Abstract
It is not currently known in what state (folded, unfolded, alternatively folded) client proteins interact with chaperone Hsp90. We show that one client, the p53 DNA-binding domain, undergoes a structural change in the presence of Hsp90 to adopt a molten globule-like state. Addition of one- and two-domain constructs of Hsp90, as well as the full-length three-domain protein, to isotopically-labeled p53 results in reduction in NMR signal intensity throughout p53, particularly its central β-sheet. This reduction appears to be associated with a change of structure of p53 without formation of a distinct complex with Hsp90. Fluorescence and hydrogen-exchange measurements support a loosening in the structure of p53 in the presence of Hsp90 and its domains. We propose that Hsp90 interacts with p53 by multiple transient interactions, forming a dynamic heterogeneous manifold of conformational states that resembles a molten globule.
Keywords: protein-protein interaction, chaperone
The ~90 kDa heat shock protein Hsp90 is a highly conserved molecular chaperone found in great abundance in eukaryotic cells (~1% of cytosolic protein)1–3. It is involved in a large number of cellular processes, mainly related to the conformational stabilization and regulation of a specific set of client proteins, including steroid hormone receptors, kinases and polymerases. The specificity of Hsp90 for its various clients appears to be mediated through the interactions of co-chaperones such as p23 and Cdc37, and the cycle of binding and release of client proteins generally requires ATP. Hsp90 is a homodimer, with three domains per monomer, the N-terminal domain (N), a middle domain (M) and a C-terminal dimerization domain (C) (Figure 1a). Structures are available for the complete dimers of Escherichia coli HtpG4 and yeast Hsp825, but the conformations of the client proteins when bound to Hsp90 are unknown. There are no published reports of high-resolution structures of Hsp90-client protein complexes; an electron microscopic reconstruction of a complex between Hsp90, the co-chaperone Cdc37 and the client kinase Cdk4 has been reported6. The complexes between Hsp90 and different clients may be quite different, and may even involve different sites on the chaperone7.
Figure 1. Schematic diagram showing the domains of Hsp90 and p53.
(a) Domains of Hsp90 showing the position of the charged linker and the C-terminal tetratricopeptide-binding sequence. (b) Domains of p53. AD: N-terminal activation domain; PRD: proline-rich domain; DBD: DNA-binding domain; TD: tetramerization domain; BD: C-terminal regulatory domain. (c) 1H-15N HSQC spectrum of 15N-labeled p53 (black), and with the addition of unlabeled Hsp90 NM at the indicated concentration ratios. All spectra are plotted at the same contour level. Two examples of cross peaks that shift as NM is added are circled. The p53 sample used in these experiments contained two mutations (Y346F and T253I)13.
One Hsp90 client that has been extensively studied is the tumor suppressor p53, a 393-amino acid transcription factor that is mutated in over 50% of human cancers8,9. Most of the mutations seen in cancers occur in the DNA-binding domain of p53 (Figure 1b), which is also the site of interaction with Hsp9010–12. The Hsp90 binding affinity of the DNA-binding domain alone is similar to that of full-length p5312. We set out to determine the state of folding of the human p53 DNA-binding domain (residues 94–312) in the presence of the chaperone human Hsp90α using spectroscopic methods in solution.
RESULTS
Design of Hsp90 Domain Constructs
A series of constructs of human Hsp90α were prepared: single-domain constructs representing the N-terminal (N) and middle (M) domains (the single C-domain construct is not well-behaved in solution) and two-domain constructs containing the N + M (NM) and M + C (MC) domains. A shortened construct of full-length Hsp90, where part of a long charged linker between the N and M domains characteristic of eukaryotic Hsp90s had been deleted (Hsp90Δ) was used in preference to the full length protein, which was much more difficult to express and purify from bacterial cell culture in quantities suitable for NMR spectroscopy. Based on size-exclusion chromatography, the N, M, and NM constructs are monomeric in solution, and the MC domain and Hsp90Δ are homodimers.
