineered disulfide bond between Cys-3 and Cys1. To address this possibility, we compared the CPGC tetrapeptide to the two naturally occurring disulfide loops with the same CPGC sequence in the Protein Data Bank22, 23. The loops from cystathionine beta-synthase (1JBQ) and S-adenosylmethionine-dependent methyltransferase (1M6Y) superimpose on each other and resemble the CPGC loop here (Fig. 2b), except at the Gly-Cys amide bond, magnified in Figure 2b. Thus, the nitrogen and oxygen atoms of the Gly-Cys amide bond in the intein precursor lie in a plane shifted by ~90° relative to the two comparator N and O atoms. Importantly, no backbone distortion is detected in either comparison loop, with typical φ and ψ angles for Gly21. Furthermore, in quantum mechanical simulations, relaxing the φ and ψ angles does not affect the integrity of the S-S bond in the redox-trapped structure, suggesting that the strained bonds are independent of the disulfide (A.K. Dearden and S. Nayak, personal communication). These observations taken together suggest that the distortion is not a property of the CPGC sequence but is instead intrinsic to the intein. This view is further supported by hydrogen bonding interactions involving the amide nitrogen of Cys1, the amide oxygen of Pro-2, and the hydroxyl of Thr69 (Fig. 2c). Thr69 is an essential catalytic residue24 and forms part of the signature TXXH motif present in > 90% of inteins. We speculate that the Thr69-mediated interactions supply binding energy that offsets the strain at Gly-1, and could predispose Gly-1 to nucleophilic attack through a destabilization mechanism.
Based on the behavior of the designed CPGCDnaE precursor, we speculated that redox-regulated protein splicing might also occur in Nature. The possibility of an intein/extein relationship runs counter to the definition of inteins as autonomous elements, and evidence for intein-based switches is lacking. Nonetheless, we identified a plausible candidate in the molybdate-biosynthesis MoaA enzyme of Pyrococcus abyssi4. The intein in MoaA, Pab MoaA, contains an N-terminal Cys1 and the N-extein contains a cysteine at the optimal −3 position. The Cys-3 (underscored) forms part of a conserved CXXXCXXC motif necessary for Fe-S formation and the oxidoreductase activity of MoaA, the activity of which, interestingly, is oxygen-labile25. From the redox experiments above, the oxygen-lability of MoaA, and the physiological responsiveness of P. abysii to oxidative stress26, we hypothesized that MoaA protein production could be post-translationally blocked by a Cys-3-to-Cys1 disulfide bond under hyperoxia, forestalling oxidative damage to the mature enzyme. While the motif at the N-extein/intein boundary of the MoaA intein, C−3W−2Y−1C1, differs from our engineered sequence, we were encouraged by the X-ray structure of a CYWC sequence in β-mannase (PDB, 1QNO), which shows the paired cysteine residues as a disulfide bond.
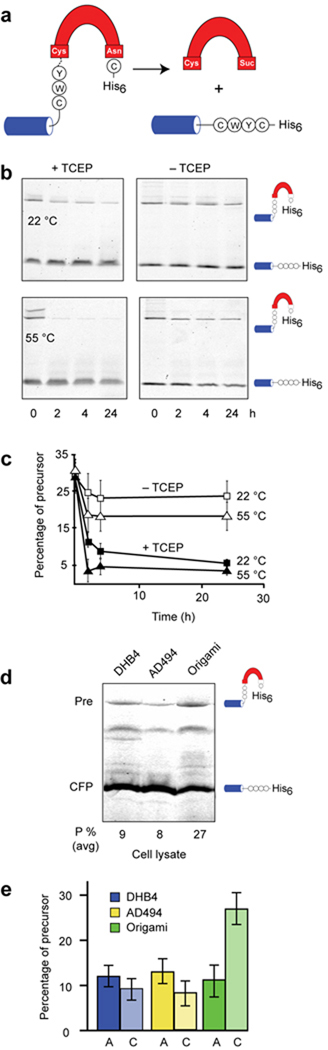
To explore the possibility of redox-sensitive protein splicing in P. abysii, we cloned Pab MoaA, flanked by native N-extein residues, into a splicing reporter construct, CWYCMoaA (CFPCWYCMoaA-His6) (Fig. 3a). The CWYCMoaA precursor was expressed in origami and although some splicing occurred intracellularly, sufficient precursor was recovered to study redox sensitivity in vitro. Processing of the CWYCMoaA precursor was monitored in phosphate buffer (0.02 M, pH 8) with and without TCEP at 22 °C and 55 °C. Activity of CWYCMoaA activity was relatively modest in the absence of a reducing agent at both 22 °C and at 55 °C (Fig. 3b, c). However, with added TCEP, at 22 °C, autoprocessing of CWYCMoaA was readily apparent by the disappearance of fluorescent precursor, and activity increased markedly at 55 °C (Fig. 3b, c).
