Abstract
Dedicated chaperones facilitate eukaryotic proteasome assembly, yet how they function remains largely unknown. Here we demonstrate that a yeast 20S proteasome assembly factor, Pba1–Pba2, requires a previously overlooked C-terminal HbYX (hydrophobic-tyrosine-X) motif for function. HbYX motifs in proteasome activators open the 20S proteasome entry pore, but Pba1–Pba2 instead binds inactive proteasomal precursors. We discovered an archaeal ortholog of this factor, here named PbaA, that also binds preferentially to proteasomal precursors in a HbYX-dependent fashion using the same proteasomal α-ring surface pockets bound by activators. Remarkably, PbaA and the related PbaB protein can be induced to bind mature 20S proteasomes if the active sites in the central chamber are occupied by inhibitors. Our data suggest an allosteric mechanism in which proteasome active-site maturation determines assembly chaperone binding, potentially shielding assembly intermediates or misassembled complexes from non-productive associations until assembly is complete.
Keywords: proteasome, yeast, archaea, assembly factor, chaperone, ubiquitin
In eukaryotes, 26S proteasomes mediate a major fraction of intracellular protein degradation1. This ~2.5 MDa protease complex consists of a 20S proteasome core capped on either one or both ends by the 19S regulatory particle (RP). Four coaxially stacked heptameric rings make up the 20S core particle. Structurally related α and β subunits comprise the outer and inner rings, respectively2. An interior chamber created by the β-subunit rings bears the protease active sites. Proteolytically active β subunits are synthesized with N-terminal propeptides, which are autocatalytically cleaved near the end of proteasome assembly, thereby exposing the catalytic N-terminal nucleophilic residue, Thr1. A narrow gated central pore in each α-subunit ring controls substrate access. N-terminal segments of the α subunits occlude the opening but are displaced upon activator binding. All proteins capable of activating 20S, including the RP, PAN (proteasome-activating nucleotidase), Blm10 and PA28/11S, insert short C-terminal peptide segments into shallow pockets on the α-ring surface, triggering conformational changes that lead to gate opening. A subset of the RP ATPases and the archaeal ortholog of the RP ATPases, PAN, have a C-terminal hydrophobic-tyrosine-X (HbYX) motif crucial for activation.
Relatively little is known about how the proteasome is assembled. Proteasome-specific assembly chaperones function in this process3–5. For the 20S, these include Ump1, the Pba1–Pba2 (for proteasome biogenesis-associated) complex (called PAC1–PAC2 in mammals), the Pba3–Pba4 (PAC3–PAC4) complex, and potentially Blm10 (PA200)6–13. Recently, four dedicated chaperones that promote assembly of the RP have been described14–18. Many mechanistic details of assembly factor function remain unknown. For example, the Pba1–Pba2 heterodimer associates with various 20S proteasome assembly intermediates in both yeast and mammals and was suggested to function in preventing off-pathway interactions between pre-formed α-rings6,7. However, even elementary information, such as the nature of the interaction between Pba1–Pba2 and the 20S proteasome, not to mention its actual role in proteasome biogenesis, is not known. The subunit complexity of the eukaryotic proteasome has thwarted efforts to create an in vitro assembly system.
Archaea often provide useful model systems to dissect the function of more complicated eukaryotic machinery. Archaeal 20S proteasomes are structurally similar to their eukaryotic counterparts but are usually composed of only one type of α and β subunit19,20 versus seven of each in eukaryotes. When expressed recombinantly in bacteria, archaeal 20S subunits yield proteasomes that are functionally and structurally indistinguishable from their native counterparts21. Their compositional simplicity and the ease of assembling them through heterologous coexpression in bacteria have led to the assumption that archaeal proteasomes do not need assembly factors. In this report we identify two archaeal proteins, from the mesophilic methanogen Methanococcus maripaludis, which we named PbaA and PbaB that are likely orthologs of the eukaryotic Pba1–Pba2 assembly chaperone. Using an in vitro system, we demonstrate their direct association with archaeal proteasome precursors, paralleling the exclusive association of yeast Pba1–Pba2 with assembly intermediates in vivo. Importantly, this association requires a C-terminal HbYX motif that is universally conserved in both archaeal and eukaryotic proteins and is essential for the function of the Pba1–Pba2 assembly factor in vivo. Proteasome maturation correlates with chaperone release. Surprisingly, PbaA and PbaB bind stably to mature 20S proteasomes if the β-subunit active sites are bound to inhibitors, implying that their binding is regulated allosterically by changes initiated at the distant active sites. We suggest that this ancient chaperone class promotes 20S proteasome assembly by sterically blocking off-pathway protein-protein interactions such as pairing of α-subunit rings and helps limit binding of activators to incompletely assembled or inhibited proteasomes.
RESULTS
Sequence Analysis of the Pba1–Pba2 Assembly Factor
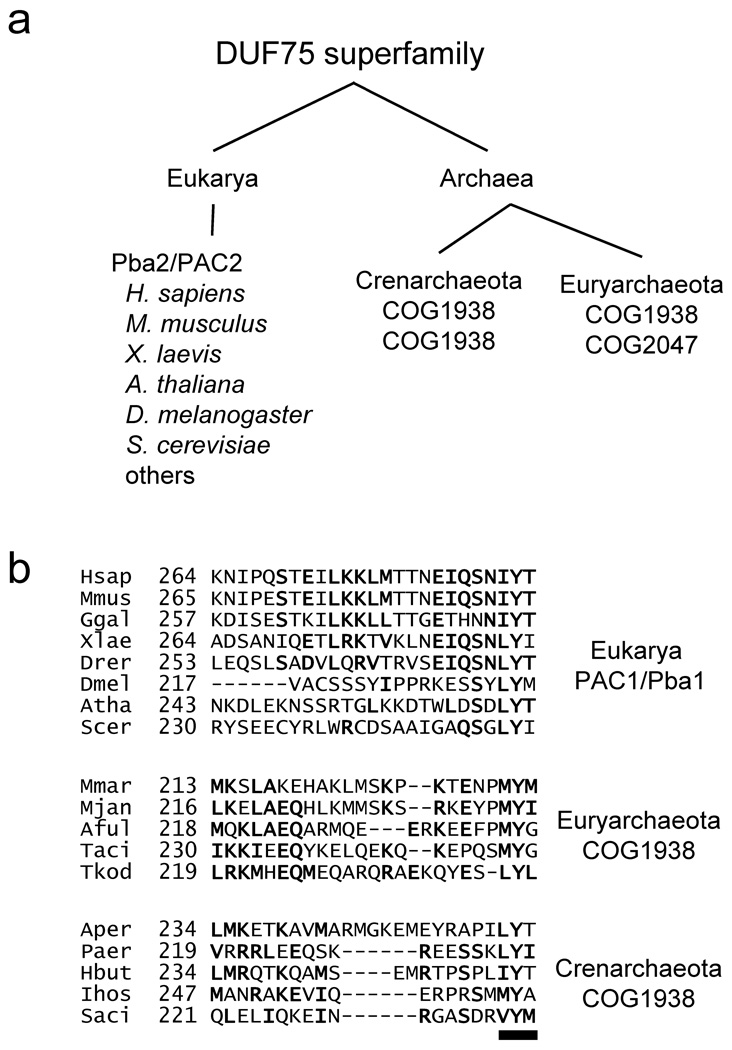
The S. cerevisiae heterodimer Pba1–Pba2, the ortholog of human PAC1–PAC2, associates with proteasome intermediates ranging from particles with a full α-subunit ring but only three β subunits to the pre-holoproteasome (PHP), which has brought together two complete half-proteasomes but has not yet processed the β-subunit propeptides6. To characterize this assembly factor further, we first analyzed its primary sequences more closely. Pba2 and PAC2 assembly chaperones are members of the Domain of Unknown Function 75 (DUF75) superfamily (Fig. 1a), and our sequence analysis indicates that Pba1/PAC1 is as well (see Supplementary Methods). DUF75 members are also found throughout the archaea and can be divided into two related orthologous groups, COG1938 and COG2047 (Fig. 1a and Suppl. Fig. 1). Intriguingly, despite poor overall sequence conservation6, all eukaryotic Pba1/PAC1 proteins terminate with a highly conserved tripeptide reminiscent of the HbYX motifs found in proteasome activators (Fig. 1b). HbYX motifs mediate activator binding to 20S proteasomes and drive gate opening in the α ring22–24. COG1938 members are found in all examined archaea and, intriguingly, also all contain a highly conserved C-terminal HbYX motif (Fig. 1b).
Figure 1. A HbYX motif in the yeast Pba1–Pba2 assembly factor and its archaeal orthologs.
(a) Distribution of DUF75 superfamily members among the eukarya and archaea. In eukaryotes, Pba2/PAC2 proteins belong to the DUF75 superfamily; PAC1 proteins are also related to this superfamily. All archaea species encode at least two DUF75 proteins, which cluster into two conserved orthologous groups (COGs).
