Abstract
Inherited mutations in the tumour suppressor BRCA2 predispose to pancreatic adenocarcinomas, which carry activating mutations in the KRAS oncogene in >95% of cases, as well as frequent TP53 inactivation. Here, we have established an RNA interference (RNAi) screen to identify genes whose depletion selectively inhibits the growth of cells lacking BRCA2, and then studied the effects of the genetic depletion or pharmacologic inhibition of one candidate, the checkpoint kinase CHK1, in the context of pancreatic cancer. Pharmacologic inhibition of CHK1 using small-molecule inhibitors (CHK1i) reduced cell growth in several cell lines depleted of BRCA2. Unexpectedly, these drugs did not suppress the growth of BRCA2-deficient pancreatic cancer cell lines from humans or gene-targeted mice expressing active Kras and trans-dominant inhibitory mutant Trp53. Remarkably, the expression of KRASG12V and TP53G154V in BRCA2-depleted HEK293 cells was sufficient to render them resistant to CHK1i (but not to Mitomycin C or inhibitors of PARP1). CHK1i sensitivity was restored by gemictabine, an S-phase genotoxin used to treat pancreatic adenocarcinoma. Thus, the growth-suppressive effect of CHK1 inhibition in BRCA2-mutant tumours can be opposed by concurrent KRAS, activation and TP53 mutations typical of pancreatic adenocarcinoma; and CHK1i resistance in this setting can be overcome by gemcitabine. Our findings exemplify that approaches to exploit potential therapeutic targets for cancer identified in ‘synthetic lethal’ RNAi screens are affected by the genetic context of specific malignancies and combination therapy with other agents. This concept should be taken into account in the ongoing and future development of targeted cancer therapies.
Keywords: pancreatic cancer, TP53, KRAS, synthetic lethality, RNAi screen
Introduction
Cancer predisposition associated with inactivation of the BRCA2 tumour suppressor is believed to arise from the loss of a critical role of BRCA2 in homology-directed DNA repair (1-2). Indeed, BRCA2-deficient cells not only exhibit chromosomal instability (3-4) but are also sensitive to various types of DNA damaging agents, such as mitomycin C (MMC) (4), ionizing radiation, topoisomerase inhibitors (5), and alkylating agents (3, 6). The DNA repair defect exhibited by BRCA2-deficient cells has also been exploited in new approaches to targeted therapy, based on the principle of ‘synthetic lethality’ first espoused by Hartwell and colleagues (7). Two genes are said to exhibit synthetic lethality when the deficiency of either gene alone is compatible with viability but the simultaneous lack of both genes causes cell death (7-8). Thus, BRCA2-deficient cells are inviable when the activity of the base excision repair (BER) pathway enzyme poly-ADP ribose polymerase (PARP) 1 is inhibited, because BRCA2 deficiency is synthetic lethal with PARP1 deficiency (9-11).
Functional genomic approaches using RNA interference (RNAi) screens are increasingly being used to identify synthetic lethal interactions targeting loss-of-function mutations in DNA repair genes, such as FANCG deficiency, associated with cancer (12). In general, these approaches utilize cell lines whose genetic make-up does not accurately reflect the genetic context of specific malignancies. It is therefore conceivable that synthetic lethal interactions identified in such screens will not accurately identify genuine therapeutic targets for the treatment of malignancy. This issue has yet to be systematically evaluated. Here, we have examined the example of familial pancreatic cancer associated with inherited BRCA2 mutations.
Besides breast and ovarian cancers, germline BRCA2 mutations also predispose to pancreatic adenocarcinomas (2). Indeed, it has been estimated that 5-20% of familial cases of pancreatic cancer may carry BRCA2 mutations (13-14). Moreover, the KRAS oncogene is activated by point mutations in over 90% of these cancers (15), and the TP53 tumor suppressor is inactivated in 50-75% of cases (16). Thus, familial pancreatic cancer associated with BRCA2 inactivation offers a unique experimental model in which to test the effect of genetic context on synthetic lethal interactions identified in RNAi screens.
In this work, we have employed a strategy that involves three steps; firstly, we established a BRCA2 synthetic lethal RNAi screen, which identified checkpoint kinase 1 (CHK1) as a potential therapeutic target; secondly, we confirmed that the pharmacologic inhibition of CHK1 replicated the effects of genetic depletion in the screening results; and thirdly, we examined the effect of CHK1 inhibitors in the context of a specific malignancy, BRCA2 deficient pancreatic cancers with associated KRAS/TP53 mutations. Unexpectedly, we report here that CHK1 inhibitors fail to suppress the growth of BRCA2-deficient cells in the context of KRAS activation and TP53 inactivation found in pancreatic cancers. Thus, our findings reveal that the utility of CHK1 as a potential therapeutic target for BRCA2-deficient tumors is dependent on the genetic context of the malignancies. The context dependence of synthetic lethality should be taken into account when extrapolating the results of synthetic lethal RNAi screens to clinical trials with targeted therapies.
