Abstract
Curcumin is a polyphenolic compound derived from the Indian spice turmeric. We used nanoparticle-encapsulated curcumin to treat medulloblastoma and glioblastoma cells. This formulation caused a dose-dependent decrease in growth of multiple brain tumor cell cultures, including the embryonal tumor derived lines DAOY and D283Med, and the glioblastoma neurosphere lines HSR-GBM1 and JHH-GBM14. The reductions in viable cell mass observed were associated with a combination of G2/M arrest and apoptotic induction. Curcumin also significantly decreased anchorage-independent clonogenic growth and reduced the CD133-positive stem-like population. Downregulation of the insulin-like growth factor pathway in DAOY medulloblastoma cells was observed, providing one possible mechanism for the changes. Levels of STAT3 were also attenuated. Hedgehog signaling was blocked in DAOY cells but Notch signaling was not inhibited. Our data suggest that curcumin nanoparticles can inhibit malignant brain tumor growth through the modulation of cell proliferation, survival and stem cell phenotype.
Key words: curcumin, hedgehog, IGF, glioblastoma, medulloblastoma, nanocurcumin, nanoCurc™
Introduction
Medulloblastoma and glioblastoma are the most common malignant cancers arising in the central nervous systems of children and adults, respectively.1 Current therapeutic strategies include surgery, radiation therapy and chemotherapy, but these are associated with significant side effects and only limited efficacy, particularly in patients with glioblastoma. More effective, less harmful therapeutic agents are urgently needed. A number of preclinical and clinical studies suggest that natural compounds such as curcumin may represent useful additions to therapeutic regimens of cancer patients.2,3
Turmeric has been historically used in Indian Ayurvedic medicine to treat a variety of disorders.4 More recently, curcumin, also known as diferuloyl methane, a polyphenolic compound derived from turmeric, has been shown to exert antitumor effects in many different cancer cell lines and animal models.5,6 The clinical effects of curcumin are currently being investigated in human clinical trials for a variety of conditions including pancreatic cancer, colorectal cancer and multiple myeloma (www.clinicaltrials.gov/ct2/show/NCT00094445, www.clinicaltrials.gov/ct2/show/NCT00118989, www.clin icaltrials.gov/ct2/show/NCT00113841).
The mechanisms by which curcumin is thought to inhibit tumorigenesis are diverse, and pro-apoptotic, anti-angiogenic, anti-inflammatory, immunomodulatory and anti-mitogenic effects have been described in various systems.3,7 Some of curcumin's potential molecular targets include insulin-like growth factor (IGF), Akt, mitogen-activated protein kinase (MAPK), signal transducer and activator of transcription 3 (STAT3), Nuclear factor kappa B (NFκB) and Notch.6,8 These pathways are all thought to be active in malignant brain tumors,9,10 raising the possibility that curcumin could be effective in treating medulloblastoma or glioblastoma.
Several groups have begun to examine the potential of curcumin in neuro-oncology. Curcumin was first found to repress the in vitro invasion of astrocytic glioma cells through inhibiting gene expression of matrix metalloproteinases.11 In another study, the ERK, JNK and MAPK/Elk-1/Egr-1 pathways were found to be required for p53-independent transcriptional activation of p21Waf/Cip in U87MG glioblastoma cells in response to curcumin treatment.12 In addition, there was evidence that curcumin induces G2/M arrest and non-apoptotic autophagic cell death in both U87MG and U373MG glioma cells.13
Most prior reports have focused on glioblastoma, and relatively little is known about the potential of curcumin to treat other brain malignancies such as medulloblastoma. Also, while several studies have suggested that curcumin can suppress clonogenicity,14,15 the direct effects of curcumin on stem-like brain tumor cells are not well understood. Past investigators have used adherent glioblastoma cells grown in high serum rather than neurosphere lines, limiting investigations into stem cell effects. We therefore investigated the effects of curcumin on both medulloblastoma and glioblastoma, and examined in particular changes in the stem-like tumor subpopulations, using neurosphere cultures when possible. NanoCurc™, a recently described polymeric nanoparticle formulation of curcumin amenable to systemic delivery,16 was used in this study in order to facilitate eventual clinical translation. We found that medulloblastoma and glioblastoma cells, including neurospheres derived from malignant gliomas, had their growth and clonogenicity significantly inhibited, with a concomitant reduction in the percentage of stem-like tumor cells. A variety of signal transduction pathways possibly mediating these effects were altered following treatment. These preliminary in vitro studies set the stage for the in vivo translation of NanoCurc™ (henceforth “nanocurcumin”) in preclinical animal models of medulloblastoma and glioblastoma, respectively.