(b) A C-terminal HbYX motif (underline) is conserved in eukaryotic Pba1/PAC1 (top) as well as archaeal DUF75 superfamily members (middle, bottom). Hsap, Homo sapiens; Mmus, Mus musculus; Ggal, Gallus gallus; Xlae, Xenopus laevis; Drer, Danio rerio; Dmel, Drosophila melanogaster; Atha, Arabidopsis thaliana; Scer, Saccharomyces cerevisiae; Mmar, Methanococcus maripaludis; Mjan, Methanocaldococcus jannaschii; Aful, Archaeoglobus fulgidus; Taci, Thermoplasma acidophilum; Tkod, Themococcus kodakarensis; Aper, Aeropyrum pernix; Paer, Pyrobaculum aerophilum; Hbut, Hyperthermus butylicus; Ihos, Ignicoccus hospitalis; Saci, Sulfolobus acidocaldarius.
These bioinformatic results suggest that proteins belonging to the COG1938 and COG2047 groups may represent archaeal orthologs of the eukaryotic Pba1and Pba2 proteins and that this family of proteins might use the conserved HbYX motif in binding to 20S proteasomes.
Functional Significance of HbYX Motifs in Yeast Pba1–Pba2
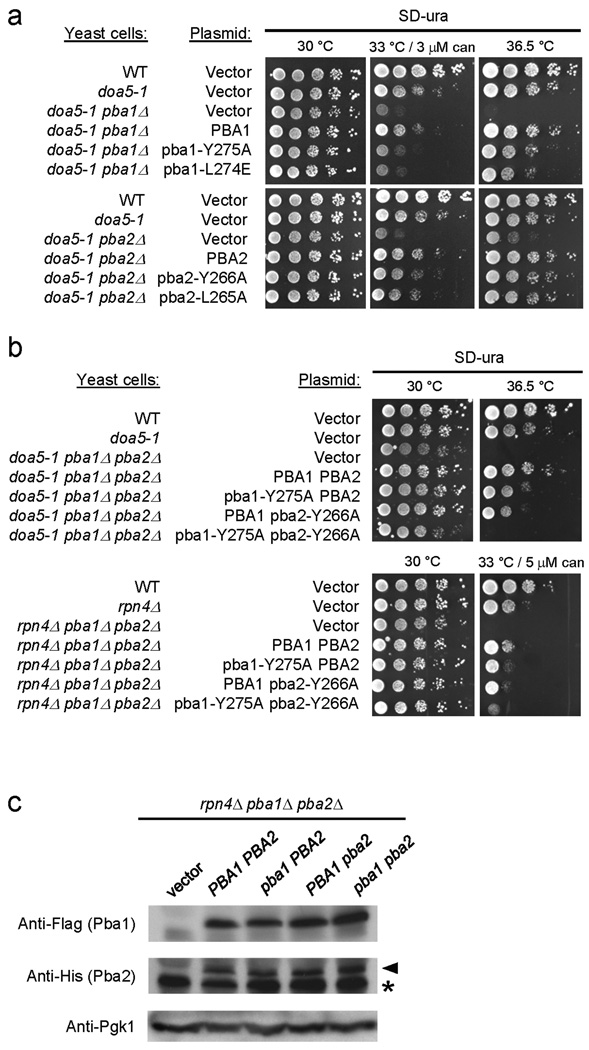
All eukaryotic Pba1/PAC1 orthologs have the highly conserved HbYX motif, as do fungal Pba2 proteins; by contrast, nonfungal Pba2/PAC2 proteins terminate with a terminal hydrophobic-tyrosine/phenylalanine (Hb-Y/F) motif (Suppl. Fig. 2a). We tested whether the HbYX motifs of yeast Pba1 and/or Pba2 contribute to Pba1–Pba2 function in vivo. Neither pba1Δ nor pba2Δ strains (nor the pba1Δ pba2Δ double mutant) exhibit any obvious growth defects by themselves (Suppl. Fig. 2b and Ref. 6). However, Pba1–Pba2 becomes important in cells in which proteasome function has been compromised6. Therefore, we used strains that included either a point mutation in the 20S α5 subunit (doa5-1) or a deletion of RPN4, which encodes a transcription factor necessary for normal proteasome expression (Fig. 2)27,28. Whereas WT PBA1 and PBA2 introduced on low-copy plasmids complemented the growth defects of the respective mutants, pba1 or pba2 alleles with HbYX point mutations only partially rescued the growth defects (Fig. 2a), suggesting that both Pba1 and Pba2 HbYX motifs contribute to in vivo activity.
Figure 2. The HbYX motifs of yeast Pba1–Pba2 are functionally important in vivo.
(a, b) The indicated yeast strains were transformed with either an empty low-copy vector or vector with the indicated PBA genes. Six-fold dilution series of liquid cultures were spotted onto various media and incubated as indicated. can, canavanine sulfate.
(c) The rpn4Δ pba1Δ pba2Δ triple mutant was transformed with a plasmid carrying the WT (PBA) or mutant (pba) alleles shown in (b). Lysates were prepared and analyzed by two immunoblots. The first was probed with anti-Flag (Pba1) antibody, and the membrane stained with Ponceau S to compare loading. The second was probed with anti-His (Pba2) antibody and reprobed with anti-Pgk1 to compare loading. Asterisk denotes non-specific band.
The modest deficiencies of the single HbYX mutants could be due to functional overlap of the Pba1 and Pba2 HbYX motifs. Therefore, we co-expressed mutant Pba1 and Pba2 proteins and tested their ability to restore normal growth to pba1Δ pba2Δ rpn4Δ and pba1Δ pba2Δ doa5-1 triple mutants. Strains with HbYX mutations in both Pba1 and Pba2 exhibited defects almost as severe as a complete loss of these proteins (Fig. 2b). The defects in the double HbYX mutants were not due to decreased protein expression (Fig. 2c, Suppl. Fig. 3, and Supplementary Methods).
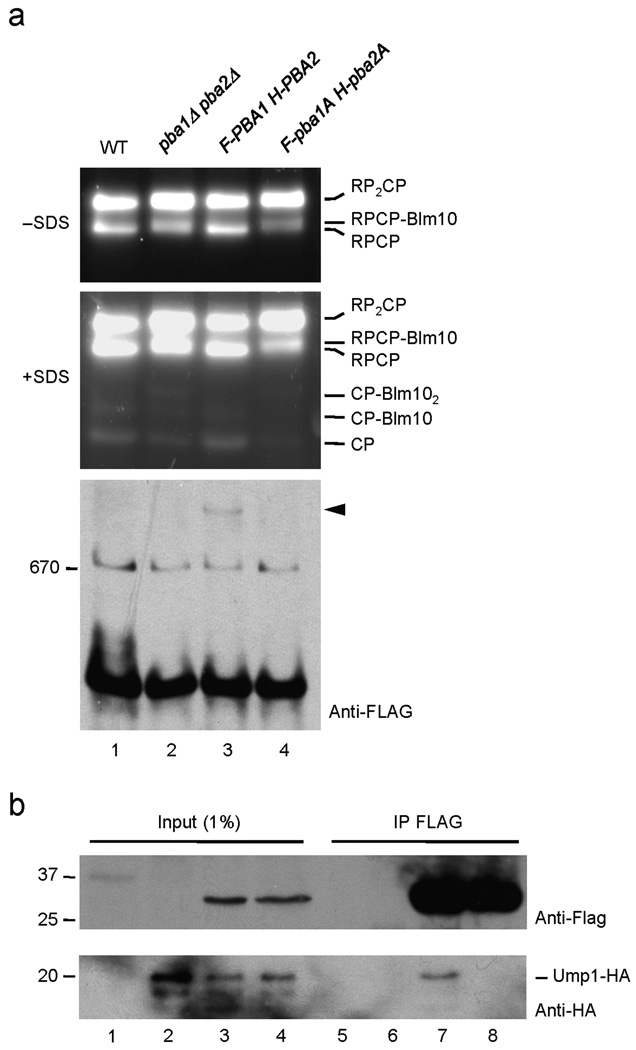
Although pba1Δ and pba2Δ mutants by themselves exhibit no obvious growth impairment, a very weak 20S proteasome assembly defect is observed9. Following fractionation of WT yeast proteins by native PAGE and incubation of the gel with a fluorogenic peptide substrate, most proteolytically active proteasomes are in the form of doubly (RP2CP) and singly (RPCP) capped 26S proteasomes with a small amount detectable as free 20S (CP; the latter requires SDS for activation) (Fig. 3a, lane 1). Elimination of Pba1–Pba2 caused a subtle reduction in free 20S (lane 2, +SDS). Moreover, the dearth of functional 20S particles led to a higher fraction of 20S in the doubly capped RP2CP form and in association with Blm10 (RPCP–Blm10 and CP–Blm102). Epitope-tagged WT Pba1 and Pba2 restored the normal profile of proteasomal particles to pba1Δ pba2Δ cells; in contrast, versions of these proteins with the critical HbYX tyrosine mutated to alanine failed to do so (Fig. 3a, lanes 3 and 4).