Materials and Methods
Cell lines
The human BRCA2 deficient fibroblast cell line, EUFA423, was a kind gift from VU University Medical Center in 2004. EUFA423, EUFA423B2 (750μg/ml of G418 was added), MRC5VA, Mia-PaCa2, 293T, HEK293 and mouse pancreatic cancer cell lines (Pdx1Cre; KrasG12D/+; Trp53R270H/+; Brca2Wt/Wt or Tr/Δ11) (17) were grown in Dulbecco’s Modified Eagle’s medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS), 100U/ml penicillin and streptomycin. Capan-1 cells were cultured in RPMI medium (Gibco) supplemented with 15% FBS, 100U/ml penicillin and streptomycin. EUFA423 was authenticated by observation of morphology and by measuring sensitivity to known agents (MMC, PARP inhibitors) in our laboratory. For, MRC5VA, 293T, HEK293 and Capan-1 cells, which were obtained from Cancer Research UK (CRUK) Cell Services before 2008, authentication was not done in our laboratory other than what was done by CRUK. All the mouse pancreatic cancer cell lines were genotyped for their conditional alleles. Western blotting using epithelial markers confirmed that they were of epithelial origin.
Antibodies and inhibitors
The antibodies used in this study were: Anti-BRCA2 mouse monoclonal (Ab-1) and rabbit polyclonal (Ab-2) antibodies (Calbiochem); Anti-FLAG (M2) and anti-β-actin (AC-15) antibodies (Sigma-Aldrich); Anti-RAD51 antibody (Ab-1) (Calbiochem). UCN-01 and MMC were purchased from Sigma. Olaparib was purchased from JS research chemicals (Schleswig Holstein, Germany). KU0058948, KU0051529 and 2e were synthesized by Nexus Discovery Solutions ltd (UK). The chemical structures of these inhibitors are listed in Supplementary Figure 1.
Plasmids and siRNAs
The p3xFLAG-BRCA2 construct was generated by excising BRCA2 cDNA from RV-BRCA2 at the Not I and Kpn I sites, and inserting it in p3xFLAG CMV-10 (Sigma-Aldrich). pcDNA3.1-KRASG12V, generously provided by Dr Patrizio Castagnola (Istituto Nazionale per la Ricerca sul Cancro, Genova, Italy), pBabe-puro-p53G154V and pRRL-BRCA2shRNA were used for co-transfection experiments. The sequence of the siRNA oligo targeting BRCA2 was (5′-GAAGAAUGCAGGUUUAAUA-3′); whereas the sequence for luciferase control siRNA oligo was (5′-CGUACGCGGAAUACUUCG A-3′).
siRNA screen
A RNAi library comprising four unique siRNA duplexes targeting each of 880 human kinases and cell cycle related genes, arrayed in a one-gene-one well format on 96-well microtitre plates, was purchased from Dharmacon (Lafayette, CO). Triplicate reverse transfections were performed on day 0 in 96-well plates using Dharmafect I (Dharmacon) with 3,500 EUFA423 or EUFA423B2 cells and smart pool siRNA library at a concentration of 25nM using a Biomek NXP liquid-handling workstation (Beckman Coulter, High Wycombe, UK) and WellMate Microplate Dispenser (Thermo Fisher Scientific, Cheshire, UK). On day 5, cell viability was measured using the Cell Titer Blue Cell Viability Assay (Promega). The validation screen was performed using the same format.
Transfection
For siRNA transfection, cells were seeded at 20-30% confluence in one day before transfection. Oligofectamine reagent (Invitrogen) was used to transfect cells with 40 nM siRNA duplex twice at 48-h intervals. For DNA transfection, Lipofectamine 2000 reagent was used according to manufacture’s instruction.
Drug sensitivity assay
BRCA2 depleted cells were plated in 96 well plates at a density of 4000 cells per well one day before adding the CHK1 inhibitors (2e or UCN-01), MMC, or Olaparib, and allowed to incubate for 3 days. BRCA2 deficient mouse pancreatic cancer cells were plated at 2000-3000/well one day before adding the drugs and allowed to incubate for 3 days. Cell viability was measured with The CellTiter-Blue Assay Platform (Promega).
Western blot and immunoprecipitation
Whole-cell extracts were made in the NP-40 lysis buffer (50 mM HEPES (pH 7.4), 100 mM NaCl, 0.5% NP-40, 10 mM EDTA, 20 mM β-glycerophosphate, 1 mM DTT, 1 mM sodium orthovanadate, 1 mM PMSF, complete protease inhibitor cocktail (Roche). BRCA2 was resolved by 3–8% gradient Tris-acetate gels (Invitrogen) and detected with anti-BRCA2 monoclonal antibody (Ab-1). FLAG-BRCA2 was transiently expressed in 293T cells and immunoprecipitated with anti-FLAG monoclonal antibody M2. Immunoprecipitates were subjected to SDS-PAGE before western blotting.
Statistical analysis
Statistical analysis for the screens was performed using Microsoft Excel. Plate median fluorescent intensity values were normalized to the mean of plate medians of the intensity for each of EUFA423 and EUFA423B2 data sets. To calculate a mean ratio between 2 cell lines for each gene, the normalized fluorescent intensity values of EUFA423 were divided by the values of EUFA423B2 for each of the three replicates for each gene. siRNAs, with the mean ratio less than the mean −2 SD, were scored as putative hits.
Statistical analysis for the effect of inhibitors was performed using GraphPad Prism version 5 (GraphPad Software Inc). Nonlinear regression analysis was used to fit a dose response curves and to calculate IC50 values, which were compared using Mann-Whitney test or extra sum of squares F test.