Results
Curcumin nanoparticles inhibit the growth of brain tumor cell lines via programmed cell death and G2/M cell cycle arrest.
Nanocurcumin treatment caused a dose-dependent decrease in cell growth as measured by MTS assay in multiple malignant brain tumor cell lines (Fig. 1). After 3 days of treatment using 10 µM curcumin, we noted a statistically significant 35% growth reduction in adherent DAOY medulloblastoma cells (Fig. 1A, p < 0.001). A second non-adherent line, D283Med, had growth inhibited by 87% over a somewhat longer period (Fig. 1B). To investigate the specificity of the observed inhibitory effects, we also examined NIH-3T3 cells. Although free curcumin concentrations above 30 µM had been previously shown by Jiang et al.17 to be toxic in NIH-3T3 cells, the doses of nanocurcumin (up to 20 µM) used in this study which slowed or arrested brain tumor growth resulted instead in increased growth rate of NIH-3T3 cells (Fig. 1C). This suggests that the effects are at least somewhat selective, and do not inhibit cellular proliferation and growth in all cells at these doses.
Figure 1.
Growth of brain tumor cell lines is selectively inhibited by nanocurcumin via programmed cell death and cell cycle arrest. MTS assays performed on brain tumor cell lines DAOY (A), D283Med (B), U87 (D), JHH-GBM14 (E) and HSR-GBM1 (F) showed significant growth reduction after treatment over the period of time indicated. This was due to both apoptotic induction (G–I) and G2/M cell cycle arrest (J–L). However, the non-neoplastic NIH-3T3 line instead showed greater growth after nanocurcumin treatment (C). Statistical significance was calculated using the final time points in (A–F). *p < 0.05, **p < 0.01, ***p < 0.001 compared to vehicle.
We also investigated the inhibitory effects of curcumin on the commonly used adherent glioblastoma cell line U87, and found that the growth of these was 32 ± 3.9% slower with 20 µM curcumin treatment over 5 days (Fig. 1D). Finally, we examined several glioblastoma neurosphere lines, which are thought to represent genetically and pathologically superior models as they stably maintain the genomic changes of primary tumors without accumulating significant additional alterations, allow the maintenance of stem-like tumor cells, and more faithfully recapitulate the invasive behavior of glioblastoma.18 These include lower-passage cells generated from a tumor recently resected at our institution (JHH-GBM14) as well as an established line cultured for several years (HSR-GBM1). Both were sensitive to the effects of nanocurcumin, with 36 ± 0.8% to 53 ± 4.3% statistically significant inhibition respectively (±SEM) in growth of the spheres over 6 days at higher doses (Fig. 1E and F, p < 0.001). All experiments were repeated at least two times with similar results.
To determine the mechanisms of growth inhibition, we examined both apoptosis and cell cycle parameters in nanocurcumin and vehicle treated cells. The percentage of apoptotic cells increased significantly from 2.3–30.5% (13-fold, p < 0.001) when DAOY cultures were treated for 1 day with 10 µM curcumin (Fig. 1G). The percentage of dead cells also dramatically increased over this time period. It is not clear why the level of apoptotic induction was higher than the overall growth suppression seen using MTS assay. A significant, dose-dependent induction in the fraction of apoptotic and dead cells was observed in the medulloblastoma line D283Med (Fig. 1H) and in HSR-GBM1 (Fig. 1I) and JHH-GBM14 glioblastoma neurospheres (data not shown). Cell cycle arrest also contributed to the slower growth in cultures exposed to nanocurcumin. In medulloblastoma (DAOY, D283Med) and glioblastoma (HSR-GBM1) cell lines, the percentage of cells in G2/M increased by 74, 63 and 25% respectively, with a roughly corresponding reduction in the G1 fraction (Fig. 1J–L).