Figure 3. The HbYX motifs of yeast Pba1–Pba2 are necessary for proteasome precursor binding.
(a) Native PAGE fractionation of yeast lysates. Samples for lanes 2–4 derive from pba1Δ pba2Δ cells transformed with plasmids that lack inserts (2); carry FLAG-tagged PBA1 and His-tagged PBA2 (3); or bear tagged versions of HbYX-mutated alleles, pba1-Y275A and pba2-L265A. The top two panels show fluorogenic substrate overlays performed as in Fig. 4b; 0.02% SDS was added after the initial (top) picture was taken to visualize free 20S proteasomes (CP; middle panel). The lower panel is a native gel anti-FLAG immunoblot. A HbYX-dependent intermediate species is indicated by the arrowhead.
(b) Co-precipitation of the 20S proteasome assembly factor Ump1 with FLAG-tagged Pba1 is HbYX-dependent.
Interestingly, the FLAG-tagged WT Pba1 protein was detected in a presumptive assembly intermediate (Fig. 3a, lower panel, arrowhead), but the HbYX-mutated version was not. This complex was of very low abundance, so its precise identity could not be ascertained. However, we had previously shown that Pba1–Pba2 associates exclusively with proteasome assembly intermediates, most of which contain the Ump1 assembly factor as well6. Consistent with this, immunoprecipitation of Flag-Pba1 co-precipitated Ump1 (Fig. 3b, lane 7). Most importantly, this interaction also depended on the HbYX motifs (lane 8).
From these results, we conclude that the HbYX motifs in Pba1 and Pba2 contribute to proteasome assembly in vivo, are required for Pba1–Pba2 binding to Ump1-containing proteasome precursors, and are partially redundant with one another.
Characterization of Archaeal PbaA and PbaB Proteins
Mechanistic analysis of the contribution of the Pba1–Pba2 HbYX motifs to proteasome binding and assembly is complicated by the complexity of the eukaryotic 20S proteasome and the lack of an in vitro assembly system. We therefore turned our attention to the putative archaeal versions of this assembly factor (Fig. 1). We cloned the genes for the COG1938 and COG2047 proteins from the archaeon Methanococcus maripaludis S2, whose genome has been sequenced25. Open reading frames Mmp0914 (COG1938) and Mmp1611 (COG2047), hereafter referred to as pbaA and pbaB, respectively, encode proteins of 26.4 and 27.5 kDa, respectively. The genes both express well in E. coli.
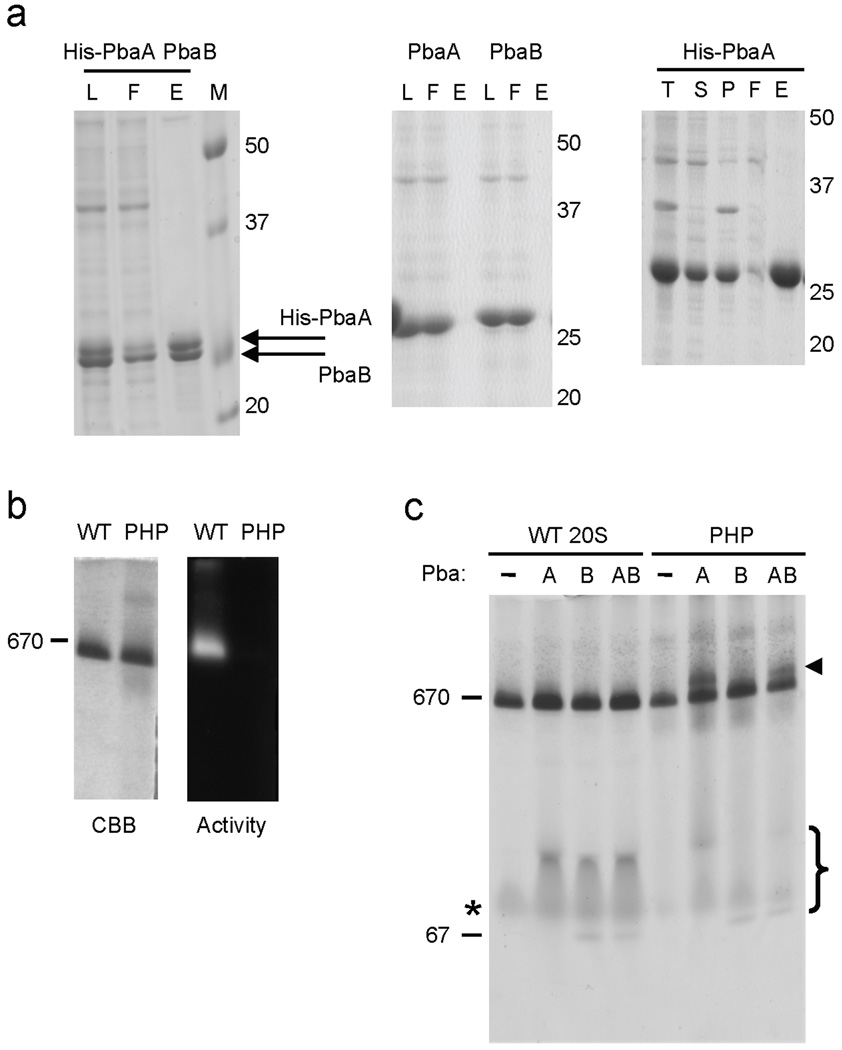
Pba1–Pba2 and PAC1–PAC2 form heterodimers6,7, so we tested whether the putative archaeal orthologs could also form complexes. When an N-terminally His6-tagged version of the PbaA protein was coexpressed with an untagged version of PbaB, the two proteins coeluted from Ni-NTA resin in equimolar amounts, as judged by protein staining (Fig. 4a, left panel). The same result was observed when lysates expressing these two proteins individually were mixed first and then applied to Ni-NTA resin (not shown). Coelution was specific because untagged versions of these proteins did not bind the resin (Fig. 4a, right panel). The PbaA–PbaB complex was no longer observed when these eluates were analyzed by native PAGE, suggesting it is not stable under the conditions used for nondenaturing electrophoresis (Suppl. Fig. 4). We conclude that PbaA and PbaB can interact in vitro.
Figure 4. Archaeal PbaA protein binds preferentially to a 20S proteasome intermediate.
(a) Archaeal PbaA and PbaB proteins form a complex. Lysates of E. coli expressing the indicated protein combinations were fractionated by Ni-NTA. L, load. F, flow-through. E, eluate. T, total lysate. S, soluble fraction (equivalent to L). P, pellet fraction.
(b) Recombinant M. maripaludis wild-type 20S proteasomes (WT, α-His+βΔpro) and preholoproteasomes (PHP, α-His+β(T1A)) were affinity purified on a Ni-NTA resin and electrophoresed on a nondenaturing 4–10% gradient gel. Proteolytic activity was visualized using a Suc-LLVY-AMC fluorogenic substrate (right panel), and the gel was then stained with GelCode Blue (CBB; left). Migration of the thyroglobulin protein standard (670 kDa) is indicated.
(c) PbaA is sufficient to bind PHP. Lysates of E. coli expressing either His-tagged wild-type archaeal proteasomes (WT) or preholoproteasomes (PHP) were mixed with lysates of E. coli expressing the indicated untagged Pba proteins. The mixtures were fractionated by Ni-NTA binding and native PAGE. Proteins were visualized with GelCode Blue. Arrowhead denotes position of a 20S–Pba protein complex. Non-assembled α subunit species are marked with an asterisk, and free PbaA and PbaB are marked with a bracket. Free Pba proteins in the WT samples may have coeluted from Ni-NTA with the non-assembled α subunits or could represent weakly bound species that did not survive native electrophoresis. Thyroglobulin (670 kDa) and BSA (67 kDa) protein standards are indicated. The gel-shifted species (arrowhead) were excised and analyzed by LC-MS/MS, resulting in the identification of the indicated proteins (bottom).
Pba1–Pba2 and PAC1–PAC2 directly bind a subset of proteasome α subunits in vitro, although the functional relevance of these specific interactions remains unknown6,7. If PbaA and PbaB are orthologs of these proteins, they might also exhibit affinity for the archaeal proteasome α subunit. We cloned the M. maripaludis proteasome α subunit gene (psmA) and coexpressed it along with either of the putative chaperones in E. coli. Both PbaA and PbaB interacted with the α subunit when the latter was expressed with a C-terminal His-tag (Suppl. Fig. 5). The low level of co-purified PbaA or PbaB suggests either weak or substoichiometric binding. These results suggest that the M. maripaludis PbaA and/or PbaB proteins may indeed be archaeal orthologs of Pba1–Pba2/PAC1–PAC2.