Results
Restoring expression of functional BRCA2 in the human EUFA423 cell line
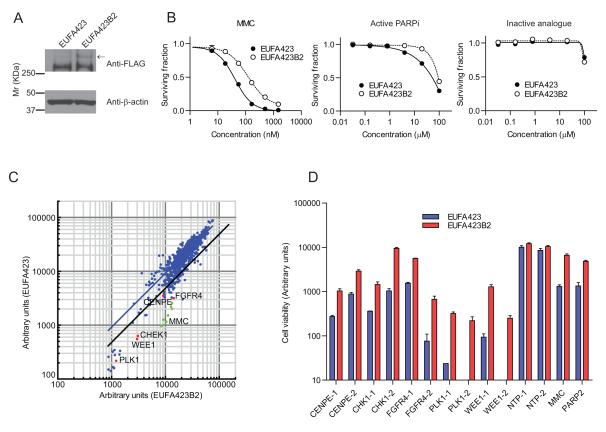
We generated a pair of stable isogenic cell lines lacking or expressing functional BRCA2 by introducing an N-terminal FLAG tagged full-length BRCA2 cDNA into the human fibroblast cell line, EUFA423. This line is derived from a patient within the D1 complementation group of Fanconi anemia, and is characterised by compound germline heterozygosity for BRCA2 mutations, which encode C-terminally truncated and functionally defective BRCA2 proteins (18). The reconstituted cell line (EUFA423B2) showed constitutive expression of FLAG-BRCA2 by western blotting with an antibody raised against the FLAG epitope (Figure 1A). We gathered several lines of evidence to show that the FLAG-tagged BRCA2 expressed in the cells is functional. EUFA423B2 cells were less sensitive than the parental line to MMC, a genotoxin known to engage BRCA2 dependent, homology-directed repair as well as to an active PARP1 inhibitor, KU0058948, but not to an inactive analogue, KU0051529 (10) (Figure 1B). Moreover, transient expression of the FLAG-tagged protein was able to restore formation of RAD51 nuclear foci in response to ionizing radiation in EUFA423 cells (Supplementary Figure 2A). Finally, immunoprecipitation with the anti-FLAG antibody confirmed that the tagged protein was able to interact with endogenous RAD51, a key partner of BRCA2, in 293T cells (Supplementary Figure 2B).
Figure 1. An RNAi screen to identify genes synthetic lethal with BRCA2 deficiency.
A, FLAG-BRCA2 expression was confirmed by immunoblot with an antibody against the FLAG epitope. B, The survival fraction is plotted for each of the indicated doses of MMC, active PARP inhibitor (KU0058948) and inactive analogue (KU0051529). Error bars represent the standard error of the mean (SEM) from three independent experiments. C, The results from the primary BRCA2 synthetic lethal RNAi screen are shown. Each dot on the XY scattered plot corresponds to the viable cell number (ie., fluorescence intensity values from the Fusion plate reader) of EUFA423 (y-axis) or EUFA423B2 (x-axis) expressed in arbitrary units. The ratio of the viable cell population of EUFA423 to EUFA423B2 was calculated and 30 siRNAs for which the ratios were at least 2 standard deviations (SD) less than the population mean (black line) were selected as candidate hits. D, Five candidate synthetic lethal targets to BRCA2 deficiency were validated using two different siRNA oligonucleotides. (Also shown in C with red dots). Y-axis values correspond to the viable cell population in EUFA423 (blue) and EUFA423B2 (red) expressed in arbitrary units. Error bars represent SEM from three independent experiments. NTP-1 and NTP-2 siRNAs are used as negative controls. PARP2 siRNA and MMC are used as positive controls.
An RNAi screen to identify genes synthetic lethal with BRCA2 deficiency
We utilised an RNAi library that targets 880 kinases and cell cycle regulated proteins to identify genes whose knockdown is synthetic lethal with BRCA2 deficiency. Cell viability was assessed in triplicate wells of 96-well plates 5 days after transfection of siRNA pools in each of the two isogenic lines and the ratio of the viable cells in EUFA423 compared to EUFA423B2 was calculated (Supplementary Figure 2C). Employing a statistical cut-off of 2 standard deviations (SD) from the mean, the primary screen identified 30 candidate genes that selectively suppressed the growth of BRCA2 deficient cells (Figure 1C). These candidates were further validated with two independent siRNA oligonucleotides of different sequence to exclude off-target effects (Figure 1D and Table). Five candidates successfully validated, however, we chose CHK1 for further investigation on the basis of the following two criteria: 1) CHK1 and centromere protein E (CENPE) were less cytotoxic to the BRCA2 proficient EUFA423B2 cell line than FGFR4, PLK1 and WEE1 and therefore their effect was more selective for BRCA2 deficient cells. 2) CHK1 is currently intensively pursued as a potential cancer specific therapeutic target, and several CHK1 inhibitors are available commercially and in the published scientific literature (19). At present, a CENPE specific inhibitor (GSK-923295) is not widely available, although it is also an intriguing candidate (20).
Table.