Effect of Hsp90 on the NMR Spectrum of p53 DNA-Binding Domain
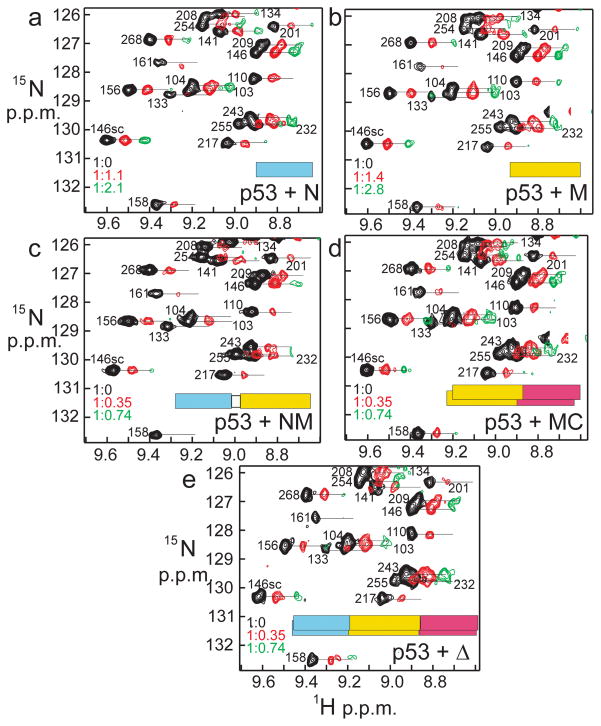
The major effect of addition of any of the Hsp90 constructs to 15N-labeled p53 DNA-binding domain is the lowered intensity of a large proportion of the cross peaks in the 1H-15N HSQC or TROSY NMR spectrum. This intensity loss occurs even for the stabilized mutant p53 that was used to obtain the solution structure13, as shown in Figure 1c. Some cross peaks (yellow in Figure 1c) remain at the same intensity in the p53 spectrum after addition of excess Hsp90 protein, indicating that the corresponding residues in the protein are likely independently mobile and not closely associated with the main body of the protein. Many of these persistent resonances correspond to the C- and N-terminal residues and to exterior mobile loops of the p53 domain. An expansion of one part of the p53 spectrum is shown in Figure 2, illustrating the effects of the various added Hsp90 proteins (full spectra are shown in Supplementary Fig. 1). The lowering of the cross peak intensity varies in extent as different Hsp90 constructs are added: addition of the two-domain and full-length constructs (Figure 2c–e) results in reduced intensity and eventual disappearance of resonances at lower titrant concentrations than for either of the single-domain constructs (Figure 2a, b). Interestingly, the same cross peaks in the p53 spectrum lose intensity with the addition of each of the Hsp90 constructs, all of which contain the N and/or M domain.
Figure 2. 1H-15N spectra of 15N-labeled p53 with added Hsp90.
Each panel a-e shows the same portion of the 1H-15N HSQC spectrum of 15N-labeled p53 (black, left cross peaks), and with the addition of unlabeled Hsp90 domains at mole ratios represented by the green (middle) and red (right) cross peaks as shown in each panel. The three spectra in each panel are plotted at the same contour level, and the levels in the various panels are adjusted to give comparable intensity for the black spectrum in each case. The three spectra are offset in each panel for clarity. The rectangles in each panel illustrate the Hsp90 construct, colored according to the scheme of Figure 1a. Concentration of proteins: a. p53 148 μM, Hsp90 N 0, 163, 311 μM; b. p53 163 μM, Hsp90 M 0, 180, 342 μM; c. p53 130 μM, Hsp90 NM 0, 46, 96 μM; d. p53 156 μM, Hsp90 MC 0, 55, 115 μM; e. p53 100 μM, Hsp90Δ 0, 35, 74 μM.
Resonance assignments have been reported for the p53 DNA-binding domain14; we have used these data as a basis for the backbone assignment of p53(94–312). The effect of addition of each of the Hsp90 constructs on the 1H-15N HSQC spectrum of the p53 DNA binding domain was quantified by measuring the volumes and intensities of the cross peaks in the spectrum. As in previous studies of this kind15, we may hypothesize that sites on p53 where resonance intensity is affected at the lowest concentrations of Hsp90 titrant may represent the binding site(s) of Hsp90 on p53. To identify such sites, we determined for each cross peak the concentration ratio of Hsp90 titrant added to the p53 that resulted in reduction of the cross peak to half its original volume or intensity. These values (conc1/2) are plotted as a function of residue number for each Hsp90 titrant in Figure 3. The average conc1/2 value for all of the affected resonances (horizontal lines in Figure 3) can also be used to derive a crude estimate of the affinity of p53 for each of the Hsp90 constructs, assuming that the remaining intensity of the cross peaks is due solely to the remaining free p53 and that the spectrum of the bound p53 is invisible. Approximate dissociation constants calculated in this way give the following values: 130μM for N, 250μM for M, 30μM for NM, 50μM for MC and 30μM for Hsp90Δ. Values for Kd could also be calculated directly from the intensity data, assuming the formation of a 1:1 complex in each case, giving average values of 133 μM for N and 232 μM for M. The NM titration could not be fitted by this method, and the MC and Hsp90Δ titrations did not fit well to the assumption of a 1:1 complex. This result is likely consistent with our observation that the MC and Hsp90Δ proteins are present as dimers, which might be expected to have two or more potential client binding sites.
Figure 3. p53 resonance attenuation by Hsp90 proteins.
(a–e) For each amino acid residue in p53(94–312) the concentration ratio (conc1/2 between p53 and added Hsp90 titrant) for which the intensity (measured as cross peak volume) of a given cross peak is halved compared to that of the free protein, is plotted for all residues for which cross peaks could be resolved in the 1H-15N HSQC spectrum. Cross peaks that show no attenuation are represented by an arbitrary value of 5. Horizontal lines show the positions of the average conc1/2 values that were used to estimate Kd values. (f) Ribbon diagram of residues 94–297 of a single structure from the family of NMR structures of the human p53 DNA-binding domain13 with the backbone colored to show the attenuation of resonances by the addition of Hsp90 M. Red color of the backbone indicates residues where conc1/2 was below the value representing (mean minus 1× standard deviation), and the orange color shows residues where conc1/2 was less than the mean. Figure prepared using MolMol34.