Curcumin inhibits clonogenicity and depletes stem-like cells from malignant brain tumor cultures.
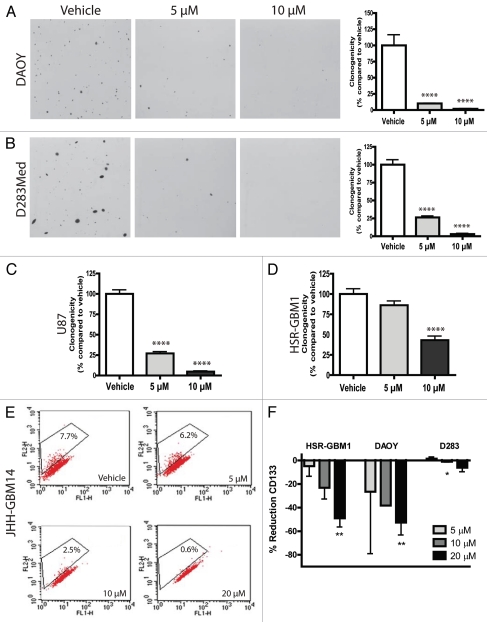
We next examined the ability of brain tumor cell lines to form anchorage-independent colonies over several weeks when treated with nanocurcumin. Equal numbers of DAOY single cells were seeded in soft agar and then grown in the presence of empty nanoparticles or increasing doses of nanocurcumin. Compared to vehicle-treated cells, 5 and 10 µM curcumin significantly reduced the normalized clonogenicity of DAOY cells from 100 to 10% (p < 0.0001) and 1.8% (p < 0.0001) respectively (Fig. 2A). Similar statistically significant reductions were observed in the D283Med and U87 cell lines (Fig. 2B and C). In the glioblastoma neurosphere cell line HSR-GBM1, statistically significant reductions were observed only with 10 µM curcumin (Fig 2D). The above findings raise the possibility that stem-like cells required for clonogenic tumor growth are depleted by nanocurcumin.
Figure 2.
Curcumin reduces clonogenicity of brain tumor cell lines DAOY (A), D283Med (B), U87 (C) and HSR-GBM1 (D) in a dose-dependent fashion. Equal number of cells were seeded and treated with the indicated nanocurcumin concentrations or void NVA622 nanoparticles (vehicle). Curcumin reduces the CD133-positive stem-like fraction in glioblastoma cell lines JHH-GBM14 (E) and HSR-GBM1, as well as D283Med medulloblastoma cell line (F). JHH-GBM14 (E) and HSR-GBM1 (F) cells were treated with void NVA622 nanoparticle (vehicle), 5, 10 or 20 µM nanocurcumin for 2 days and collected for flow cytometry analysis with CD133 antibodies. DAOY and D283Med (F) cells were treated with the same doses for only 1 day. *p < 0.05, **p < 0.01, ****p < 0.0001 compared to vehicle control.
To more directly address the effects of curcumin on stem-like cancer cells, we examined changes in the percentage of cells expressing the marker CD133. Glioblastoma cells expressing this marker have been shown by some investigators to be more clonogenic in vitro, and are more effective in establishing xenografts in vivo.19–21 However, it has recently been shown that CD133-negative cells can also be tumorigenic,22–24 and may contain an alternative subpopulation of stem-like cells. In our hands, CD133 expression in HSR-GBM1 neurospheres is associated with an approximately 2-fold increase in clonogenic potential.25 Using flow cytometric analysis, we found that the CD133-positive population in the JHH-GBM14 and HSR-GBM1 neurosphere lines decreased markedly over 2 days following addition of nanocurcumin (Fig. 2E and F). In JHH-GBM14 neurospheres, a step-wise 92% decrease from 7.7-0.6% was seen with increasing doses of curcumin (Fig. 2E). In HSR-GBM1, we observed an approximately 5, 23 and 49% reduction in the percentage of CD133-positive stem-like cells following 5, 10 and 20 µM doses of curcumin respectively (Fig. 2F). While the biological significance of CD133 expression in DAOY and D283Med is less clear, we have previously shown that CD133 marks cells with increased xenograft initiating potential in these lines,26 and observed decreases of 6 and 53% in the proportion of D283Med and DAOY medulloblastoma cultures expressing this stem cell marker when treated with 20 µM curcumin as compared to vehicle (Fig. 2F).