Validated candidates in secondary screen
| Gene | Ratio of EUFA423/EUFA423B2 cell viability Gene (Ave.± S.D.) |
|---|---|
| WEE1 | 0.10±0.02*** |
| PLK1 | 0.10±0.14*** |
| RAPGEF3 | 0.19±0.02 |
| CDC27 | 0.19±0.04 |
| COASY | 0.22±0.07 |
| MMC | 0.29±0.04* |
| DUSP7 | 0.32±0.05 |
| TSSK3 | 0.35±0.13 |
| CHEK1 | 0.35±0.05*** |
| EEF2K | 0.36±0.05 |
| CENPE | 0.37±0.04*** |
| PARP2 | 0.38±0.05* |
| FGFR4 | 0.38±0.02*** |
| NTpool-1 | 1.18±0.08** |
| NTpool-2 | 1.17±0.05** |
Positive control,
Negative control (non-target siRNA)
Validated by 2 siRNAs
BRCA2-deficient cells are sensitive to CHK1 inhibitors
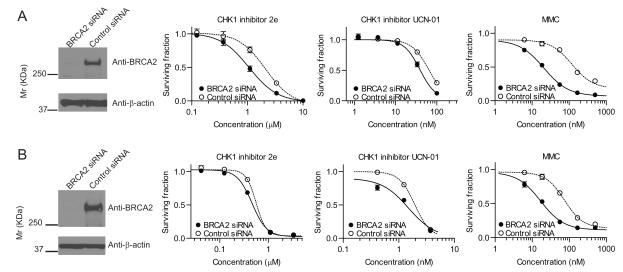
The phenotypic effects of genetic depletion of protein kinases by siRNA are not always recapitulated by small-molecule kinase inhibitors (21). Therefore, we tested if pharmacological inhibition of CHK1 kinase activity could selectively suppress the growth of human fibroblast (MRC5VA) and cancers (U2OS) depleted of BRCA2 protein using siRNA. We used two different ATP competitive CHK1 inhibitors, UCN-01 (22) and 2e (more selective for CHK1 over CHK2) (23). BRCA2 depleted MRC5VA cells were more sensitive to both CHK1 inhibitors (2e and UCN-01) as well as MMC compared to control cells (Figure 2A). (2e; IC50=1.1μM for BRCA2-depleted and 2.1μM for control, p=0.0004; UCN-01, IC50=39nM for BRCA2-depleted and 63nM for control, p<0.0001). BRCA2 depleted U2OS cells also demonstrated significantly higher sensitivity to CHK1 inhibition compared to controls (Figure 2B) (2e; IC50=0.46μM for BRCA2-depleted and 0.56μM for control, p=0.009; UCN-01, 1.5nM for BRCA2-depleted and 1.9nM for control, p=0.0003). In addition to these BRCA2 depleted cells, EUFA423B2 demonstrated less sensitivity to 2e compared to EUFA423 (Supplementary Figure 2D). On the basis of these experiments, we conclude that pharmacological inhibition of CHK1 kinase activity was able to replicate the RNAi screening result. Our data are in agreement with a previous study that demonstrated the specific effect of CHK1 inhibition on BRCA2 depleted cells using a different CHK1 inhibitor, Gö6976 (24).
Figure 2. Pharmacologic CHK1 inhibition suppresses the growth of BRCA2-deficient cell lines.
MRC5VA (A) and U2OS (B) cells were transfected with BRCA2 siRNA or Luciferase siRNA. Western blots were performed with antibodies against BRCA2 (top row) and β-actin (bottom row) to confirm efficient BRCA2 knockdown in siRNA treated cells. Survival fractions represent mean ±SEM from three independent experiments.
The extra sum-of-squares F test was used for the comparison of the IC50 values between BRCA2 siRNA and control siRNA transfected cells.
Chk1 inhibitors do not selectively suppress the growth of murine and human BRCA2 deficient pancreatic cancer cell lines
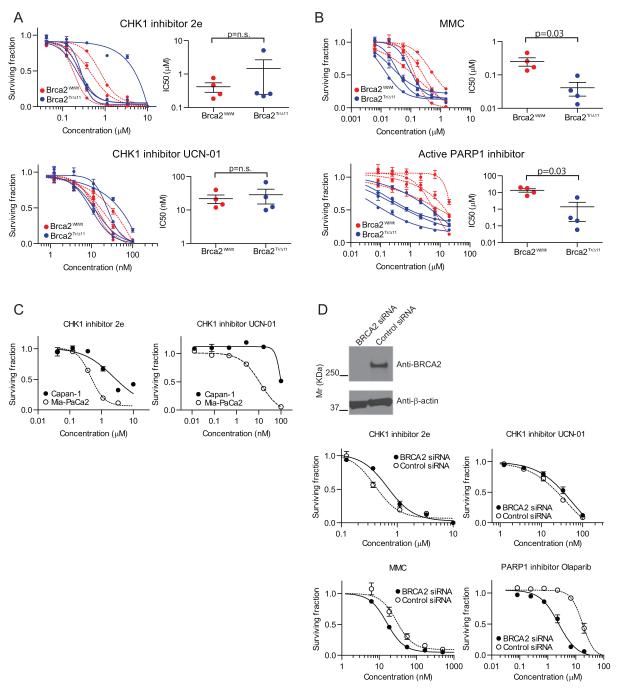
BRCA2 deficient epithelial cell lines from tissues with a high risk of transformation in carriers of germline BRCA2 mutations are scarce, and often not directly comparable. In order to explore whether the synthetic lethal interaction with CHK1 inhibition is dependent on cellular context, we exploited a set of BRCA2-deficient mouse pancreatic cancer cell lines, generated from a novel transgenic model of familial, BRCA2-related pancreatic cancer (17). This model emulates heterozygosity for Brca2 in the somatic cells using the germline, prematurely truncating mutant Brca2Tr1492, coupled with inactivation of the second Brca2 allele specifically in Pdx1-expressing pancreatic progenitor cells, by means of Cre-loxP recombination. Fully penetrant pancreatic adenocarcinoma develops in mice with conditional activation of the Kras oncogene (KrasG12D/+) coupled with inactivation of Trp53 by means of the Trp53R270H/+ dominant negative contact mutant and either wild-type Brca2 (Brca2wt/wt) or bi-allelic Brca2 inactivation (Brca2Tr/Δ11). Eight Pdx1-Cre;KrasG12D/+; Trp53R270H/+ mouse pancreatic cancer cell lines, 4 with wild type Brca2 and 4 with bi-allelic Brca2 inactivation, were treated with CHK1 inhibitors at concentrations previously determines from a dose-response titration (Supplementary Figure 2E).