Mapping of the sites where cross peaks were preferentially affected onto the published structure of the p53 DNA-binding domain13 showed that no matter which titrant is used, the affected resonances are distributed throughout the folded core of the protein, including the central β-sheet (Figure 3f). If the affected sites represent contact sites between the Hsp90 and the p53 DNA-binding domain, we would expect to see the major effects on the surface, rather than on the interior of the protein. We thus conclude that the observation of reduced resonance intensity in the NMR spectrum of p53 upon addition of Hsp90 and its component domains indicates a general change in the environment of protons and nitrogens throughout the folded portion of the p53 DNA-binding domain, an effect that occurs as a result of the presence of domains of Hsp90.
A close examination of the spectra reveals the presence of small shifts on certain surface resonances that can be used to locate points of contact between the Hsp90 domains and p53 (examples are circled in Figure 1c) and such observations have been used as a basis for the elucidation of a binding surface between the Hsp90 M domain and the p53 DNA-binding domain (SJP and HJD, unpublished). However, the major effect of Hsp90 on the spectrum of p53 remains the intensity reduction and disappearance of p53 cross peaks. We suggest that this effect is due to the formation of a structurally heterogeneous molten globule-like state in p53 in the presence of Hsp90 or its domains. The intensity and linewidth of NMR resonances of biological macromolecules can be affected by the molecular size and by exchange between states (for example between free p53 and a complex with Hsp90) at rates that cause resonance broadening. We believe that these factors do not provide the major source of the intensity reduction we observe for p53 in the presence of Hsp90, primarily because the spectra of Hsp90 and its domains are not affected in the same way as those of p53 (shown in Supplementary Fig. 2). A detailed explanation of the reasoning leading to this conclusion is given in a Supplementary Discussion.
Molten Globule-Like Behavior of p53 in the Presence of Hsp90
Lowered resonance intensity in protein NMR spectra can be caused by the presence of a heterogeneous ensemble of structures, such as a molten globule. Although a number of molten globules have been characterized by NMR16–21, sample preparation and spectroscopy are invariably extremely challenging. The defining characteristic of molten globule NMR spectra is the presence of broadened and low-intensity resonances, which are thought to arise due to the fluid nature of the interior of the molten globule. Most if not all of the native-state secondary structure is present in a molten globule state, but the tertiary structure is not fixed; this state can be thought of as an ensemble of conformations that interconvert on the intermediate time scale that causes broadening of NMR resonances, sometimes to the point of disappearance. The formation of a state resembling a molten globule in p53 in the presence of Hsp90 could explain why the p53 resonances are affected in the presence of Hsp90 (Figure 1, 2, Supplementary Fig. 1) but the Hsp90 domains are relatively unaffected when p53 is added (Supplementary Fig. 2). It also provides a rationale for the wide extent of the effect over the entire structured core of the p53 DNA binding domain. This hypothesis assumes that the intensity observed in any of the p53 spectra in the presence of Hsp90 domains arises almost solely from the remaining free p53, and that the spectrum of the “bound” form (apart from the residues in the termini, which appear to be unstructured) is invisible. A schematic diagram illustrating this hypothesis is shown in Figure 4, specifically in Fig. 4a.
Figure 4. Schematic diagram of the model for the NMR titration results.
(a) Cross peaks are observed in the 1H-15N HSQC spectrum of free p53(94–312) for both surface and core residues (blue dots). Upon addition of Hsp90 or its constituent 1- and 2-domain constructs, the core resonances disappear (green dots), but many of the flexible surface resonances remain. The complex exchanges with any remaining free p53 in the solution. Therefore, at sub-maximal concentration ratio of Hsp90 titrant, the cross peaks of the core residues are visible but lowered in intensity, since they correspond to resonances of the remaining free protein. (b) When the buffer is exchanged for D2O, the surface amide protons of free p53 are exchanged for D, resulting in disappearance of the corresponding cross peaks (red dots). Core amides that are hydrogen bonded and sequestered from bulk solvent are exchanged more slowly, and their cross peaks therefore appear in the HSQC spectrum in D2O. (c) If p53 resembles a molten globule in the presence of Hsp90, buffer exchange of p53 to which Hsp90 has been added should result in a faster H/D exchange of the core amides than is observed for the free protein because the loosened structure of p53 promotes faster exchange of the core amide H for D while the p53 is bound to Hsp90. Since p53 is exchanging on and off the Hsp90, the core amide cross peaks in the HSQC spectrum (observed as for part (a), for the remaining free p53) should disappear more rapidly following D2O exchange. In effect, the addition of a small amount of Hsp90 acts as a catalyst to speed up H/D exchange in the core of p53.
Evidence for the p53 molten globule: ANS Fluorescence
The hypothesis that p53 forms a molten globule-like structure consisting of an ensemble of loosely folded structures is difficult to test directly by NMR, since we postulate that the spectrum of this form is largely invisible. Indeed, our NMR observations are problematic in terms of standard ideas of “fast exchange” and “slow exchange” (described more fully in a Supplementary Discussion). Alternatively, the hypothesis can be tested in two other ways: by observing the effect of the protein on the fluorescence spectrum of a dye, 1-anilinonaphthalene-8-sulfonic acid (ANS) and by observing the effect of Hsp90 on the H/D exchange rates of amide protons in p53.