Archaeal Pba Proteins Associate with 20S Intermediates
If PbaA and/or PbaB are functionally similar to Pba1–Pba2, they might also associate preferentially with proteasome assembly intermediates. We generated one such intermediate, the pre-holoproteasome (PHP), by coexpressing the archaeal α subunit along with a β subunit (β-T1A) that cannot cleave off its 14-residue propeptide21,26. For comparison, “wild-type” (WT) 20S proteasome was generated by coexpressing WT α subunit with a β-subunit version(βΔpro) that lacked its propeptide, ensuring fully mature 20S proteasomes (see Supplemental Methods and Suppl. Fig. 6). We confirmed assembly by subjecting the Ni-NTA-purified proteasomes to native PAGE and both protein and activity staining (Fig. 4b). The PHP migrated at the same position as the WT proteasome but, unlike WT, did not exhibit any peptidase activity (Fig. 4b).
Lysates of E. coli expressing His-tagged versions of WT 20S or PHP were mixed with lysates expressing untagged versions of PbaA, PbaB, or both proteins followed by proteasome isolation using Ni-NTA chromatography. Resolution of the eluates by native PAGE revealed a novel species migrating above free PHP after PHP incubation with PbaA or PbaA plus PbaB (but not PbaB alone) (Fig. 4c, arrowhead). In contrast, no gel-shifted species were observed with any combination of WT 20S and Pba proteins. The gel-shifted complexes from PHP-mixed samples were excised and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Multiple peptides corresponding to proteasome α and β subunits as well as PbaA were identified (Table 1). In the PbaA plus PbaB sample (last lane), a single peptide corresponding to PbaB was also observed. It is possible that this reflects a complex of PbaA–PbaB binding to the PHP from which PbaB is stripped during electrophoresis (Suppl. Fig. 5), but this cannot be concluded from this result alone (see below).
Table 1.
| PHP + PbaA |
||
| Protein | No. of peptides | Coverage (%) |
| α subunit | 72 | 65 |
| β subunit | 43 | 39 |
| PbaA | 32 | 54 |
| PHP + PbaA-PbaB |
||
| Protein | No. of peptides | Coverage (%) |
| α subunit | 52 | 65 |
| β subunit | 32 | 37 |
| PbaA | 9 | 34 |
| PbaB | 1 | 6 |
HbYX-Dependent Binding of PbaA to 20S Intermediates
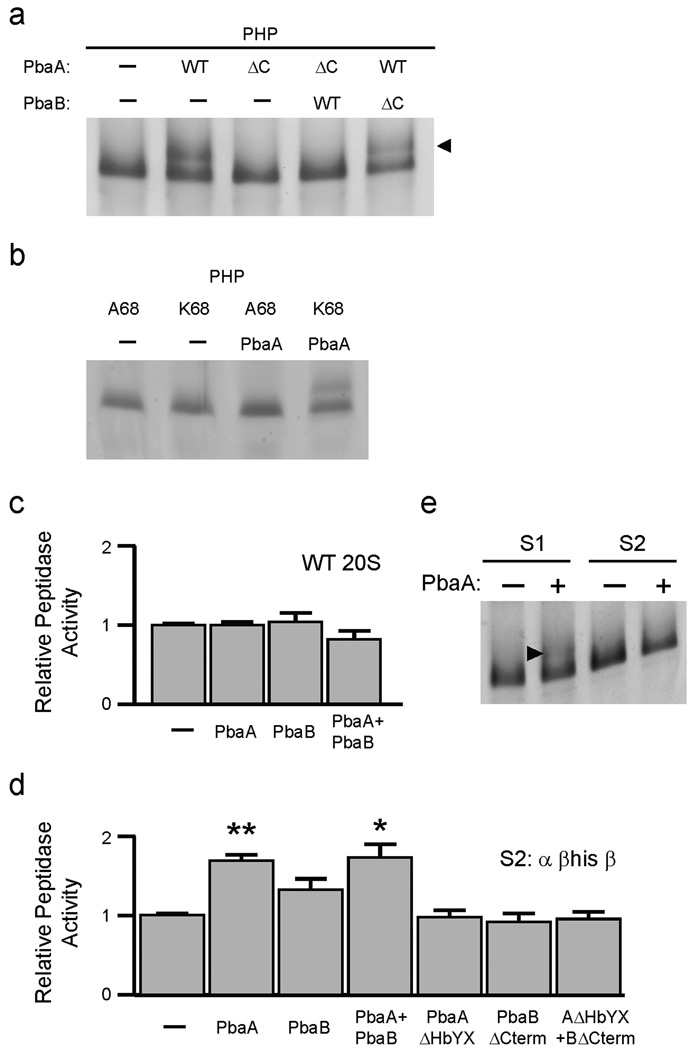
Inasmuch as PbaA alone, but not PbaB, associates stably with the PHP, and only PbaA contains a conserved HbYX motif (Fig. 1b), we asked if the PbaA–PHP association was HbYX-dependent. We repeated the PHP binding analysis using mutant versions (ΔC) of PbaA and PbaB lacking their three C-terminal residues (Fig. 5a). Deleting the PbaA HbYX motif prevented formation of the PbaA–PHP complex. The same PbaA HbYX deletion also abrogated PHP binding in the presence of PbaB, but mixing PbaB-ΔC with wild-type PbaA did not block binding of the latter to the PHP. Therefore, PbaA requires its HbYX motif to associate stably with the PHP.
Figure 5. PbaA HbYX motif mediates binding to the α ring of 20S proteasome intermediates.
(a) Deletion of the HbYX motif of archaeal PbaA (ΔC) abrogates binding to the PHP (β-T1A) intermediate. Proteins were analyzed as in Fig. 4. ΔC denotes Pba protein lacking last 3 residues.
(b) Mutation of proteasome α-subunit lysine-68 to alanine prevents PHP binding to PbaA.
(c) Archaeal Pba proteins do not stimulate mature (WT) 20S proteasome activity. Purified recombinant 20S archaeal proteasomes (α+βΔpro) were incubated with the indicated recombinant Pba proteins. Activity in the absence of Pba proteins was set to unity. Error bars represent standard errors.
(d) Archaeal Pba proteins stimulate peptidase activity of S2 proteasome intermediates (α+βHis+β). Asterisks denote statistically significant differences from the S2-only sample (**, p = 0.003; *, p = 0.049; n = 3). Activity of S2 in the absence of Pba proteins was set to unity.
(e) PbaA causes gel-shift of S1 (α+β (T1A)His+β) but not S2 (α+βHis+β). Lysates of E. coli expressing either His-tagged S1 or S2 were mixed with lysates of E. coli expressing untagged PbaA. The mixtures were fractionated as in Fig. 4c, and proteins were visualized with GelCode Blue. Arrowhead denotes the position of PbaA-shifted proteasomal S1 species.
We then asked whether the PbaA HbYX motif bound to the same proteasome sites as the HbYX motifs in activators such as PAN. Activators bind to specific 20S surface pockets formed between the α subunits, which contain a highly conserved lysine residue22; when this lysine is mutated, PAN binding is lost. We generated the analogous mutation in the M. maripaludis α subunit (K68A) in the context of the PHP. Wild-type PbaA did not bind detectably to the K68A PHP (Fig. 5b and Suppl. Fig. 7). Together, these results strongly suggest that the PbaA HbYX motif docks into the same proteasome α-ring pocket as the HbYX motifs of known proteasome activators, although PbaA binds preferentially to assembly intermediates (the PHP) rather than mature 20S proteasomes.
Pba Proteins and 20S Activation
Because previously characterized HbYX-motif proteins such as PAN or Blm10 act as proteasome activators, we asked if Pba proteins were activators as well. Recombinant yeast Pba1–Pba2 did not stimulate the peptidase activity of purified yeast 20S proteasomes even though the latter were activation-competent (Suppl. Fig. 8a). We also did not detect binding between purified recombinant Pba1–Pba2 and purified yeast 20S proteasomes (not shown). Others have reported that PAC1–PAC2 is degraded by 20S proteasomes upon completion of assembly7, so the absence of 20S binding and activation by purified Pba1–Pba2 might be due to its degradation during co-incubation. However, we saw no degradation of Pba1–Pba2 after prolonged incubation with yeast proteasomes (Suppl. Fig. 8b). Thus, Pba1–Pba2 does not appear to function as an activator, at least not in vitro, despite both subunits having HbYX motifs.
Similarly, we saw no significant increase in the peptidase activity of WT archaeal 20S in the presence of archaeal Pba proteins (Fig. 5c), consistent with our failure to detect binding between these factors and holo-20S by native PAGE (Fig. 4c). PbaA and PbaB were also stable in the presence of archaeal 20S (Suppl. Fig. 9). This suggests that the archaeal Pba proteins also do not function as activators.