Unexpectedly, the CHK1 inhibitors did not suppress the growth of BRCA2 deficient cells compared to that of the BRCA2 proficient cells (2e; IC50=1.5±1.2 μM (mean+/−SEM) for BRCA2 deficient and 0.42±0.13μM for BRCA2 proficient, p=n.s.; UCN-01, IC50=29±13nM for BRCA2 deficient and 22±6.3nM for BRCA2 proficient, p=n.s.) (Figure 3A). Interestingly, BRCA2 deficient murine pancreatic cancer cells remained significantly more sensitive to MMC (p=0.03) and the active PARP inhibitor (Olaparib) (p=0.03) compared to the cells with wild type Brca2 (Figure 3B), suggesting that the lack of sensitivity to the CHK1 inhibitor occurs in the setting of impaired homology-directed DNA repair, which underpins sensitivity to MMC and PARP1. These results were further substantiated in the setting of human pancreatic cancer, because Capan-1, a BRCA2 deficient human pancreatic cancer cell line, was less sensitive to both Chk1 inhibitors compared to Mia-PaCa-2, a pancreatic cancer cell line with wild-type BRCA2 status (Figure 3C). In addition, human pancreatic cancer cells (Mia-PaCa2) carrying activated KRAS and mutant TP53 exhibited a statistically significant increase in resistance to both CHK1 inhibitors after BRCA2 depletion using RNA interference, despite their enhanced sensitivity to Olaparib and MMC (Figure 3D).
Figure 3. Chk1 inhibitors do not selectively suppress the growth of murine and human BRCA2 deficient pancreatic cancer cell lines.
Four Brca2 wild type (red) and 4 Brca2 deficient (blue) murine pancreatic cancer cell lines were used. These cells were treated with CHK1 inhibitors, 2e or UCN-01(A), MMC or PARP1 inhibitor, Olaparib (B). The left column shows the surviving fraction for each drug treatment. The fit curves show Brca2 wild type cells (red) and Brca2 deficient cells (blue). Right columns showed the variation of IC50 value between Brca2 genotypes. The differences of the IC50 values for each drug between the two populations were compared using the Mann-Whitney test. C, Capan-1 (BRCA2 deficient human pancreatic cancer cell line) and Mia-PaCa2 (BRCA2 wild type human pancreatic cancer cell line) were treated with CHK1 inhibitors and the cell viability was measured. The IC50 (2e) for Capan-1 (2.4μM) was significantly higher than that for Mia-PaCa-2 (0.46μM) (p<0.0001). UCN-01 also showed higher IC50 (98nM) in Capan-1 than in Mia-PaCa-2 (12nM), respectively (p<0.0001). D, Mia-PaCa2 cells were transfected with BRCA2 siRNA or Luciferase siRNA, and cell viability after exposure to the indicated drugs was measured. The x-axis shows the concentration of drugs and the y-axis depicts the surviving fraction. Western blots were performed with antibodies against BRCA2 (top row) and β-actin (bottom row) to confirm efficient BRCA2 knockdown in siRNA-treated cells. The IC50 (2e) for BRCA2 knockdown (0.67μM) was significantly higher than that for the control (0.40μM) (p<0.002). UCN-01 also showed a higher IC50 (56nM) for BRCA2 knockdown than control (41nM) (p<0.03).