In the presence of fully folded or fully unfolded proteins, the fluorescence of ANS is unchanged from that of the free dye, but in the presence of molten globule structures the fluorescence is increased and the emission maximum is shifted towards the low-wavelength (blue) end of the spectrum. If p53 forms a molten globule-like state when it interacts with Hsp90, we should observe an increased and blue-shifted fluorescence of ANS when both proteins are present, compared to its behavior in the presence of the free proteins. The result of such an experiment are shown in Figure 5a. The fluorescence emission of ANS alone is relatively weak, with an emission maximum at 522 nm. In the presence of p53 alone, the fluorescence emission is barely changed, indicating that the p53 DNA-binding domain construct that we have employed is well folded in solution. Somewhat unexpectedly, since they appear to be fully folded by NMR and CD spectroscopies, each of the Hsp90 N, M, NM and MC constructs, as well as the full-length protein Hsp90Δ, shows increased intensity and blue-shift of the ANS fluorescence (emission maximum ~ 480 nm). Such an increase can arise due to the binding of ANS in hydrophobic pockets that may be present in folded proteins, and has been observed, for example, with bovine serum albumin22. A solution containing a 1:1 mixture of p53 and Hsp90 shows a further marked blue shift of the emission maximum (to 472 nm) compared to the Hsp90 protein alone (Figure 5a). The change in fluorescence intensity from free Hsp90 to the complex is quite small at pH 7.1, but the intensity is markedly increased at a slightly lower pH, 6.0 (Figure 5a), with a further blue shift of the emission maximum relative to that of free Hsp90 (475 nm for Hsp90 vs 467 for the complex at this pH). NMR spectra of the component proteins are unchanged with the change of pH from 7.1 to 6.0 (Supplementary Fig. 3), indicating that there is no major change in the overall protein structure. We conclude that the slightly lowered pH intensifies the loosening of the structure of p53 in the presence of Hsp90, allowing the binding of more ANS and the subsequent greater increase in fluorescence intensity. The blue shift of the spectra of the mixture compared to those of the free Hsp90 at both pH 7.1 and pH 6 is consistent with the binding of additional ANS to p53, over and above the amount bound to the free Hsp90, suggesting that p53 contains molten globule-like structure over and above that of the Hsp90.
Figure 5. ANS Fluorescence Spectra and H/D Exchange.
(a) Fluorescence spectrum of 11.3 μM 1-anilinonaphthalene-8-sulfonic acid (ANS) in 25 mM phosphate buffer pH 7.1 (blue dotted line) or at pH 6.0 (black dotted line), with addition of 5 μM p53 DNA-binding domain (pH 7.1, blue line; pH 6.0, pink line), with addition of 5 μM Hsp90Δ (pH 7.1, green line; pH 6.0, orange line) and with further addition of 5 μM p53 DNA-binding domain to form a 1:1 complex (pH 7.1, dark blue; pH 6.0, red). The arrows indicate the extent of blue shift and increase of the emission maximum at each pH after the addition of p53 to Hsp90Δ. (b) Plot of the normalized peak volume difference (calculated according to the equation shown) between the cross peak intensities of amides of p53 that are persistent (slowly-exchanging) in D2O. (Inset) Selected sets of cross peaks from four 1H-15N HSQC spectra overlaid and then offset for clarity, for p53 (123μM) in H2O buffer (black), in D2O buffer (orange), in the presence of a 1:1 concentration ratio of Hsp90 M (green) and in the presence of a 1:1 concentration ratio of Hsp90 M and subsequently exchanged into D2O buffer (pink). All spectra were acquired under otherwise identical conditions, for an identical length of time and are plotted at the same contour level. The complete plot is shown in Supplementary Figure 4.
