Abstract
FusionDB (http://igs-server.cnrs-mrs.fr/FusionDB/) constitutes a resource dedicated to in-depth analysis of bacterial and archaeal gene fusion events. Such events can provide the ‘Rosetta stone’ in the search for potential protein–protein interactions, as well as metabolic and regulatory networks. However, the false positive rate of this approach may be quite high, prompting a detailed scrutiny of putative gene fusion events. FusionDB readily provides much of the information required for that task. Moreover, FusionDB extends the notion of gene fusion from that of a single gene to that of a family of genes by assembling pairs of genes from different genomes that belong to the same Cluster of Orthogonal Groups (COG). Multiple sequence alignments and phylogenetic tree reconstruction for the N- and C-terminal parts of these ‘COG fusion’ events are provided to distinguish single and multiple fusion events from cases of gene fission, pseudogenes and other false positives. Finally, gene fusion events with matches to known structures of heterodimers in the Protein Data Bank (PDB) are identified and may be visualized. FusionDB is fully searchable with access to sequence and alignment data at all levels. A number of different scores are provided to easily differentiate ‘real’ from ‘questionable’ cases, especially when larger database searches are performed. FusionDB is cross-linked with the ‘Phylogenomic Display of Bacterial Genes’ (PhydBac) online web server. Together, these servers provide the complete set of information required for in-depth analysis of non-homology-based gene function attribution.
INTRODUCTION
Gene fusion events have been proposed to represent valuable ‘Rosetta stone’ information for the identification of potential protein–protein interactions and metabolic or regulatory networks (1,2). More generally, information on gene fusion events can be combined with other non-homology-based approaches, such as phylogenomic profiling and identification of conserved chromosomal localization, to provide hypotheses for the characterization of proteins of unknown function (3–5). A number of web-based databases, such as AllFuse (5), STRING (6) and Predictome (7), implement this idea already. However, most of the available databases limit the definition of a gene fusion event to simple non-overlapping side-by-side BLAST (8) matches of two genes from a reference genome to a single open reading frame (ORF) in a target genome, but without providing much information for further in-depth analysis. Searches based on these databases give good starting points for hypothesis building, but the false positive rate may be quite high (in particular in cases where genes evolved through gene duplication and where the identification of gene orthology is hence difficult). The user is then left with the task of assembling the data required for more extensive case analysis.
Here we present a database that is based on a more strict definition of a gene fusion event, applying a mutual best match criteria [(9), see Fig. 1 and methods]. It drastically reduces the number of false positives, at the expense of a potentially similarly high number of false negatives. To recover from this drawback, gene fusion events between genes from different genomes that belong to the same Cluster of Orthologous Groups (COG) (10) are pulled together in what we call ‘COG fusion events’. Analysis of these COG fusion events then allows for the investigation of gene fusion in its phylogenomic context, using multiple alignments and phylogenetic tree reconstruction. Questions on the history of individual gene fusion events, such as whether a particular event occurred only once or many times during evolution, or whether more complex processes such as horizontal gene transfer, gene fission and gene decay are involved may be addressed using the information provided by FusionDB. The extension to ‘COG fusion events’ also provides information on general gene fusion tendencies in a whole bacterial genomic context to address questions such as ‘Which type of genes are most likely to fuse?’ FusionDB thereby complements our phylogenetic profiling web server PhydBac (http://igs-server.cnrs-mrs.fr/phydbac/) (11), which is based on the same philosophy: providing detailed non-homology-based information for in-depth analysis of potential protein–protein interactions. FusionDB is thus complementary to the databases cited above (5–7).
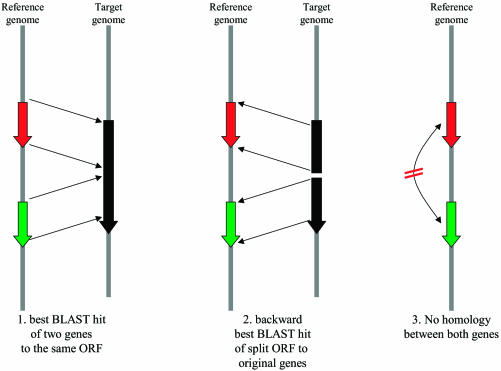
Figure 1.
Criteria for a putative gene fusion event based on a mutual best match criteria (see text for details).
SOURCES OF GENOMIC DATA AND METHODS
All available 89 fully sequenced non-redundant bacterial and archaeal genomes (see http://igs-server.cnrs-mrs.fr/FusionDB/methods/ for a full list) were downloaded from NCBI RefSeq. Those genomes for which a COG annotation of their genes was available (51 genomes) were checked for putative gene fusion (PFE) events in all 89 genomes as follows: a PFE between two genes from a given reference genome in a given target genome is subject to three criteria (Fig. 1):
(i) Each of the two reference genes must match the same ORF in the target genome as their highest scoring BLAST hit. The overlap between the BLAST hits of both genes must not exceed 10% of the size of the smaller of the two target genes.