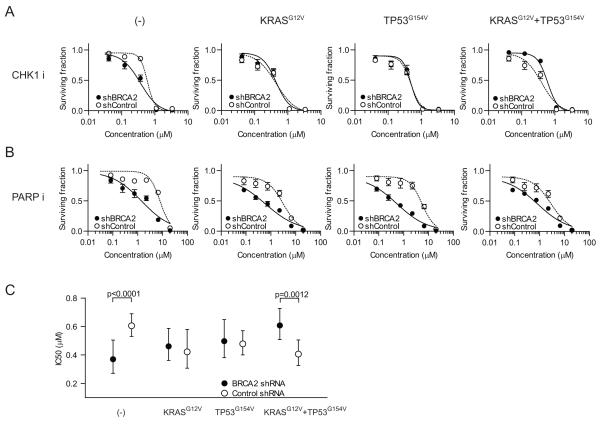
Oncogenic KRAS and dominant negative TP53 render BRCA2 depleted HEK293 cells resistant to CHK1 inhibition
We considered that the genetic constitution of Brca2-deficient pancreatic cancer cells accounted for their lack of sensitivity to CHK1 inhibitors. Pancreatic adenocarcinoma is driven by activating mutations in the KRAS oncogene, which are found in >90% of all cases (15), coupled with inactivation of TP53 in 50-75% of tumours (16). Given the dominant role of these two events, we hypothesized that they could account for the observed lack of sensitivity to CHK1 inhibition in the pancreatic cancer lines. In order to test this directly, we transfected HEK293 cells with KRASG12V and TP53G154V and simultaneously knocked down endogenous BRCA2 with shRNA. Then, we checked if the oncogenic KRAS activation may facilitate the cell survival in the BRCA2 deficient background to CHK1 inhibition. In support of our earlier results, BRCA2 knockdown alone sensitized HEK293 cells to the CHK1 inhibitor (Figure 4A, (-) and 4C). Expression of KRASG12V and TP53G154V, on the other hand, was sufficient to overcome this synthetic lethal interaction (Figure 4A, KRASG12V+TP53G154V, and 4C). The effect of combined expression of KRASG12V and TP53G154V was specific to CHK1 inhibition because BRCA2 depleted cells remained sensitive to the PARP1 inhibitor in this context. (Figure 4B). These results suggest that the combination of oncogenic KrasG12D and dominant negative Trp53R270H may, at least in part, be responsible for the lack of sensitivity to CHK1 inhibitors of BRCA2 deficient mouse pancreatic cancer cells.
Figure 4. Expression of oncogenic KRASG12V and dominant negative TP53G154V opposes the sensitivity of BRCA2-depleted cells to CHK1 inhibition.
BRCA2 or control shRNA-transfected HEK293 cells, co-transfected with different combinations of KRASG12V and TP53G154V, were treated with either CHK1 inhibitor 2e (A) or a PARP inhibitor (B). C, IC50 values for 2e in HEK293 with different combinations of mutant KRAS and dominant negative p53. The mean IC50 values (closed circle, BRCA2 shRNA; open circle, control shRNA) and 95% confidence intervals (bars for each side) are calculated based on curve fits with the nonlinear regression method. The extra sum-of-squares F test was used for the comparison of the IC50 values between BRCA2 shRNA and control shRNA transfected cells.
Gemcitabine restores sensitivity to CHK1 inhibition in Brca2 deficient pancreatic cancer cells with Kras/Trp53 mutations
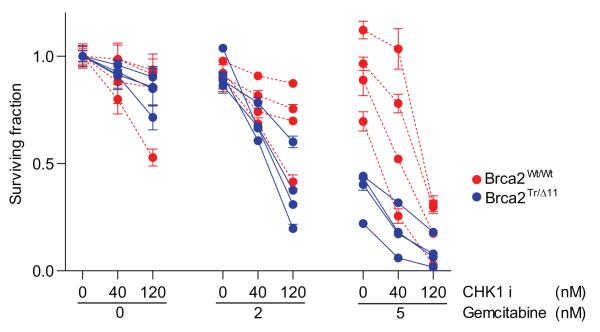
We hypothesized that low level S-phase dependent genotoxic stress might overcome the ability of oncogenic KRAS and dominant negative TP53 to dampen the response of BRCA2-deficient cells to CHK1 inhibition. For these experiments, we chose a deoxycytidine analogue, gemcitabine (25-26), which is the mainstay of human pancreatic cancer treatment, and has recently been shown to be synergistic with CHK1 inhibition in suppressing the growth of a pancreatic cancer cell line (27). The result was striking. No difference in cell viability was observed between Brca2 deficient and proficient pancreatic cancer cell lines at the lower range of 2e concentration in the absence of gemcitabine. Addition of gemcitabine at a low concentration, 2nM, specifically sensitised Brca2 deficient- but not Brca2 proficient- cells to the CHK1 inhibitor. This selectivity was less apparent when the concentration of gemcitabine was increased to 5nM. Indeed, this concentration of gemcitabine suppressed BRCA2-deficient cell growth even in the absence of the CHK1i 2e (Figure 5), in agreement with a previous study in BRCA2-deficient Chinese Hamster Ovary (CHO) cells (28). Thus, our results suggest that the combination of low doses of gemcitabine with CHK1i may be useful in the treatment of familial pancreatic cancer associated with BRCA2 mutations.
Figure 5. Gemcitabine restores sensitivity to CHK1 inhibition in Brca2 deficient pancreatic cancer cells with Kras/Trp53 mutations.
Four Brca2-wild type (red) and 4 Brca2 deficient (blue) mouse pancreatic cancer cell lines were treated with the combination of CHK1i and gemcitabine. The y-axis depicts the surviving fraction for each drug combination. The x-axis shows the concentration of the drugs used in each combination of CHK1i and gemcitabine. Error bars represent the mean +/− SEM from three independent experiments.
Discussion
We show here that the growth suppressive effect of CHK1 inhibitors on BRCA2-deficient cells identified in an RNAi screen is opposed by the genetic changes (KRAS activation and TP53 inactivation) commonly found in familial pancreatic adenocarcinoma associated with BRCA2 mutations (15-16). The precise mechanism underlying CHK1i resistance in this setting is unclear, but we speculate that it may be related to alterations in the pathways for DNA double-strand break repair that KRAS activation is suggested to engender (29). The expression of ras oncogenes correlates with tumor resistance to ionizing radiation (30-31). Intriguingly, signalling downstream of oncogenic KRAS may affect the efficacy of cellular mechanisms used for double-strand break repair (29, 32). Furthermore, these signals appear to limit the DNA damage induced by a CHK1 inhibitor in human multiple myeloma cells (33). Together, these considerations also raise the possibility that oncogenic ras activation may accompany poor therapeutic responsiveness to CHK1i in settings other than familial pancreatic cancer associated with BRCA2 mutations.