The apparent lack of binding between archaeal Pba proteins and mature 20S proteasomes prompted us to create 20S intermediates containing mixtures of processed and unprocessed β subunit propeptides. Such particles might retain detectable binding affinity for Pba (as seen with inactive PHP) but also contain some mature β subunits whose peptidase activity can be monitored. The aim was to find a sensitive and quantitative fluorometric assay for Pba–proteasome interaction. We constructed two operons for recombinant protein expression (see Supplementary Methods) that give rise to two sets of 20S particles (S1 and S2) of intermediate maturity between PHP and fully mature 20S (Suppl. Fig. 10a, b). The S1 (“earlier”) intermediates had, on average, fewer matured particles than S2. Using purified S1 and S2 (Fig. 5d and Suppl. Fig. 10c, d), we found a small (<2-fold) but significant stimulatory effect on peptidase activity in the presence of PbaA or PbaA plus PbaB. A slight effect was also observed with PbaB alone, though it did not rise to statistical significance. Stimulation by PbaA was dependent on its HbYX motif, suggesting the assay was specifically reporting on the binding of archaeal Pba proteins to these species. Although PbaA stimulated the peptidase activity of both S1 and S2, a gel shift was only detected with S1 (Fig. 5e and Suppl. Fig. 11).
Our results support a number of conclusions. First, the peptidase assay is sensitive enough to detect weak binding of archaeal Pba proteins to proteasomes, including binding to species that are (S1), or are not (S2), detectably bound by PbaA as determined by native PAGE. Second, the archaeal Pba proteins, like eukaryotic Pba1–Pba2, are unlikely to function as appreciable activators of the mature 20S proteasome. Finally, during maturation from PHP to holo-20S, a β-subunit propeptide processing threshold is apparently reached whereupon PbaA binding to the maturing 20S particle is substantially weakened.
Archaeal Pba Binding to Proteasomes with Engaged Active Sites
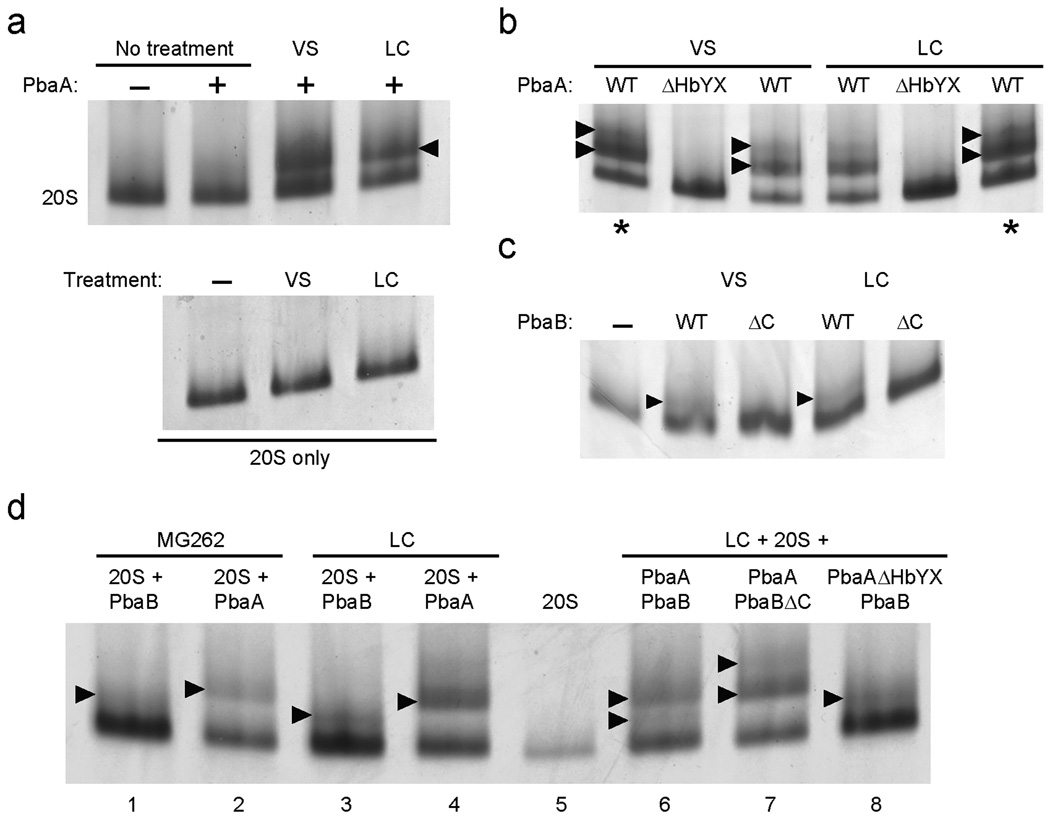
Crystal structures of mature archaeal 20S proteasomes and a 20S mutant (β-T1G), equivalent to the PHP described here, reveal a less intimate and more disordered interface between β-subunit rings in the PHP29. The α rings in holo-20S and PHP look essentially identical, which raises the question of why archaeal PbaA and eukaryotic Pba1–Pba2 proteins bind preferentially to the PHP. The mature 20S is a dynamic complex that alternates between (at least) two states, open and closed30,31, due to motions of the α subunit N-terminal tails32,33. A recent study uncovered a shift in the balance between closed and open conformations for yeast 20S proteasomes that were idle (i.e. substrate-free) or had their active sites engaged (by substrates, inhibitors, or propeptides in T1A mutants)34. As observed above (Fig. 4c), PbaA failed to induce a gel-shift of the untreated WT 20S (Fig. 6a, top). Remarkably, however, if prior to the Ni-NTA-binding step we treated the lysate mixtures with an inhibitor (peptide vinyl sulfone (VS) or lactacystin (LC)), we observed clear gel-shifts (Fig. 6a). A second, slower-migrating species above 20S was also detected (Fig. 6b; more easily visualized in the overloaded lanes, denoted by asterisks). We observed this second species in experiments with PHP too, but its detection was more variable (not shown). The simplest explanation for the upper gel-shifted band is that it represents PbaA proteins associated with both ends of the 20S cylinder. The gel-shifts were not due to 20S modification by inhibitor, as treatment of 20S proteasomes in the absence of PbaA did not alter their migration (Fig. 6a, bottom). Inhibitor treatment abolished the peptidase activity of the 20S, indicating (near) complete modification of all active sites (not shown). The analysis was repeated with C-terminally truncated PbaA (ΔHbYX) (Fig. 6b). As observed with PHP, the gel-shifts of VS-treated and LC-treated mature proteasomes were eliminated when PbaA lacked the HbYX motif. These data strongly suggest that PbaA can associate stably with the α ring of mature 20S proteasomes when the proteasomes are bound to active-site inhibitors.
Figure 6. Binding of archaeal Pba proteins to inhibitor-treated mature 20S proteasomes.
(a) Lysates of E. coli expressing WT (α-His+βΔpro) proteasomes were mixed with lysates of E. coli expressing untagged PbaA or with buffer. Where indicated, Z-Leu3-vinyl sulfone (VS) or clasto-lactacystin (LC) were added to 50 µM. For the control experiments in the lower panel, no PbaA was added. Samples were fractionated by native PAGE; gels were stained with GelCode Blue. Arrowhead denotes position of PbaA–20S species.
(b) E. coli lysates containing WT proteasomes and lysates with untagged full-length PbaA (WT) or PbaA lacking its last 3 residues (ΔHbYX) were mixed and analyzed as in (a).
(c) Lysates of E. coli expressing WT proteasomes were mixed with lysates of E. coli expressing untagged full-length PbaB (WT) or PbaB lacking its last 3 residues (ΔC) Samples were processed as in (a) except a 4–7% gradient gel was employed.
(d) E. coli lysates with WT proteasomes and lysates with the indicated untagged archaeal Pba proteins were mixed and analyzed as in (c). Where indicated, LC (50 µM) or MG-262 (5 µM) was added. For MG-262, the inhibitor was also added to all buffers during the Ni-NTA fractionation.
Because PbaB failed to bind detectably to either PHP or mature 20S proteasomes using the native-PAGE assay, we were surprised that PbaB produced a gel-shifted species in VS- and LC-treated 20S proteasomes (Fig. 6c). The presence of PbaB in the gel-shifted bands was verified by LC-MS/MS (not shown). As with PbaB-stimulated peptidase activity, the PbaB-induced gel-shifts required an intact PbaB C-terminus (Fig. 6c). These findings are consistent with the weak stimulation by PbaB of peptidase activity in the S1 and S2 “precursors” (Fig. 5). Finally, treatment with a proteasome inhibitor, MG-262, that has different structural34 and functional35 effects on yeast 20S proteasomes, nevertheless led to both PbaA- and PbaB-induced mobility shifts of the archaeal proteasome (Fig. 6d).