Curcumin inhibits IGF and STAT3 signaling.
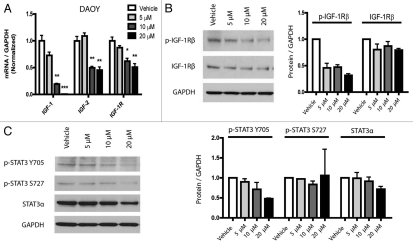
The IGF-1 receptor pathway is aberrantly regulated in tumors of the nervous system, including both medulloblastoma and glioblastoma.27,28 These tumors often overexpress IGF-1R and secrete both IGF-1 and IGF-2 ligands.27–30 We identified a remarkable 18-fold downregulation of IGF-1 expression following curcumin treatment in a preliminary oligonucleotide microarray analysis experiment in DAOY cells (data not shown). We therefore examined the effects of curcumin in this pathway using quantitative PCR and protein analysis. IGF-1, IGF-2 and IGF-1R levels were measured in RNA extracted from cells treated with nanocurcumin or void NVA622 nanoparticle (vehicle), revealing a dose-dependent reduction in expression of both the IGF ligands and IGF-1R in DAOY cells which was statistically significant at 10 µM and above (Fig. 3A). A significant reduction in IGF-1 mRNA levels, but not in IGF-2 or IGF-1R, was also observed in D283Med cells (data not shown). Western blot analysis showed a modest reduction in overall IGF-1Rβ protein levels, as well as a more pronounced decrease in the phosphorylated, active form of the receptor (p-IGF-1Rβ, Tyr 1135/1136) after curcumin treatment in DAOY cells (Fig. 3B). Normalized intensities are depicted as histograms in the right sub-panel.
Figure 3.
The IGF-1R pathway is attenuated after nanocurcumin treatment in medulloblastoma cells. DAOY (A) cells treated with the indicated concentrations of curcumin showed decreased transcript levels of IGF-1, IGF-2 and IGF-1R. *p < 0.05, **p < 0.01, ***p < 0.001 compared to vehicle control. Lysates from DAOY (B) cells treated for 26 hours with vehicle or the indicated nanocurcumin concentrations show reduction in p-IGF-1Rβ (Tyr 1135/1136) and IGF-1Rβ protein levels. (C) p-STAT3 (Tyr 705) levels were reduced following curcumin nanoparticle treatment in DAOY cells. Normalized intensities measured by densitometry of two independent experiments are depicted as bar graphs (B and C, right graphs).
IGF signaling activates two key downstream signal-transduction cascades, the lipase kinase PI3K/Akt pathway and the GTPase Ras-Raf-ERK/MAPK pathway. To determine if curcumin can modulate Akt signaling in medulloblastomas, we treated DAOY cells with vehicle or 20 µM curcumin for 6 or 26 hours and carried out western blot analysis for total and phospho-Akt (Ser 473). Supplemental Figure 1 shows a reduction of phospho-Akt levels after 6 hours of nanocurcumin treatment by 34%, but this had begun to normalize by 26 hours (normalized intensities are depicted as histograms in the bottom sub-panel). Total Akt levels were also reduced by curcumin treatment, and could account for at least a portion of the decrease in the phosphorylated fraction. This experiment was repeated two times with similar results. We also examined MAPK pathway activity after curcumin treatment. Most p42/p44 MAPK, p42/p44 phospho-MAPK, ERK1/2 and phospho-ERK1/2 proteins appeared unchanged following treatment, although some phosphoproteins were present at levels too low to be detected (data not shown). We also examined the effects of curcumin on STAT signaling, as this pathway has been reported to act downstream of IGF in some contexts,31,32 and is also known to promote a stem-like fate in brain tumors.33,34 STAT3α proteins were expressed in both medulloblastoma (Fig. 3C) and glioblastoma cells (data not shown). Immunoblotting with phospho-specific antibodies confirmed that STAT3α is phosphorylated on both Tyr 705 and Ser 727 residues, suggesting that the pathway is active. Treatment of the cells with dose-escalating nanocurcumin reduced total STAT3α protein levels, and to a greater extent, phosphorylation of STAT3 at Tyr 705 in DAOY cells (Fig. 3C). There was also a very slight reduction of the same phospho-protein in HSR-GBM1 (data not shown). In contrast, while in some experiments Ser 727 p-STAT3 levels appear to be reduced when grown in both serum-free media (Fig. 3C, left part) and 2% FBS (data not shown), in others they remained relatively constant or increased slightly, as reflected in the combined serum-free data depicted in the right part.