Evidence for the p53 molten globule: H/D Exchange
Further evidence that the structure of the p53 DBD is loosened in the presence of Hsp90 comes from hydrogen exchange measurements. The p53 DNA binding domain forms a well-structured folded domain in isolation, with a central β-sheet. Many of the amide protons in this central β-sheet form stable cross-sheet hydrogen bonds, which, combined with the buried nature of the sheet, serve to protect the amide protons from exchange with solvent water13. In an experiment where the solvent is changed to deuterium oxide (D2O), these protected amides will be exchanged slowly for D, while unprotected amides, for example on the surface of the molecule, will be rapidly exchanged, resulting in the loss of the corresponding cross peaks from the 1H-15N HSQC spectrum (illustrated in Figure 4b). The effect can be quantitated site-specifically by observing the intensity of the signal that remains in the 1H-15N HSQC spectrum after buffer exchange into D2O23. Amide proton exchange has also been used to demonstrate the formation of intermolecular complexes24: the usual observation is that the exchange rates of surface amides in the vicinity of the binding site are slowed when the complex is formed, as the site becomes protected from solvent upon complex formation. In the case of the p53–Hsp90 complex, if the p53 structure is loosened upon complex formation we should observe the reverse effect: the protected amides in the center of the molecule should actually exchange faster in the complex than in the free state, as illustrated in Figure 4c. The HD exchange experiment is difficult to design for the p53–Hsp90 system. Common methods of measuring HD exchange involve the rapid accumulation of multiple 2D spectra to determine exchange rates by analysis of the peak intensities of specific cross peaks in successive spectra. This approach is not ideal for the p53–Hsp90 system: because the sample concentrations are low, the pulse power to accumulate the multiple NMR spectra (using the SOFAST method25) must be high, resulting in sample heating, which invalidates the results. We are most interested in the exchange rates of the persistent amides, many of which are also those cross peaks whose intensity is lowered in the spectrum of p53 upon addition of Hsp90. Therefore, the H/D exchange experiment must be carefully controlled to determine any change in the intensity of these amide cross peaks that might be due to HD exchange rather than to the effect of the added Hsp90. In order to design this experiment, we took advantage of the fact that many of the persistent amides in the p53 DBD persist for a substantial time in D2O without a major change in intensity13. It was therefore possible to estimate the differences in the exchange rates under various conditions by acquiring a single NMR spectrum on four p53 samples, variously containing H2O, D2O and an Hsp90 domain, keeping all other factors exactly the same. A selection of corresponding cross peaks from the four spectra is shown as an inset in Figure 5b. The complete spectra are shown in Supplementary Fig. 4. The amount of M domain added to the p53 (a 1:1 ratio, about equivalent to that of the red spectrum in Figure 2b) is not sufficient to cause severe intensity loss (comparing black and green cross peaks in Figure 5b). Upon exchange into D2O buffer, cross peaks corresponding to solvent-exposed amides disappear from the spectrum of the free p53, but a substantial number remain, some at intensities comparable to those of the free protein in H2O (orange cross peaks). The intensity of many of these cross peaks is further diminished, sometimes almost completely, when the D2O buffer exchange is performed in the presence of Hsp90 M (pink cross peaks). The differences in the intensities and volumes of the cross peaks representing the persistent amide proton signals are plotted in Figure 5b. There are a few resonances that show an apparent small increase in intensity in the presence of Hsp90 M, but the vast majority of the cross peaks show a decrease in intensity. This is an important result, consistent with the outcome expected if our hypothesis about the molten globule-like state of p53 in the presence of Hsp90 is correct (Figure 4c), and completely the opposite of the increased protection that would normally be expected upon formation of a complex.
DISCUSSION
We conclude from the NMR, H/D exchange and fluorescence data that a loosened heterogeneous molten globule-like state is formed by the p53 DNA-binding domain in the presence of Hsp90 and its N and M domains. (Since we cannot study the effect of the isolated C domain, we have no information on whether this domain can also participate in the interaction.) The interaction between p53 and Hsp90 has been reported previously, but the results from different groups have been interpreted inconsistently. A fluorescence and NMR study26 concluded on the basis of the temperature dependence of the NMR spectra of the p53 DNA-binding domain in the absence and presence of full-length Hsp90, that the p53 domain was bound to Hsp90 in an unfolded state, and that the folded p53 DBD would not interact with Hsp90. On the other hand, extensive studies from other groups10–12 suggested that folded p53 was also capable of interacting with Hsp90. The fundamental difference between the conclusions of Rüdiger et al26 and our own observations may be a result of a difference in the Hsp90 protein used (Hsp90β26 vs Hsp90α (present work)) or in salt concentrations. An examination of the published NMR spectrum26 reveals results that are consistent with the data presented here: the NMR spectrum of the bound p53 DBD contains far fewer resonances than would be expected for a protein of this size, and, as pointed out by the authors26, those that are present likely correspond to the residues in the unstructured N- and C-termini of the domain. Lowered intensity for the majority of the resonances in the spectrum is exactly the same as we observe for p53 in the presence of Hsp90, but we also observe similar effects in the presence of smaller proteins representing the constituent domains of Hsp90. These results are not consistent with the presence of unfolded p53 in the complex, for which one might expect to see sharper resonances with chemical shifts different from those of the (folded) free protein, if only in the low molecular-weight complexes of the N and M domains. Instead, we observe loss of intensity, directly proportional to the amount of added N, M, NM, MC or Hsp90Δ, consistent with the formation of a heterogeneous conformational ensemble resembling a molten globule in the p53 client.
Our results are not consistent with the formation of a unique complex between the Hsp90 domains and p53. We see none of the effects, such as resonance shifts, that are expected upon complex formation, yet it is clear that p53 is affected by the presence of Hsp90, and that the effect increases as more Hsp90 is added. Our observations are consistent with many of the known attributes of Hsp90 – its relatively wide stable of client proteins, the effects on specificity and affinity of added extrinsic molecules, co-chaperones and ATP, as well as the absence of any reports of crystallization of Hsp90–client complexes. We suggest that Hsp90 affects p53 to give the NMR behavior that we observe by undergoing multiple transient interactions, primarily using the N and M domains. Such a manifold of chaperone-client “complexes” could be modified and tuned to various tasks, depending on the presence of ATP and specific co-chaperones. However, we suggest that the basic underlying interaction of Hsp90 with its client proteins is heterogeneous and transient. In the case of p53, these interactions result in the loosening of the structure of the protein, to a state resembling a molten globule.