We also tested whether a PbaA–PbaB complex might bind to mature proteasomes in the presence of active-site inhibitors. To this end, we incubated lysates containing His-tagged mature 20S proteasomes with: i) lysates containing both wild-type PbaA and PbaB, ii) lysates with PbaAΔHbYX and wild-type PbaB, or iii) lysates with PbaA and PbaBΔC (Fig 6d, lanes 6–8). All samples were treated with LC prior to the Ni-NTA step. Coexpression of PbaA and PbaB (lane 6) resulted in two gel-shifted species; a lower band similar to that produced by PbaB alone (compare to lane 3) and an upper one similar to that seen with PbaA alone (compare to lane 4). In the PbaAΔHbYX plus PbaB sample, only the lower band was present (lane 8). By contrast, in the PbaA plus PbaBΔC sample, this lower band was absent (lane 7). These data are consistent with the identification of this species as a 20S–PbaB complex. The two slower migrating species in the PbaA plus PbaBΔC sample (lane 7) were analyzed by LC-MS/MS. For each, many peptides derived from PbaA as well as the proteasome α and β subunits were identified but none from PbaB (not shown), suggesting that PbaBΔC is not able to stably associate with PbaA (under conditions of native PAGE) when the latter binds to the proteasome.
In summary, the results suggest that occupation of the archaeal proteasome active sites by inhibitors (or propeptides) triggers an allosteric signal that modulates binding of PbaA and PbaB to the outer surface of the α-ring.
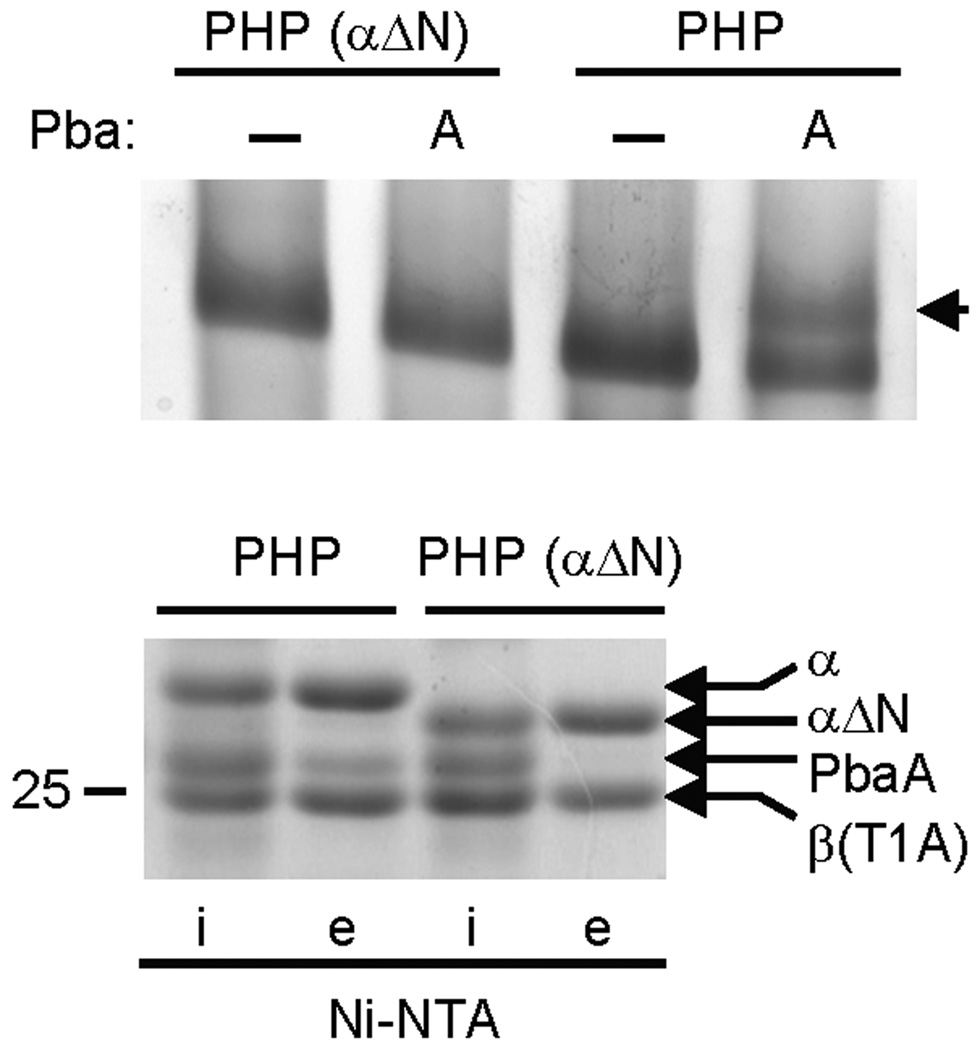
PbaA Binding to Proteasomes Requires the α-ring N-termini
That archaeal Pba proteins discriminate between fully-matured 20S proteasomes and assembly intermediates (or inhibitor-treated proteasomes) is consistent with previous observations, confirmed here (Fig. 3), showing that eukaryotic Pba1–Pba2 and PAC1–PAC2 associate with assembly intermediates such as the PHP but not mature proteasomes6,7. This presumably reflects an allosteric signal originating in the β ring that is transmitted to the α ring. The best-documented conformational change that occurs in the α ring is the opening and closing of the central gate32,36 constituted by the N-termini of the α subunits. To explore whether the α ring gate is relevant to Pba binding, we generated a permanently open PHP33 by deleting 13 residues from the α subunit N-terminus (αΔN PHP) and tested it for its ability to bind PbaA. The αΔN mutation abolished the gel shift normally observed in the presence of PbaA (Fig. 7, top), and this was confirmed by the absence of PbaA in Ni-NTA eluates of His-tagged αΔN PHP (Fig. 7, bottom). We conclude that the residues comprising the α-ring gate must be present in order for PbaA to interact with the PHP.
Figure 7. Binding of PbaA to PHP requires the α-subunit N-termini.
Lysates of E. coli expressing His-tagged PHPs (α+β(T1A)His) containing either wild-type α subunits (PHP) or mutant α subunits lacking their 13-residue N-terminal tails (PHP αΔN) were mixed with buffer (−) or with lysates of E. coli expressing untagged PbaA. The mixtures were fractionated by Ni-NTA and the eluates subjected to native PAGE on 4–10% gradient gels (top gel). Arrowhead denotes PbaA–PHP species. The input (i) and eluate (e) fractions from the Ni-NTA resin were also separated by 11% SDS-PAGE (bottom gel). Migration of the 25 kDa protein standard is indicated. For both gels, proteins were stained with GelCode Blue.
DISCUSSION
Considerable gaps remain in our understanding of basic functional properties of proteasome assembly chaperones, including Pba1–Pba2/PAC1–PAC2. Here we demonstrate that the yeast Pba1–Pba2 chaperone utilizes C-terminal HbYX motifs to bind to proteasome precursors and promote 20S proteasome assembly. In proteasome activators, these motifs function in opening the α-ring gate to the 20S interior. In contrast, Pba1–Pba2 only binds detectably to 20S precursors and is not an appreciable 20S activator. Surprisingly, Pba1–Pba2 is conserved even in archaea despite the compositional simplicity of archaeal proteasomes. The related M. maripaludis PbaA and PbaB proteins also form a complex, and PbaA (and possibly a PbaA–PbaB complex) also preferentially associates with proteasome assembly intermediates containing unprocessed catalytic β subunits. The conserved PbaA HbYX motif binds to the same surface pocket in the α-ring that is bound by proteasomal activators. Strikingly, mature archaeal proteasomes can be induced to bind PbaA and PbaB when the β subunits are bound to active-site inhibitors. This supports a model in which PbaA (and PbaB) binding is allosterically regulated by occupation of the active sites by β-subunit propeptides. When these are processed at the end of proteasome assembly, chaperones dissociate from the distal α-ring surface.
Our results identify for the first time the region on the proteasome to which Pba1–Pba2 binds, namely, the outer surface of the α-ring. This explains the failure to observe a physical interaction between PAC1–PAC2 and hUmp17, inasmuch as the two assembly factors bind to opposite faces of α-ring-containing intermediates, and the observation that C-terminally peptide-tagged versions of Pba1 and Pba2 are not able to fully complement the respective yeast deletion strains8. Neither Pba1–Pba2 nor archaeal Pba proteins function as proteasome activators, at least in vitro, consistent with our previous work showing Pba1–Pba2 only associates detectably with proteasome assembly intermediates in vivo6. Interestingly, a metazoan proteasome inhibitor protein (PI31) that can compete with activators for binding to the core particle also contains a C-terminal HbYX motif37,38. Although it has not yet been shown that PI31 utilizes this motif for 20S binding, it would appear that HbYX motifs are not utilized exclusively by proteasome activators. The C-termini of activators share a number of functional similarities in the manner in which they bind 20S39. It will be interesting to determine whether marked differences exist between how HbYX motifs of activators and non-activators bind to the α-ring pockets. PbaA-proteasome association also requires the N-terminal α-subunit gate segments (Fig. 7), which might also bind PbaA directly.