(ii) When split between the two BLAST hits, the two halves of the target ORF must match back to the original two reference genes as their best BLAST hit to the reference genome.
(iii) The reference genes must not be homologous to each other.
Note that the search for PFEs is done on the basis of the annotated genes from a given reference genome, but against all possible ORFs in the target genome (including overlapping ORFs). This increases the chances of finding a gene fusion event that might have been discarded by a human annotator. Every PFE is then subjected to a scoring scheme based on different evaluations of its pairwise and multiple (triple) alignments by calculating the following five scores.
(i) The separation index (sep) is a measurement of the mix between the domains from the two reference genes when they are placed in a triple alignment with the target ORF. This index varies between 0 (total mix) and 1 (complete separation).
(ii) The fusion index (fus) is the fraction of residues in the concatenated reference genes that have similar properties to their aligned counterparts in the target ORF. This index may vary between 0 (virtually no homology between the reference genes and the target ORF) and 1 (strong homology).
(iii) The gene coverage (cov) is the fraction of the two reference genes that is alignable with the target ORF in a triple alignment. This index varies between 0 (no relationship at all between the reference genes and the target ORF) and 1 (all domains of the reference genes have a counterpart in the target ORF).
(iv) The size ratio (ratio) between the size of the reference genes and the target ORF indicates possible domain gain or loss after the gene fusion event has occurred.
(v) The ‘baditude’ (bad) is the fraction of residues that are aligned between the reference genes when placed in a triple alignment with the target ORF. This index varies between 0 (both reference genes are evolutionarily unrelated) and 1 (both reference genes are homologues). A high ‘baditude’ is an indicator of genes with paralogous domains.
QUERYING THE DATABASE
FusionDB may be searched by gene name, gene annotation, gene function, COG identifier or simply by entering an amino acid sequence in FASTA format. Queries may be confined to specific reference and target genomes, and limits on the different scores can be imposed. Output in full-page mode contains visualization of the different alignments that were used for scoring, and in the case of gene pairs that both belong to a COG a special COG-analysis page is provided. This COG-analysis page contains different types of multiple alignments and related phylogenetic trees, as well as information on related COG fusion events (networks) (Fig. 2). Extension of the research results, e.g. to all hits to a given fusion ORF is possible. In cases where a gene fusion event has a match to a heterodimer in the Protein Data Bank (PDB), a special PDB analysis page is available, providing a scored multiple alignment between the reference genes, the fusion gene and the sequences of the heterodimer in the PDB file. Output in tabulated mode or limitation to only the best hit for each gene pair may be requested if a large number of hits is expected. On each page a cross-link to PhydBac gives direct access to the phylogenetic profiles and eventual conserved chromosomal proximity of the two fusion genes.
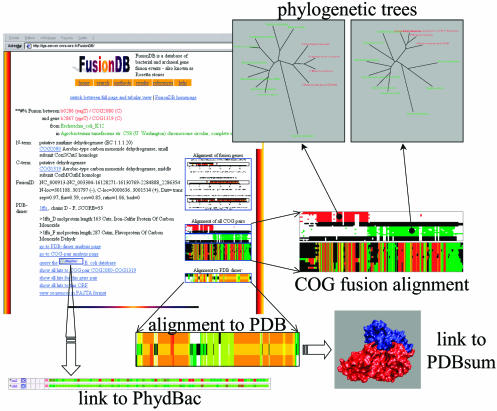
Figure 2.
Screenshot of FusionDB full-page output for a query to COG2080 and examples of some related information that can be obtained through this page. PhydBac (http://igs-server.cnrs-mrs.fr/phydbac/) is the ‘Phylogenomic Display of Bacterial Genes’ online web tool. In the top of the ‘COG fusion alignment’, N- and C- terminal genes are presented in red and green, respectively, fusion ORFs are in black. The alignment of the merged genes with the fusion genes is presented below. A colour scale ranging from green over yellow to red represents the EMBOSS plotcon score for this ‘merged alignment’. The ‘phylogenetic trees’ are based on the N- and the C-terminal ‘COG fusion alignments’, respectively. Genomes in which fusion events occurred are highlighted in red in the trees. The ‘alignment to the PDB’ is a representation of the T-Coffee alignment core index of the reference genes (top row), the fusion ORF (middle row) and the sequence of the heterodimer (bottom row), warmer colours indicating a higher confidence in the alignment quality. PDBsum (http://www.biochem.ucl.ac.uk/bsm/pdbsum/) is a database of the known 3D structures of proteins and nucleic acids.