If this observation proves general, it has important implications for the understanding of Hsp90 function. Hsp90 is present in cells at up to 1% of the total cellular protein, which argues for a central role of this protein in cellular metabolism. A general propensity on the part of Hsp90 to cause loosening of the structures of its client proteins would be consistent with a number of the known functions of Hsp90. For example, Hsp90 is thought to sequester the ligand binding domains (LBDs) of nuclear hormone receptors in a binding-competent state in the absence of the hormone signal, and to release the protein once the hormone is present27. Since the structures of LBDs in the presence of their hormone ligands show the hydrophobic hormone molecules deeply buried in the core of the domain, it is tempting to speculate that the LBD may also form a molten globule-like state in the presence of Hsp90, a state that would likely be loose enough for the hormone to access the hydrophobic interior. Once the hormone is bound, the LBD becomes more structured, providing a mechanism for the release of the protein from the chaperone.
METHODS
Preparation of Proteins
Constructs of human Hsp90α (1–732) were the N-terminal domain (N; 1–235), middle domain (M; 293–554), the two-domain constructs (NM; 1–554 and MC; 293–732), and the full-length protein without part of the N–M linker (Hsp90Δ; 1–732, 241–268 deleted). Proteins were expressed in M9 minimal medium in E. coli BL21 (DE3) [DNAY] with induction at 15°C for 12–20 h. Cells were lysed by sonication in 25 mM Tris buffer, pH 8.0, 5 mM DTT, Protease Inhibitor Cocktail (Roche), 4 mM EDTA. The soluble fraction of the cell lysate was applied to a 60 ml Sepharose Q FF column equilibrated with 25 mM Tris, pH 7.5, 2 mM EDTA, 2 mM DTT and proteins eluted with a linear gradient to 1 M NaCl, concentrated using Centriprep10 (Amicon) and purified by gel filtration on a 350 ml Sephacryl S100HR or S300HR (2.6 × 65 cm) in 20 mM Tris, pH 7.2, 0.1 M KCl, 2 mM EDTA, 2 mM DTT.
The DNA-binding domain (DBD) of human p53 (94–312), (wild-type or double mutant Y346F/T253I)13 was expressed in M9 minimal medium in E. coli BL21 (DE3) [DNAY] with induction at 15°C for 12–20 h. Cells were lysed by sonication in 25 mM Tris buffer, pH 7.0, 5 mM DTT, Protease Inhibitor Cocktail (Roche), 10 μM ZnSO4, 40 mM NaCl. The soluble fraction of the cell lysate was applied to a 10 ml Hitrap Q column equilibrated with 25 mM Tris, pH 7.0, 10 μM ZnSO4, 5 mM DTT and proteins eluted with a linear gradient to 1 M NaCl, and purified by Heparin column in 25 mM Tris, pH 7.0, 40 mM NaCl, 5mM DTT. We chose to work in buffers containing NaCl in order to avoid oligomerization of p53 DBD observed in low salt conditions28.
Expression of [70% 2H, 13C, 15N]-labeled proteins for resonance assignment was carried out in M9 minimal medium in D2O containing 15NH4Cl (0.5 g L−1), 15NH4SO4 (0.5 g L−1) and 13C-labeled glucose (2 g L−1), and [2H, 15N]-labeled proteins were obtained using 12C-deuterated glucose (2 g L−1). The culture was induced by 1 mM IPTG at 15°C with 16-h incubation.
NMR spectroscopy
The triply labeled [70% 2H, 13C, 15N] p53 DBD sample was exchanged into NMR buffer (25 mM sodium phosphate (pH 7.0), 50 mM NaCl, 100 mM glycine, 5 mM DTT, 10 μM ZnSO4 in 90% H2O/10% D2O). NMR spectra were acquired at 20°C on a Bruker DRX800 or Avance900 spectrometer with a cryoprobe. Backbone resonances were assigned using HNCA29,30, HNCO31, and NOESY-HSQC (mixing time, 150 ms) spectra and compared to the published assignments32,14.
NMR titrations of 15N labeled p53 DBD (160μM) with each Hsp90 construct were performed at 10°C on a Bruker DRX800 or Avance900 spectrometer. All samples were exchanged into binding buffer (25 mM sodium phosphate pH 7.0, 100 mM NaCl, 5 mM DTT) before titration. No appreciable difference in the behavior of the p53 DBD was observed upon addition of ATP or ATP analogs AMP-PNP or ATP-γ-S. A control experiment where bovine serum albumin was added to 15N-labeled p53 DBD showed a small amount of resonance attenuation throughout the molecule, both qualitatively and quantitatively different from that observed for the Hsp90 constructs (Supplementary Fig. 5).
Data Analysis
For each cross peak in a series of 1H-15N HSQC spectra containing ratios p53: Hsp90 of 1:0 up to 1:3.6, the intensity and volume were evaluated using NMRView33 and plotted as a function of concentration ratio. The points were fitted to a parabola, consistent with the assumption that the intensity or volume of the signal at any given concentration ratio reflects the concentration of free p53 DNA binding domain in the solution. To normalize for differences in intensity or volume between different cross peaks in the spectrum, the degree of attenuation was evaluated as the concentration ratio at which the intensity or volume was at 50% of the initial value in the absence of added Hsp90.