Pba1–Pba2 and PAC1–PAC2 function as heterodimers6,7. Additional experiments are required to ascertain the nature of the functional Pba species in archaea. Our gel-shift experiments indicate a tight, HbYX-based association of PbaA with the PHP. This specificity of binding, and its persistence throughout our Ni-NTA-based fractionation steps in high salt buffers, strongly suggests physiological relevance. By contrast, PbaB, which lacks a HbYX motif, was unable to cause a similar gel-shift with PHP. Nevertheless, PbaB was able to cause a weak gel shift of inhibitor-treated WT proteasomes, leaving open the nature of the functional species (see Supplementary Notes).
The precise role of Pba1–Pba2/PAC1–PAC2 and archaeal Pba function in proteasome assembly remains to be determined, but one can envision several possibilities based on our findings. By preferentially binding to the outer α-ring surface of intermediates such as the PHP, these factors could prevent activators from prematurely binding to nascent assembly structures or may help sequester incorrectly formed species. Consistent with these models, we find that PbaA can distinguish different states of maturing archaeal 20S proteasomes. While peptidase assays indicate that PbaA interacts with both the partially matured S1 and S2 intermediate species, it only causes a detectable gel-shift of the 20S with the less completely matured S1 intermediates (Fig. 5e and Suppl. Fig. 11). The drop in apparent affinity for S2 relative to S1 suggests that PbaA can “sense” the ratio of processed and unprocessed β subunits.
The ability of archaeal Pba proteins to bind preferentially to the α rings of assembly intermediates indicates that these factors recognize a feature(s) present in these α rings but not in those of mature 20S. Crystal structures of the PHP-equivalent T1G mutant and mature 20S did not reveal any obvious differences between α ring surfaces29. However, dense packing typical of protein crystals can alter the gated structure of the α ring34. Strong support for the idea that the binding of PbaA (and PbaB) to the 20S is regulated allosterically by changes in the β-subunit active sites comes from binding analysis of inhibitor-treated mature 20S proteasomes (Fig. 6). Unlike untreated 20S, inhibitor-bound 20S particles form discrete complexes with both PbaA and PbaB. We propose that covalent modification of active sites with inhibitors mimics the presence of the propeptides found in immature proteasomes. During assembly, the presence of the intact propeptides of the β subunits sends an allosteric signal to the α-subunit N termini, resulting in an altered conformation that can be decoded by Pba proteins. Allosteric signaling from engaged active sites has been postulated previously to explain the stability of 19S RP-20S core particle complexes under conditions of active protein degradation35. We have not been able to detect Pba1–Pba2 binding to yeast 20S proteasomes treated with inhibitors (not shown). The salient point from the current study is that propeptide-dependent signaling occurs between the β-subunit active sites and the α-subunit ring, and this causes the α rings of immature or aberrant β-subunit-containing intermediates to adopt a conformation(s) recognized by Pba assembly chaperones.
20S proteasomes are also found in actinomycete bacteria and, intriguingly, so are DUF75 proteins. Genome linkage has been used to argue that actinomycete DUF75 proteins may somehow function in proteasome assembly40. However, structural features of actinomycete DUF75 proteins, and the mechanism of actinomycete 20S assembly41, make it unlikely that they function in the same manner as eukaryotic and archaeal Pba proteins (see Supplementary Notes and Suppl. Fig. 12).
Alternative functions for Pba proteins, consistent with their preferential binding to proteasome precursors, can also be proposed. They may promote β-subunit maturation, a role analogous to that proposed for Blm1012; however, we found no evidence for this (not shown). Pba proteins could also function during degradation. Peptide hydrolysis proceeds through intermediates that include covalent modification of the active site Thr1 residue, and an alternative view of inhibitor-treated 20S is that these species mimic such hydrolysis intermediates34. Consequently, Pba proteins could be recruited to actively hydrolyzing proteasomes, perhaps forming hybrid complexes with other activators, although experimental evidence for this is lacking. Finally, archaeal Pba and eukaryotic Pba1–Pba2 proteins may have roles in proteasome quality control. Proteasomes that have not properly matured or have their active sites blocked might be recognized by the Pba factors, thereby preventing activator association and ultimately eliminating them from the pool of available 20S proteasomes.
METHODS
Yeast Strains and Media
Yeast manipulations were carried out according to standard protocols42. Strains are listed in Supplementary Table I online. Serial dilutions were carried out as described9.
Plasmids
Plasmids are listed in Supplementary Table II online. DNA fragments encoding archaeal genes were amplified by PCR using Methanococcus maripaludis S2 genomic DNA as template (a kind gift of John Leigh, U. Washington). Yeast genes were amplified by PCR using S. cerevisiae genomic DNA as template, and included sequences extending 500 bp upstream and 500 bp downstream of the start and stop codons, respectively. Where required, primers incorporated N- or C-terminal epitope tags. Generation of multi-protein operons for bacterial expression was previously described9. Required mutations were introduced by Quikchange mutagenesis (Stratagene). Gene sequences were verified by automated DNA sequencing.
Bacterial Protein Expression
PbaA, PbaB, and Pba1–Pba2 were produced recombinantly in E. coli as described9. Frozen cell pellets were thawed on ice and resuspended in 0.6 ml of buffer A (50 mM HEPES-NaOH, pH 7.6, 0.5 M NaCl, 5 mM MgCl2) supplemented with 40 mM imidazole, 2 mM Pefabloc, 0.3 mg ml−1 lysozyme, 10 mg ml−1 DNase I and 0.1% (v/v) Triton X-100. The suspensions were lysed, centrifuged, and the soluble material applied to a Ni-NTA His SpinTrap (GE Healthcare) column as described9. The columns were washed twice with 0.6 ml buffer A supplemented with 40 mM imidazole and the bound material was eluted with 0.5 ml buffer B (50 mM HEPES-NaOH, pH 7.6, 0.5 M NaCl, 0.5 M imidazole, 5 mM MgCl2). Aliquots of total crude lysate as well as soluble, insoluble, flow-through, and partially purified eluate fractions were mixed with 5× SDS sample buffer and electrophoresed on 11% polyacrylamide gels. Nondenaturing gel analysis was carried out as described9 except 4–10% and 4–7% gradient gels were used instead, and these were run at 4 °C at constant voltages of 50 V and 80 V, respectively. Prior to nondenaturing PAGE, samples were desalted by serial centrifugation using Vivaspin 500 columns (Vivascience) with a molecular weight cut-off of 10 kDa in order to remove imidazole and lower the salt to below 150 mM. The T:C acrylamide ratio for nondenaturing gels was 30:1. Proteins were visualized using GelCode Blue (ThermoScientific).
Native Gel-shift Assays for Protein Binding
Frozen bacterial cell pellets were lysed and lysates centrifuged as above. Soluble fractions of various lysates were combined as described in the figure legends. Where indicated, proteasome inhibitors Z-LLL-vinyl sulfone, MG-262 (Enzo) and clasto-lactacystin β-lactone (Sigma) were added and the lysates incubated at 37 °C for 30 min. Lysates were then applied to Ni-NTA and processed as above. Eluates were concentrated five-to-ten fold using Vivaspin 500 columns, and the concentrates diluted in 500 µl of 50 mM Hepes-NaOH pH 7.6. Diluted samples were reconcentrated and rediluted in buffer C (50 mM HEPES-NaOH, pH 7.6, 0.15 M NaCl, 5 mM MgCl2). Protein concentrations were determined using the BCA Assay (ThermoScientific). Concentrated samples were electrophoresed on nondenaturing gels as above. The gels were subjected to substrate overlay and/or stained with GelCode Blue. Gel shifted bands were excised and sent to Midwest Bio Services LLC for analysis by Nano-LC-MS/MS.
Peptidase Activity Assays
Yeast 20S was purified as described43 from MHY1659 cells, which express FLAG-His6-Pre1 (β4) subunit from the normal chromosomal locus. Briefly, cells grown in YPD to an OD600 of ~1.8 were pelleted and ground to a powder under liquid nitrogen. The powder was thawed in one pellet volume of Buffer AY (50 mM Tris HCl pH 7.5, 150 mM NaCl, 10% glycerol, 5 mM MgCl2). The extract was centrifuged for 20 min at 34,310g in a Sorvall RC-5B Refrigerated Superspeed Centrifuge, and the supernatant was applied to 300 µl of anti-FLAG M2 agarose beads for 2 h on a rotating wheel at 4 °C. The beads were then collected in a column and washed with 50 volumes of Buffer AY containing 0.2% Triton X-100 (v/v). Bound proteins were eluted for 3 h at 4 °C with Buffer AY containing 100 µg ml−1 FLAG peptide. To remove FLAG peptide, the eluate was dialyzed in 4 L of Buffer AY overnight using a Slide-A-Lyzer cassette (100,000 MWCO ThermoScientific). Proteasome concentrations were determined as described below, and aliquots were stored at −80 °C.
For peptidase assays with archaeal proteins, recombinant archaeal 20S and Pba proteins were partially purified from bacteria as above. Proteins were separated by 12% SDS-PAGE and stained with GelCode Blue to assess purity. A BSA standard curve was run simultaneously to measure the amount of purified protein by quantifying a digital image of the gel using a G-Box system (Syngene).