Figure 3.
Protein splicing by the P. abssyi MoaA intein is redox sensitive (a). Schematic of CWYCMoaA precursor. Splicing of the MoaA intein (red) with native N-extein residues (circles) ligates the cyan fluorescent protein to a hexahistidine tag (His6). (b) and (c). Suppressed activity of the CWYCMoaA intein in the absence of a reducing agent. Intein autoprocessing was monitored in the presence and absence of the reducing agent, TCEP, at 22 °C (squares) and 55 °C (triangles). Precursor (CFPCWYCMoaA-His6) and splicing product (CFP-His6) were separated by nonreducing SDS-PAGE and detected by in-gel fluorescence (excitation 457 nm, emission 526 nm). Plot of precursor disappearance in panel c, using data of panel b, shows that TCEP is required for efficient CWYCMoaA processing at 22 °C and at 55 °C. Error bars represent s.d., n=3. (d). CWYCMoaA displays redox regulation in E. coli. Representative gel images showing CFP-fused MoaA precursor with wild-type Cys-3 (CFPCWYCMoaA-His6), and CFP-products remaining after 3 h of induction in DHB4 (left lane), AD494 (middle lane), and origami (right lane). (e). Cys-3 is required for MoaA precursor accumulation in origami. Graph was derived from images as in panel (a) and shows percent of unspliced CFPCWYCMoaA-His6 (C, light shaded) and CFPAWYCMoaA-His6 (A, dark shaded) precursors after expression in the indicated strains, averaged over three independent trials. Error bars represent s.d.
Next, we determined the redox sensitivity of CWYCMoaA in vivo in the oxidizing origami strain relative to the E. coli AD494 and DHB4 strains (Fig. 3d). CWYCMoaA spliced rapidly in DHB4 and in AD494, with ~90% conversion to products. However, processing of CWYCMoaA was attenuated when expressed in origami, with only ~70% conversion. Thus, the normalized quantity of CWYCMoaA precursor after expression in the oxidizing strain was increased 2- to 3-fold relative to that of the CWYCMoaA precursor isolated in parallel from DHB4 and AD494. It is important to emphasize that the magnitude of this redox effect is similar to that exhibited in vivo by the CPGCDnaE precursor, which we crystallized. Together with the in vitro experiments, the inhibition of CWYCMoaA activity in the origami strain indicates redox responsiveness, consistent with our hypothesis for protein splicing regulation.
To explicitly test the involvement of a Cys-3-to-Cys1 disulfide bond in redox-regulated splicing, we generated a AWYCMoaA control using the same reporter construct (CFPAWYCMoaA-His6), with a Cys-3Ala N-extein mutation. We then compared processing of the mutant with CWYCMoaA during bacterial expression in DHB4, AD494, and origami (Fig 3e). Extensive in vivo processing was observed with the mutant AWYCMoaA construct, similar to the level of the CWYCMoaA construct when expressed in the DHB4 and AD494 strains. Importantly, the robust processing of AWYCMoaA persisted in the origami strain, in contrast with the buildup of CWYCMoaA precursor in this oxidizing host. Moreover, the CWYCMoaA precursor is catalytically competent, as apparent from the TCEP-activation experiments (Figure 3b). These results are fully consistent with precursor trapping by a Cys-3-to-Cys1 disulfide bond of a naturally occurring intein in a bacterial cell.