Fluorescence Spectroscopy
Fluorescence spectra were recorded in a Fluorolog-3 spectrofluorometer (JOBIN YVON INC., NJ, USA), using the fluorescent probe 1-anilinonaphthalene-8-sulfonic acid (ANS). Fluorescence emission spectra between 400 and 600 nm were obtained with a fixed excitation wavelength of 370 nm. The p53 DNA-binding domain and Hsp90 domains were prepared in the same buffer (25 mM sodium phosphate pH 7.1 or pH 6, 100 mM NaCl, 5 mM DTT). For each titration experiment, 5μM solution of p53 DNA-binding domain containing 11.3 μM ANS were prepared and a solution of one of the Hsp90 proteins added. As a control experiment, a solution containing only 11.3 μM ANS was titrated with each Hsp90 construct. The dilution effect upon adding the Hsp90 constructs into the solution was negligible.
H/D Exchange Experiments
Four samples were prepared: p53 free in H2O, p53 free in D2O, p53 in H2O in the presence of an amount of Hsp90 protein that would result in less than complete attenuation of cross peaks and p53 in D2O in the presence of the same concentration of Hsp90 protein. The p53 concentrations were kept exactly the same in all four cases. A stock solution of p53 in H2O buffer (353 μM p53 in 25 mM Na2HPO4, pH 7.0. 100 mM NaCl, 5 mM DTT) was first divided into four equal 0.5 ml volumes, two of which were added to 176.5 μl of buffer and the others to 176.5 μl of buffer containing 1 mM Hsp90 M. All samples were exchanged using NAP-5 columns, one free p53 and one complex exchanged to H2O buffer and the other free p53 and complex samples to D2O buffer. The pH of the D2O buffer was adjusted to 6.6 to account for the deuterium isotope effect. After NAP-5 exchange, equal volumes of a D2O solution of 2H-DTT and additional D2O (to provide a spectrometer lock in the H2O samples) to a final concentration of 5% were added to all samples. The final concentration of p53 in all samples was 123 μM. Samples were equilibrated for exactly 20 minutes before being placed in the probe of the 800 MHz spectrometer, where they were equilibrate at 10°C for a further 15 minutes before acquisition of an HSQC spectrum for 47 minutes.
Supplementary Material
Acknowledgments
We thank Milka Kostic for preparation of p53 and Hsp90 expression constructs and preliminary binding assays. We thank Peter Wright and members of the Wright and Dyson groups for helpful comments, Gerard Kroon for help with NMR experiments and Euvel Manlapaz for technical assistance. The original clone used to prepare the domains of human Hsp90α was kindly provided by David Toft of Mayo Clinic, Rochester, Minnesota.
Funding: This work was supported by grant GM57374 from the National Institutes of Health, and by a grant from the Korea Research Foundation, funded by the Korean Government (MOEHRD), (KRF- 2006-214-E0009).
Footnotes
Author contributions: SJP and HJD designed experiments, SJP carried out NMR and fluorescence experiments, BNB carried out H/D exchange experiments, SJP, MAY and HJD analyzed data, SJP, MAY and HJD wrote the paper.
Reference List
- 1.Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 2.Richter K, Buchner J. Hsp90: chaperoning signal transduction. J Cell Physiol. 2001;188:281–290. doi: 10.1002/jcp.1131. [DOI] [PubMed] [Google Scholar]
- 3.Terasawa K, Minami M, Minami Y. Constantly updated knowledge of Hsp90. J Biochem (Tokyo) 2005;137:443–447. doi: 10.1093/jb/mvi056. [DOI] [PubMed] [Google Scholar]
- 4.Shiau AK, Harris SF, Southworth DR, Agard DA. Structural analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 5.Ali MM, et al. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaughan CK, et al. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol Cell. 2006;23:697–707. doi: 10.1016/j.molcel.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawle P, et al. The middle domain of Hsp90 acts as a discriminator between different types of client proteins. Mol Cell Biol. 2006;26:8385–8395. doi: 10.1128/MCB.02188-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soussi T, Legros Y, Lubin R, Ory K, Schlichtholz B. Multifactorial analysis of p53 alteration in human cancer: a review. Int J Cancer. 1994;57:1–9. doi: 10.1002/ijc.2910570102. [DOI] [PubMed] [Google Scholar]
- 9.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 10.Blagosklonny MV, Toretsky J, Bohen S, Neckers L. Mutant conformation of p53 translated in vitro or in vivo requires functional HSP90. Proc Natl Acad Sci USA. 1996;93:8379–8383. doi: 10.1073/pnas.93.16.8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitesell L, Sutphin PD, Pulcini EJ, Martinez JD, Cook PH. The physical association of multiple molecular chaperone proteins with mutant p53 is altered by geldanamycin, an hsp90-binding agent. Mol Cell Biol. 1998;18:1517–1524. doi: 10.1128/mcb.18.3.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Müller L, Schaupp A, Walerych D, Wegele H, Büchner J. Hsp90 regulates the activity of wild type p53 under physiological and elevated temperatures. J Biol Chem. 2004;279:48846–48854. doi: 10.1074/jbc.M407687200. [DOI] [PubMed] [Google Scholar]