Suc-LLVY-AMC in DMSO was used as a fluorogenic substrate in peptidase assays (50 µM final concentration). The amount of DMSO never exceeded 1% (v/v). After testing for optimal ratios, proteasomes (1.54 pM) with PbaA–PbaB (77 pM) or Pba1–Pba2 (77 pM) were incubated in Buffer AY (150 µl reaction volume) at 37 °C for archaeal proteins, or at 30 °C for yeast proteins. Suc-LLVY-AMC hydrolysis was monitored at λex 360 nm and λem 430 nm for 10 min, measured every minute for 5 seconds, using an 814 Photonmultiplier (PTI) to detect fluorescence under constant voltage of 75 V in 3 mm black Suprasil quartz cuvettes. Excitations and emissions were read with a slit width of 2 nm. Cuvette stage temperature was maintained at 37 °C or 30 °C.
Statistical Analysis
Peptidase assay results were analyzed using two-tailed paired t-test. The means of proteasome activity, with and without the activator, were compared by this method. The fold difference of activity as compared to the untreated (without activator) was calculated. Since these data follow a normal distribution, a two-tailed paired t-test is appropriate to determine if the change observed is significant. We assumed a threshold value (alpha) for p to be 0.05 for each case. Values falling below this threshold were interpreted as being significant, and are indicated with an asterisk. Those that fell below a threshold of 0.01 are indicated by a double asterisk.
Supplementary Material
Acknowledgements
We thank Robert J. Tomko Jr. for comments on the manuscript. We also thank John Leigh (U. of Washington) for providing the archaeal genomic DNA. This work was supported by NIH grant GM083050 to M.H.
Footnotes
Author contributions
A.R.K. and M.H. developed the experimental approach. A.R.K., M.J.K., and R.Y.K carried out experiments. A.R.K. and M.H. wrote the paper.
References
- 1.Tanaka K. The proteasome: overview of structure and functions. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:12–36. doi: 10.2183/pjab.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Groll M, et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 3.Kusmierczyk AR, Hochstrasser M. Some assembly required: dedicated chaperones in eukaryotic proteasome biogenesis. Biol Chem. 2008;389:1143–1151. doi: 10.1515/BC.2008.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramos PC, Dohmen RJ. PACemakers of proteasome core particle assembly. Structure. 2008;16:1296–1304. doi: 10.1016/j.str.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- 6.Li X, Kusmierczyk AR, Wong P, Emili A, Hochstrasser M. beta-Subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint. Embo J. 2007;26:2339–2349. doi: 10.1038/sj.emboj.7601681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirano Y, et al. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature. 2005;437:1381–1385. doi: 10.1038/nature04106. [DOI] [PubMed] [Google Scholar]
- 8.Le Tallec B, et al. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol Cell. 2007;27:660–674. doi: 10.1016/j.molcel.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 9.Kusmierczyk AR, Kunjappu MJ, Funakoshi M, Hochstrasser M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat Struct Mol Biol. 2008;15:237–244. doi: 10.1038/nsmb.1389. [DOI] [PubMed] [Google Scholar]
- 10.Yashiroda H, et al. Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes. Nat Struct Mol Biol. 2008;15:228–236. doi: 10.1038/nsmb.1386. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt M, et al. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat Struct Mol Biol. 2005;12:294–303. doi: 10.1038/nsmb914. [DOI] [PubMed] [Google Scholar]
- 12.Marques AJ, Glanemann C, Ramos PC, Dohmen RJ. The C-terminal Extension of the β7 Subunit and Activator Complexes Stabilize Nascent 20 S Proteasomes and Promote Their Maturation. J Biol Chem. 2007;282:34869–34876. doi: 10.1074/jbc.M705836200. [DOI] [PubMed] [Google Scholar]
- 13.Fehlker M, Wendler P, Lehmann A, Enenkel C. Blm3 is part of nascent proteasomes and is involved in a late stage of nuclear proteasome assembly. EMBO Rep. 2003;4:959–963. doi: 10.1038/sj.embor.embor938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaneko T, et al. Assembly pathway of the Mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell. 2009;137:914–925. doi: 10.1016/j.cell.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Saeki Y, Toh EA, Kudo T, Kawamura H, Tanaka K. Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell. 2009;137:900–913. doi: 10.1016/j.cell.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Funakoshi M, Tomko RJ, Jr, Kobayashi H, Hochstrasser M. Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell. 2009;137:887–899. doi: 10.1016/j.cell.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roelofs J, et al. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature. 2009;459:861–865. doi: 10.1038/nature08063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Tallec B, Barrault MB, Guerois R, Carre T, Peyroche A. Hsm3/S5b participates in the assembly pathway of the 19S regulatory particle of the proteasome. Mol Cell. 2009;33:389–399. doi: 10.1016/j.molcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Lowe J, et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 20.Maupin-Furlow JA, et al. Proteasomes from structure to function: perspectives from Archaea. Curr Top Dev Biol. 2006;75:125–169. doi: 10.1016/S0070-2153(06)75005-0. [DOI] [PubMed] [Google Scholar]
- 21.Zwickl P, Kleinz J, Baumeister W. Critical elements in proteasome assembly. Nat Struct Biol. 1994;1:765–770. doi: 10.1038/nsb1194-765. [DOI] [PubMed] [Google Scholar]
- 22.Smith DM, et al. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rabl J, et al. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gillette TG, Kumar B, Thompson D, Slaughter CA, DeMartino GN. Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J Biol Chem. 2008;283:31813–31822. doi: 10.1074/jbc.M805935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendrickson EL, et al. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J Bacteriol. 2004;186:6956–6969. doi: 10.1128/JB.186.20.6956-6969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen P, Hochstrasser M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell. 1996;86:961–972. doi: 10.1016/s0092-8674(00)80171-3. [DOI] [PubMed] [Google Scholar]
- 27.Chen P, Hochstrasser M. Biogenesis, structure and function of the yeast 20S proteasome. Embo J. 1995;14:2620–2630. doi: 10.1002/j.1460-2075.1995.tb07260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie Y, Varshavsky A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc Natl Acad Sci U S A. 2001;98:3056–3061. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Groll M, Brandstetter H, Bartunik H, Bourenkow G, Huber R. Investigations on the maturation and regulation of archaebacterial proteasomes. J Mol Biol. 2003;327:75–83. doi: 10.1016/s0022-2836(03)00080-9. [DOI] [PubMed] [Google Scholar]
- 30.Osmulski PA, Gaczynska M. Atomic force microscopy reveals two conformations of the 20 S proteasome from fission yeast. J Biol Chem. 2000;275:13171–13174. doi: 10.1074/jbc.c901035199. [DOI] [PubMed] [Google Scholar]
- 31.Osmulski PA, Gaczynska M. Nanoenzymology of the 20S proteasome: proteasomal actions are controlled by the allosteric transition. Biochemistry. 2002;41:7047–7053. doi: 10.1021/bi0159130. [DOI] [PubMed] [Google Scholar]
- 32.Whitby FG, et al. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- 33.Groll M, et al. A gated channel into the proteasome core particle. Nat Struct Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 34.Osmulski PA, Hochstrasser M, Gaczynska M. A tetrahedral transition state at the active sites of the 20S proteasome is coupled to opening of the alpha-ring channel. Structure. 2009;17:1137–1147. doi: 10.1016/j.str.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleijnen MF, et al. Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nat Struct Mol Biol. 2007 doi: 10.1038/nsmb1335. [DOI] [PubMed] [Google Scholar]
- 36.Forster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 37.McCutchen-Maloney SL, et al. cDNA cloning, expression, and functional characterization of PI31, a proline-rich inhibitor of the proteasome. J Biol Chem. 2000;275:18557–18565. doi: 10.1074/jbc.M001697200. [DOI] [PubMed] [Google Scholar]
- 38.Zaiss DM, Standera S, Holzhutter H, Kloetzel P, Sijts AJ. The proteasome inhibitor PI31 competes with PA28 for binding to 20S proteasomes. FEBS Lett. 1999;457:333–338. doi: 10.1016/s0014-5793(99)01072-8. [DOI] [PubMed] [Google Scholar]
- 39.Stadtmueller BM, et al. Structural models for interactions between the 20S proteasome and its PAN/19S activators. J Biol Chem. 2009 doi: 10.1074/jbc.C109.070425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iyer LM, Burroughs AM, Aravind L. Unraveling the biochemistry and provenance of pupylation: a prokaryotic analog of ubiquitination. Biol Direct. 2008;3:45. doi: 10.1186/1745-6150-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharon M, Witt S, Glasmacher E, Baumeister W, Robinson CV. Mass spectrometry reveals the missing links in the assembly pathway of the bacterial 20 S proteasome. J Biol Chem. 2007;282:18448–18457. doi: 10.1074/jbc.M701534200. [DOI] [PubMed] [Google Scholar]
- 42.Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press; 1991. [Google Scholar]
- 43.Verma R, et al. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.