By default, all queries are limited to a separation index (sep) of 0.6. This is found to be the most robust indicator of a ‘true’ gene fusion event (K. Suhre et al., in preparation; see also FusionDB/results/). Note that the fusion index (fus) is dependent on the evolutionary distance between the reference and the target genome. Values of the gene coverage (cov) and the size ratio (ratio) that differ significantly from 1 are indicators of domains that have been lost or added in the process of evolution. Such cases should be inspected carefully. In some cases this can give rise to a high ‘baditude’ (bad) score when the added domains are homologous. If for a given query gene no fusion event is found, the user may try to extend the search to the COG family to which this gene belongs (or use the sequence search option, note also that genes with a high degree of paralogy in most genomes may not be identified as a fusion event). In situations where both genes of a PFE are associated with a COG and where several fusion events are identified by FusionDB, coherence between the phylogenetic trees of the N- and C-terminal genes as well as the history of the gene fusion can be used as indicators of ‘real’ fusion events and true functional orthology between the implicated genes in the different genomes. This kind of key information is not readily available on other existing database servers.
CONCLUDING REMARKS AND FUTURE PLANS
FusionDB presents significant additions to other gene-fusion-related databases. The extension of the concept of a gene fusion to a ‘COG fusion’ event and the application of a mutual best match criteria not only reduces the number of false positives, but also makes the use of gene fusion events as ‘Rosetta stones’ applicable at a genome-independent level, where the common gene pool of all prokaryotes is viewed as the sum of all identified (and still to be discovered) COGs. The wealth of pre-calculated multiple alignments and phylogenetic trees will be welcomed by many biological analysts and annotators, as FusionDB currently covers ∼20 000 potentially ‘real’ gene fusion events (having a separation index > 0.6), which correspond to 1355 different fused COG pairs. A more detailed analysis of these cases is underway (K. Suhre et al., in preparation). FusionDB will be updated regularly as the number of publicly available fully sequenced genomes increases, and lower eukaryotes should be added in a future version. This will be particularly beneficial for obtaining more complete phylogenetic trees, which is still the best way to evaluate the ‘reality’ of the gene fusion events. Ultimately, FusionDB is designated to prioritize and record the experimental validity of the molecular or functional interaction of the genes involved in gene fusion events.
Acknowledgments
ACKNOWLEDGEMENTS
Sequence data was downloaded from NCBI RefSeq (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/). Multiple alignments were computed using T-Coffee (http://igs-server.cnrs-mrs.fr/Tcoffee/) (12). Phylogenetic tree reconstruction was done using Phylip (http://evolution.genetics.washington.edu/phylip.html). Multiple alignments were scored with EMBOSS plotcon (http://www.hgmp.mrc.ac.uk/Software/EMBOSS/). Mode data for protein structures are from the PDB (http://www.rcsb.org/pdb/) (13).
REFERENCES
- 1.Galperin M.Y. and Koonin,E.V. (2000) Who’s your neighbor? New computational approaches for functional genomics. Nat. Biotechnol., 18, 609–613. [DOI] [PubMed] [Google Scholar]
- 2.Sali A. (1999) Functional links between proteins. Nature, 402, 23–26. [DOI] [PubMed] [Google Scholar]
- 3.Marcotte E.M. (2000) Computational genetics: finding protein function by nonhomology methods. Curr. Opin. Struct. Biol., 10, 359–365. [DOI] [PubMed] [Google Scholar]
- 4.Marcotte E.M., Pellegrini,M., Thompson,M.J., Yeates,T.O. and Eisenberg,D. (1999) A combined algorithm for genome-wide prediction of protein function. Nature, 402, 83–86. [DOI] [PubMed] [Google Scholar]
- 5.Enright A.J. and Ouzounis,C.A. (2001) Functional associations of proteins in entire genomes via exhaustive detection of gene fusion. Genome Biol., 2, 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Mering C., Huynen,M., Jaeggi,D., Schmidt,S., Bork,P. and Snel,B. (2003) STRING: a database of predicted functional associations between proteins. Nucleic Acids Res., 31, 258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mellor J.C., Yanai,I., Clodfelter,K.H., Mintseris,J. and DeLisi,C. (2002) Predictome: a database of putative functional links between proteins. Nucleic Acids Res., 30, 306–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul S.F., Madden,T.L., Schaeffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tatusov R.L., Koonin,E.V. and Lipman,D.J. (1997) A genomic perspective on protein families. Science, 278, 631–637. [DOI] [PubMed] [Google Scholar]
- 10.Tatusov R.L., Natale,D.A., Garkavtsev,I.V., Tatusova,T.A., Shankavaram,U.T., Rao,B.S., Kiryutin,B., Galperin,M.Y., Fedorova,N.D. and Koonin,E.V. (2001) The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res., 29, 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enault F., Suhre,K., Poirot,O., Abergel,C. and Claverie,J.M. (2003) Phydbac (phylogenomic display of bacterial genes): an interactive resource for the annotation of bacterial genomes. Nucleic Acids Res., 31, 3720–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Notredame C., Higgins,D.G. and Heringa,J. (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol., 302, 205–217. [DOI] [PubMed] [Google Scholar]
- 13.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]