We find that the insensitiveness of BRCA2-deficient pancreatic cancer cells to CHK1i can be overcome by combination with gemcitabine, a genotoxic agent already used in the treatment of pancreatic adenocarcinoma. Indeed, it has been suggested that CHK1 inhibition may potentiate the replication-associated DNA damage induced by gemcitabine exposure (27), or by defects in the cellular pathways that respond to replication stress (28). We therefore speculate that the combination of CHK1i with gemcitabine may also be effective in pancreatic cancers lacking Fanconi anemia proteins like FANCG, components of a functional network that involves BRCA2 (34); such cancers have previously been reported to be resistant to gemcitabine alone (35).
Notably, our findings highlight the concept that synthetic lethal interactions identified by in vitro screens may not be valid in the genetic context of specific forms of cancer. Thus, the exploitation of synthetic lethality in cancer therapy is likely to require careful consideration not only of cancer genotypes, but also of potential drug combinations, if the high attrition rate observed in early phase clinical trials is to be tackled (8). In particular, our demonstration here that the sensitivity of BRCA2-deficient pancreatic cancer cells to CHK1i is modulated by cancer-associated alterations in KRAS and TP53 suggests that the validity of other potential therapeutic targets for cancer identified in ‘synthetic lethal’ RNAi screens will depend on the genetic context of specific malignancies. This proposal here warrants further investigation.
Supplementary Material
Acknowledgements
We thank Ko Sato (Venkitaraman laboratory) and Bob Boyle (Sentinel Oncology, Cambridge) for helpful discussions and the generous sharing of reagents.
Grant support, HH was supported by a fellowship from the Uehara Memorial Foundation, and grants to ARV from the Breast Cancer Campaign and UK Medical Research Council. FS received a Clinical Training Fellowship from Cancer Research UK and was supported by a grant to ARV from the UK Medical Research Council. PR was supported by a grant to ARV from the UK Medical Research Council.
References
- 1.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–72. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 2.Venkitaraman AR. Linking the cellular functions of BRCA genes to cancer pathogenesis and treatment. Annu Rev Pathol. 2009;4:461–87. doi: 10.1146/annurev.pathol.3.121806.151422. [DOI] [PubMed] [Google Scholar]
- 3.Patel KJ, Yu VP, Lee H, Corcoran A, Thistlethwaite FC, Evans MJ, et al. Involvement of Brca2 in DNA repair. Mol Cell. 1998;1:347–57. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 4.Yu VP, Koehler M, Steinlein C, Schmid M, Hanakahi LA, van Gool AJ, et al. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 2000;14:1400–6. [PMC free article] [PubMed] [Google Scholar]
- 5.Abbott DW, Freeman ML, Holt JT. Double-strand break repair deficiency and radiation sensitivity in BRCA2 mutant cancer cells. J Natl Cancer Inst. 1998;90:978–85. doi: 10.1093/jnci/90.13.978. [DOI] [PubMed] [Google Scholar]
- 6.Evers B, Schut E, van der Burg E, Braumuller TM, Egan DA, Holstege H, et al. A high-throughput pharmaceutical screen identifies compounds with specific toxicity against BRCA2-deficient tumors. Clin Cancer Res. 2010;16:99–108. doi: 10.1158/1078-0432.CCR-09-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064–8. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 8.Kaelin WG., Jr. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–98. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 9.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 10.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 11.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 12.Kennedy RD, Chen CC, Stuckert P, Archila EM, De la Vega MA, Moreau LA, et al. Fanconi anemia pathway-deficient tumor cells are hypersensitive to inhibition of ataxia telangiectasia mutated. J Clin Invest. 2007;117:1440–9. doi: 10.1172/JCI31245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn SA, Greenhalf B, Ellis I, Sina-Frey M, Rieder H, Korte B, et al. BRCA2 germline mutations in familial pancreatic carcinoma. J Natl Cancer Inst. 2003;95:214–21. doi: 10.1093/jnci/95.3.214. [DOI] [PubMed] [Google Scholar]
- 14.Couch FJ, Johnson MR, Rabe KG, Brune K, de Andrade M, Goggins M, et al. The prevalence of BRCA2 mutations in familial pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:342–6. doi: 10.1158/1055-9965.EPI-06-0783. [DOI] [PubMed] [Google Scholar]
- 15.Caldas C, Kern SE. K-ras mutation and pancreatic adenocarcinoma. Int J Pancreatol. 1995;18:1–6. doi: 10.1007/BF02825415. [DOI] [PubMed] [Google Scholar]