Inhibition of the hedgehog (Hh) pathway, but not notch, by curcumin in a subset of cell lines.
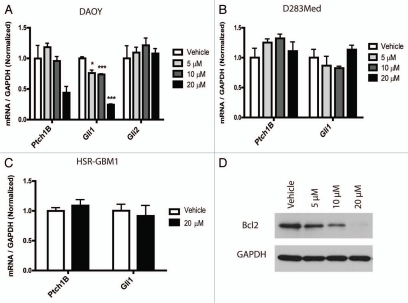
Both Hh and Notch have previously been implicated in medulloblastoma and GBM biology.21,35–37 Our microarray data revealed a 2.4-fold downregulation of Gli1 expression after 20 µM curcumin treatment in DAOY cells. Gli1 is a key target and effector of the Hedgehog (Hh) pathway, which has been associated in the initiation and growth of both medulloblastoma and glioblastoma.35,37,38 To validate our microarray results, we used quantitative real-time PCR to evaluate Gli1 transcripts and a second marker of Hh activity, Ptch1B. These targets were found to be reduced by 76 and 55% respectively in DAOY cells after treatment with 20 µM curcumin (Fig. 4A). In contrast, Gli2 transcript levels, which are not thought to reflect pathway activity, did not decrease. However, nanocurcumin did not inhibit Hh signaling in a second medulloblastoma cell line (D283Med) or in glioblastoma neurospheres (Fig. 4B and C). Because we have previously shown that Hh can control Bcl2 transcription in DAOY medulloblastoma cells and in primary tumors,39 we measured levels of this key antiapoptotic protein, and found reductions which corresponded to reductions in Gli1 (Fig. 4D).
Figure 4.
The Hh pathway is downregulated after curcumin treatment. Transcript levels of Hh pathway targets (Gli1 and Ptch1B) are reduced following treatment with nanocurcumin in DAOY (A) cells, but not in D283Med (B) or HSR-GBM1 (C). Protein levels of Bcl2 are downregulated following nanocurcumin treatment in DAOY cells (D). *p < 0.05, ***p < 0.001 compared to vehicle.
As Notch activity has been implicated in the propagation of the stem-like phenotype in glioblastoma,21 and has been proposed as a potential target of curcumin,40,41 we also measured targets of this pathway. However, expression levels of transcripts of the Notch target genes Hes1, Hes5 and Hey2 were not suppressed after nanocurcumin treatment in DAOY or HSR-GBM1 cells (Sup. Fig. 2), suggesting that curcumin does not block pathway activity in these cells.
Discussion
We investigated if nanocurcumin, a formulation that has significantly greater aqueous solubility and systemic bioavailability than free curcumin,16 can effectively inhibit the proliferation and clonogenicity of medulloblastoma and glioblastoma cell lines. Nanocurcumin was highly effective in blocking growth of the DAOY and D283Med medulloblastoma cultures, with a more modest inhibition of glioblastoma neurospheres. Both apoptotic cell death and G2/M cell cycle arrest contributed to the antitumor effects. While nothing was known about the effects of curcumin on medulloblastoma until recently, two other groups have now reported growth inhibition and the induction of caspase-mediated cell death in medulloblastoma cells following free curcumin treatment.14,42
This curcumin formulation also effectively inhibited the clonogenic potential of both medulloblastoma and glioblastoma lines, raising questions regarding its effects on stem-like tumor initiating cells. Recently, curcumin was found to target the stem-like side population in the adherent rat C6 glioma cells.43 We used a different marker, CD133 and neurospheres grown in serum-free conditions thought to help maintain stem cell populations for our glioma studies. In our tumor-derived neurospheres, we found that 20 µM curcumin induced a remarkable 49% decrease in the percentage of CD133 positive GBM cells. It also reduced this population in the D283Med medulloblastoma line. Consistent with the notion that stem-like tumor cells were depleted by nanocurcumin, soft agar clonogenic assays (Fig. 2) revealed much more pronounced effects than short term growth assays (Fig. 1). It remains to be seen, however, whether curcumin might also deplete non-neoplastic stem cells in the brain, which would have potentially significant side effects.