In summary, we have demonstrated that a disulfide bond between the catalytic cysteine of an intein and a cysteine at a specific position in the flanking N-extein sequence can be made and broken to control precursor activity in vitro and in E. coli. As in other engineered redox traps, the intein remains catalytically competent27, but incapable of functioning normally until activated by a reducing agent. Using the redox trap, we obtained the first high-resolution structure of an ordinarily transient N-extein-intein precursor. The structure suggests a previously unrecognized role for a conserved threonine in substrate destabilization, manifesting in a high-energy backbone conformation at the splice junction. This result appears to add validity to the atypical −1 N-extein residue configurations observed in the structures of catalytically inert precursors of the Mxe GyrA and Sce Vma inteins, although there, perturbations at the splice junction were characterized by changes in the amide bond angle, τ28–30. Further experiments will ascertain the source of this variation, but the prevailing view is that these structural perturbations are enforced by the intein as a means to accelerate chemistry.
The controllability of the designed DnaE precursor questioned the autonomy assigned to self-splicing inteins and thereby motivated searches for naturally occurring regulatory inteins in genome databases. We identified a candidate in an intein-containing protein of a barophilic, thermophilic, deep-sea archaeon, P. abyssi. The existence of a redox effect in Pab MoaA and its attribution to the N-extein Cys-3 residue point to the remarkable possibility of inteins being used as adaptive switches or rheostats in a biological context. Our speculation is that the disulfide bond forms during hyperoxia, building up a reservoir of MoaA precursor; which can then be spliced under reducing conditions into the mature, oxygen-labile, Fe-S enzyme.
Since identifying the MoaA intein with Cys-3, data-mining has yielded additional candidates in the pyruvate-formate lyase-activating enzyme (PFL-AE) from an uncultured archaeon, and in a radical SAM domain protein of Archaeoglobus profundus4. Interestingly, all three mature proteins are predicted to catalyze redox chemistry and each of the three inteins resides in a conserved CXXXCXXC motif. Also pertinent is that PFL-AE precursor harbors a proline at −2, resembling the designed CPGCDnaE precursor. Although further tests will be required to prove biological functions for these inteins, our findings provide an intriguing and plausible scenario whereby an intein, in collaboration with its extein, could transition from parasite to mutualist.
Supplementary Material
Acknowledgments
We thank Zhong Li for performing crystallization experiments and Dorie Smith for technical assistance; Albert Dearden and Saroj Nayak (Rensselaer Polytechnic Institute) for sharing the results of their QM-MM simulations; Brian Pereira and Gil Amitai for useful discussions; John Dansereau for preparing figures and for useful comments; and Maryellen Carl for manuscript preparation. We acknowledge the Wadsworth Center's Molecular Genetics Core for DNA sequencing and the Macromolecular Crystallography Core for equipment use. This work was supported by NIH grants GM39422 and GM44844 to MB.
Footnotes
Competing Interests The authors have no competing financial interests to declare.
Author Contributions B.P.C. conceived the study, B.P.C., N.I.T., P. VR, and M.B. designed research; B.P.C., N.I.T., M.J.S., and P.VR. performed research; B.P.C., P.VR., N.I.T. and M.B. analyzed data; and B.P.C., P.VR., N.I.T. and M.B. wrote the paper.
Accession codes. Protein Data Bank. The atomic coordinates and structure factors have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ, under accession code 3NZM.