- 13.Cañadillas JMP, et al. Solution structure of p53 core domain: Structural basis for its instability. Proc Natl Acad Sci USA. 2006;103:2109–2114. doi: 10.1073/pnas.0510941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulder FAA, Ayed A, Yang DW, Arrowsmith CH, Kay LE. Assignment of 1HN, 15N, 13Cα, 13CO and 13Cβ resonances in a 67 kDa p53 dimer using 4D-TROSY NMR spectroscopy. J Biomol NMR. 2000;18:173–176. doi: 10.1023/a:1008317825976. [DOI] [PubMed] [Google Scholar]
- 15.Martinez-Yamout MA, et al. Localization of sites of interaction between p23 and Hsp90 in solution. J Biol Chem. 2006;281:14457–14464. doi: 10.1074/jbc.M601759200. [DOI] [PubMed] [Google Scholar]
- 16.Eliezer D, Jennings PA, Dyson HJ, Wright PE. Populating the equilibrium molten globule state of apomyoglobin under conditions suitable for characterization by NMR. FEBS Lett. 1997;417:92–96. doi: 10.1016/s0014-5793(97)01256-8. [DOI] [PubMed] [Google Scholar]
- 17.Eliezer D, Chung J, Dyson HJ, Wright PE. Native and non-native structure and dynamics in the pH 4 intermediate of apomyoglobin. Biochemistry. 2000;39:2894–2901. doi: 10.1021/bi992545f. [DOI] [PubMed] [Google Scholar]
- 18.Baum J, Dobson CM, Evans PA, Hanley C. Characterization of a partly folded protein by NMR methods: studies on the molten globule state of guinea pig α-lactalbumin. Biochemistry. 1989;28:7–13. doi: 10.1021/bi00427a002. [DOI] [PubMed] [Google Scholar]
- 19.Redfield C, Smith RAG, Dobson CM. Structural characterization of a highly-ordered ‘molten globule’ at low pH. Nature Struct Biol. 1994;1:23–29. doi: 10.1038/nsb0194-23. [DOI] [PubMed] [Google Scholar]
- 20.Schulman BA, Kim PS, Dobson CM, Redfield C. A residue-specific NMR view of the non-cooperative unfolding of a molten globule. Nature Struct Biol. 1997;4:630–634. doi: 10.1038/nsb0897-630. [DOI] [PubMed] [Google Scholar]
- 21.Greene LH, Wijesinha-Bettoni R, Redfield C. Characterization of the Molten Globule of Human Serum Retinol-Binding Protein Using NMR Spectroscopy. Biochemistry. 2006;45:9475–9484. doi: 10.1021/bi060229c. [DOI] [PubMed] [Google Scholar]
- 22.Cattoni DI, Kaufman SB, Gonzalez Flecha FL. Kinetics and thermodynamics of the interaction of 1-anilino-naphthalene-8-sulfonate with proteins. Biochim Biophys Acta. 2009;1794:1700–1708. doi: 10.1016/j.bbapap.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Bai Y, Sosnick TR, Mayne L, Englander SW. Protein folding intermediates: Native-state hydrogen exchange. Science. 1995;269:192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paterson Y, Englander SW, Roder H. An antibody binding site on cytochrome c defined by hydrogen exchange and two-dimensional NMR. Science. 1990;249:755–760. doi: 10.1126/science.1697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schanda P, Kupce E, Brutscher B. SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J Biomol NMR. 2005;33:199–211. doi: 10.1007/s10858-005-4425-x. [DOI] [PubMed] [Google Scholar]
- 26.Rüdiger S, Freund SM, Veprintsev DB, Fersht AR. CRINEPT-TROSY NMR reveals p53 core domain bound in an unfolded form to the chaperone Hsp90. Proc Natl Acad Sci USA. 2002;99:11085–11090. doi: 10.1073/pnas.132393699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pratt WB, Morishima Y, Murphy M, Harrell M. Chaperoning of glucocorticoid receptors. Handb Exp Pharmacol. 2006;2006:111–138. doi: 10.1007/3-540-29717-0_5. [DOI] [PubMed] [Google Scholar]
- 28.Rippin TM, Freund SM, Veprintsev DB, Fersht AR. Recognition of DNA by p53 core domain and location of intermolecular contacts of cooperative binding. J Mol Biol. 2002;319:351–358. doi: 10.1016/S0022-2836(02)00326-1. [DOI] [PubMed] [Google Scholar]
- 29.Yamazaki T, Lee W, Arrowsmith CH, Muhandiram DR, Kay LE. A suite of triple-resonance NMR experiments for the backbone assignment of 15N, 13C, 2H labeled proteins with high sensitivity. J Am Chem Soc. 1994;116:11655–11666. [Google Scholar]
- 30.Grzesiek S, Bax A. Improved 3D triple-resonance NMR techniques applied to a 31 kDa protein. J Magn Reson. 1992;96:432–440. [Google Scholar]
- 31.Wittekind M, Mueller L. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha- and beta-carbon resonances in proteins. J Magn Reson. 1993;101:201–205. [Google Scholar]
- 32.Wong KB, et al. Hot-spot mutants of p53 core domain evince characteristic local structural changes. Proc Natl Acad Sci USA. 1999;96:8438–8442. doi: 10.1073/pnas.96.15.8438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson BA, Blevins RA. NMRView: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:604–613. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 34.Koradi R, Billeter M, Wüthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.