- 16.Redston MS, Caldas C, Seymour AB, Hruban RH, da Costa L, Yeo CJ, et al. p53 mutations in pancreatic carcinoma and evidence of common involvement of homocopolymer tracts in DNA microdeletions. Cancer Res. 1994;54:3025–33. [PubMed] [Google Scholar]
- 17.Skoulidis F, Cassidy LD, Pisupati V, Jonasson JG, Bjarnason H, Eyfjord JE, et al. Germline Brca2 heterozygosity promotes Kras(G12D) -driven carcinogenesis in a murine model of familial pancreatic cancer. Cancer Cell. 2010;18:499–509. doi: 10.1016/j.ccr.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 18.Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–9. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 19.Dai Y, Grant S. New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin Cancer Res. 2010;16:376–83. doi: 10.1158/1078-0432.CCR-09-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson JR, Patrick DR, Dar MM, Huang PS. Targeted anti-mitotic therapies: can we improve on tubulin agents? Nat Rev Cancer. 2007;7:107–17. doi: 10.1038/nrc2049. [DOI] [PubMed] [Google Scholar]
- 21.Knight ZA, Shokat KM. Chemical genetics: where genetics and pharmacology meet. Cell. 2007;128:425–30. doi: 10.1016/j.cell.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 22.Wang Q, Fan S, Eastman A, Worland PJ, Sausville EA, O’Connor PM. UCN-01: a potent abrogator of G2 checkpoint function in cancer cells with disrupted p53. J Natl Cancer Inst. 1996;88:956–65. doi: 10.1093/jnci/88.14.956. [DOI] [PubMed] [Google Scholar]
- 23.Wang GT, Li G, Mantei RA, Chen Z, Kovar P, Gu W, et al. 1-(5-Chloro-2-alkoxyphenyl)-3-(5-cyanopyrazin-2-yl)ureas as potent and selective inhibitors of Chk1 kinase: synthesis, preliminary SAR, and biological activities. J Med Chem. 2005;48:3118–21. doi: 10.1021/jm048989d. [DOI] [PubMed] [Google Scholar]
- 24.Chen CC, Kennedy RD, Sidi S, Look AT, D’Andrea A. CHK1 inhibition as a strategy for targeting Fanconi Anemia (FA) DNA repair pathway deficient tumors. Mol Cancer. 2009;8:24. doi: 10.1186/1476-4598-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang P, Chubb S, Hertel LW, Grindey GB, Plunkett W. Action of 2′,2′-difluorodeoxycytidine on DNA synthesis. Cancer Res. 1991;51:6110–7. [PubMed] [Google Scholar]
- 26.Ostruszka LJ, Shewach DS. The role of cell cycle progression in radiosensitization by 2′,2′-difluoro-2′-deoxycytidine. Cancer Res. 2000;60:6080–8. [PubMed] [Google Scholar]
- 27.Azorsa DO, Gonzales IM, Basu GD, Choudhary A, Arora S, Bisanz KM, et al. Synthetic lethal RNAi screening identifies sensitizing targets for gemcitabine therapy in pancreatic cancer. J Transl Med. 2009;7:43. doi: 10.1186/1479-5876-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McNeely S, Conti C, Sheikh T, Patel H, Zabludoff S, Pommier Y, et al. Chk1 inhibition after replicative stress activates a double strand break response mediated by ATM and DNA-dependent protein kinase. Cell Cycle. 2010;9 doi: 10.4161/cc.9.5.10935. [DOI] [PubMed] [Google Scholar]
- 29.Toulany M, Kehlbach R, Florczak U, Sak A, Wang S, Chen J, et al. Targeting of AKT1 enhances radiation toxicity of human tumor cells by inhibiting DNA-PKcs-dependent DNA double-strand break repair. Mol Cancer Ther. 2008;7:1772–81. doi: 10.1158/1535-7163.MCT-07-2200. [DOI] [PubMed] [Google Scholar]
- 30.Sklar MD. The ras oncogenes increase the intrinsic resistance of NIH 3T3 cells to ionizing radiation. Science. 1988;239:645–7. doi: 10.1126/science.3277276. [DOI] [PubMed] [Google Scholar]
- 31.Bernhard EJ, Stanbridge EJ, Gupta S, Gupta AK, Soto D, Bakanauskas VJ, et al. Direct evidence for the contribution of activated N-ras and K-ras oncogenes to increased intrinsic radiation resistance in human tumor cell lines. Cancer Res. 2000;60:6597–600. [PubMed] [Google Scholar]
- 32.Chang IY, Youn CK, Kim HB, Kim MH, Cho HJ, Yoon Y, et al. Oncogenic H-Ras up-regulates expression of Ku80 to protect cells from gamma-ray irradiation in NIH3T3 cells. Cancer Res. 2005;65:6811–9. doi: 10.1158/0008-5472.CAN-04-4065. [DOI] [PubMed] [Google Scholar]
- 33.Dai Y, Chen S, Pei XY, Almenara JA, Kramer LB, Venditti CA, et al. Interruption of the Ras/MEK/ERK signaling cascade enhances Chk1 inhibitor-induced DNA damage in vitro and in vivo in human multiple myeloma cells. Blood. 2008;112:2439–49. doi: 10.1182/blood-2008-05-159392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venkitaraman AR. Tracing the network connecting BRCA and Fanconi anaemia proteins. Nat Rev Cancer. 2004;4:266–76. doi: 10.1038/nrc1321. [DOI] [PubMed] [Google Scholar]
- 35.van der Heijden MS, Brody JR, Dezentje DA, Gallmeier E, Cunningham SC, Swartz MJ, et al. In vivo therapeutic responses contingent on Fanconi anemia/BRCA2 status of the tumor. Clin Cancer Res. 2005;11:7508–15. doi: 10.1158/1078-0432.CCR-05-1048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.