REFERENCES
- 1.Kane PM, et al. Science (New York, N.Y. 1990;250:651–657. doi: 10.1126/science.2146742. [DOI] [PubMed] [Google Scholar]
- 2.Hirata R, et al. J. Biol. Chem. 1990;265:6726–6733. [PubMed] [Google Scholar]
- 3.Duan X, Gimble FS, Quiocho FA. Cell. 1997;89:555–564. doi: 10.1016/s0092-8674(00)80237-8. [DOI] [PubMed] [Google Scholar]
- 4.Perler FB. Nucleic Acids Res. 2002;30:383–384. doi: 10.1093/nar/30.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pietrokovski S. Trends in Genetics. 2001;17:465–472. doi: 10.1016/s0168-9525(01)02365-4. [DOI] [PubMed] [Google Scholar]
- 6.Paulus H. Annu. Rev. Biochem. 2000;69:447–496. doi: 10.1146/annurev.biochem.69.1.447. [DOI] [PubMed] [Google Scholar]
- 7.Pearl EJ, Bokor AA, Butler MI, Poulter RT, Wilbanks SM. Biochim. Biophys. Acta. 2007;1774:995–1001. doi: 10.1016/j.bbapap.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Kerrigan AM, Powers TL, Dorval DM, Reitter JN, Mills KV. Biochem. Biophys. Res. Commun. 2009;387:153–157. doi: 10.1016/j.bbrc.2009.06.145. [DOI] [PubMed] [Google Scholar]
- 9.Amitai G, Callahan BP, Stanger MJ, Belfort G, Belfort M. Proc. Natl. Acad. Sci. USA. 2009;106:11005–11010. doi: 10.1073/pnas.0904366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frutos EA, Goger M, Giovani B, Cowburn D, Muir TW. Nat. Chem. Biol. 2010 doi: 10.1038/nchembio.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mills KV, Lew BM, Jiang S, Paulus H. Proc. Natl. Acad. Sci. USA. 1998;95:3543–3548. doi: 10.1073/pnas.95.7.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi J, Muir TW. J. Am. Chem. Soc. 2005;127:6198–6206. doi: 10.1021/ja042287w. [DOI] [PubMed] [Google Scholar]
- 13.Wu H, Hu Z, Liu XQ. Proc Natl Acad Sci USA. 1998;95:9226–9231. doi: 10.1073/pnas.95.16.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans TC, Jr., et al. J. Biol. Chem. 2000;275:9091–9094. doi: 10.1074/jbc.275.13.9091. [DOI] [PubMed] [Google Scholar]
- 15.Xu M-Q, Southworth MW, Mersha FB, Hornstra LJ, Perler FB. Cell. 1993;75:1371–1377. doi: 10.1016/0092-8674(93)90623-x. [DOI] [PubMed] [Google Scholar]
- 16.Bessette PH, Aslund F, Beckwith J, Georgiou G. Proc. Natl. Acad. Sci. USA. 1999;96:13703–13708. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chivers PT, Prehoda KE, Raines RT. Biochemistry. 1997;36:4061–4066. doi: 10.1021/bi9628580. [DOI] [PubMed] [Google Scholar]
- 18.Katti SK, LeMaster DM, Eklund H. Journal of molecular biology. 1990;212:167–184. doi: 10.1016/0022-2836(90)90313-B. [DOI] [PubMed] [Google Scholar]
- 19.Evans TCJ, Benner J, Xu MQ. J. Biol. Chem. 1999;274:3923–3926. doi: 10.1074/jbc.274.7.3923. [DOI] [PubMed] [Google Scholar]
- 20.Wood DW, Wu W, Belfort G, Derbyshire V, Belfort M. Nat. Biotechnol. 1999;17:889–892. doi: 10.1038/12879. [DOI] [PubMed] [Google Scholar]
- 21.Davis IW, et al. Nucleic Acids Res. 2007;35:W3753–W3783. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller DJ, et al. Protein Sci. 2003;12:1432–1442. doi: 10.1110/ps.0302403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. EMBO J. 2001;20:3910–3916. doi: 10.1093/emboj/20.15.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh I, Sun L, Xu M-Q. J. Biol. Chem. 2001;276:24051–24058. doi: 10.1074/jbc.M011049200. [DOI] [PubMed] [Google Scholar]
- 25.Hanzelmann P, et al. J. Biol. Chem. 2004;279:34721–34732. doi: 10.1074/jbc.M313398200. [DOI] [PubMed] [Google Scholar]
- 26.Marteinsson VT, et al. Appl. Environ. Microbiol. 1997;63:1230–1236. doi: 10.1128/aem.63.4.1230-1236.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumura M, Matthews BW. Science (New York, N.Y. 1989;243:792–794. doi: 10.1126/science.2916125. [DOI] [PubMed] [Google Scholar]
- 28.Klabunde T, Sharma S, Telenti A, Jacobs WR, Jr., Sacchettini JC. Nat. Struct. Biol. 1998;5:31–36. doi: 10.1038/nsb0198-31. [DOI] [PubMed] [Google Scholar]
- 29.Poland BW, Xu MQ, Quiocho FA. J. Biol. Chem. 2000;275:16408–16413. doi: 10.1074/jbc.275.22.16408. [DOI] [PubMed] [Google Scholar]
- 30.Romanelli A, Shekhtman A, Cowburn D, Muir TW. Proc. Natl. Acad. Sci. USA. 2004;101:6397–6402. doi: 10.1073/pnas.0306616101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.