If curcumin is to be most effectively used therapeutically, it will be necessary to understand which signaling cascades it modulates. We therefore examined the molecular pathway(s) curcumin alters in brain tumors. Preliminary gene expression array analysis suggested that curcumin downregulates the IGF pathway in medulloblastoma via reduction of IGF-1 and 2 ligands, and we were able to confirm suppression of IGF-1Rβ receptor expression and activity using phospho-specific antibodies. Curcumin has been previously shown to suppress IGF-1 expression in breast cancer cells,44 suggesting that this may be a common target in multiple tumor types, although to our knowledge it has not been previously identified in brain tumors. A number of prior studies have also shown that IGF-1, IGF-2 and IGF-1R play an active role in the formation and growth of medulloblastoma and other brain tumors,45,46 supporting the biological relevance of their downregulation by curcumin.
In some contexts, the STAT pathway can be activated by IGF signaling.31,32 STAT has also been implicated in modulating stem cell phenotype in non-neoplastic cells47,48 and in several types of cancer, including brain tumors.33,34 Given the suppression of IGF activity and stem cell markers observed, we examined if STAT3 was also modulated by nanocurcumin. Indeed, the phosphorylation of Tyr 705 residue on STAT3, which induces dimerization, nuclear translocation and DNA binding,49 was reduced in DAOY cells (Fig. 3C). This suggests that suppression of IGF and STAT3 by curcumin could play a role in its effects on growth and stem cell phenotype in brain tumors. We also observed less pronounced effects of curcumin on Akt expression and phosphorylation. Akt has been implicated in the survival and differentiation of brain tumors,50,51 and could also play a role in the modulation of tumor growth and clonogenicity by nanocurcumin.
Two other pathways known to play critical roles in the stem cell phenotype of brain tumors are Notch21,36 and Hh.38,52 Both have also been previously implicated as targets of curcumin. Wang et al.41 showed that curcumin could downregulate Notch1 in pancreatic cancer cells, but we did not find any suppression of Notch targets in our tumor lines following curcumin treatment. Elamin et al.42 recently found that curcumin had inhibitory effects on the Hh pathway in medulloblastoma cells. Using the MED-5 cell line, they showed more than 5-fold reductions in Gli1 and 2-fold reductions in Ptch1, after treatment with 40 µM curcumin. Our findings were similar, with 20 µM curcumin reducing expression of Ptch1B and Gli1 by 55 and 76% respectively in DAOY cells (Fig. 4A). We have previously shown that Gli1 can regulate Bcl2 levels in this line,39 and this may therefore explain the changes we observed in that anti-apoptotic protein (Fig. 4D). Also, reductions in Bcl2 protein levels may lead to the induction of apoptosis observed in Figure 1G. However, we did not find pathway suppression in a second medulloblastoma cell line D283Med (Fig. 4B), indicating that Hh inhibition by curcumin is not universal for all medulloblastomas. We also did not find down-regulation of Gli1 and Ptch1B expression in treated HSR-GBM1 cells.
In summary, we have found that a nanoparticle formulation of curcumin NanoCurc™ can reduce the growth and clonogenicity of medulloblastoma and glioblastoma cell lines, and deplete the subpopulation of cells expressing the stem cell marker CD133. Downstream signaling pathways affected by curcumin in our models included IGF, STAT3, Akt and Hh. These data provide further support for the development of curcumin as a new therapy for brain tumors, and indicate that pathways affecting both survival and neoplastic stem cell phenotype can be modulated by this natural compound. The availability of nanocurcumin, which is readily amenable to systemic delivery and biodistribution16 should facilitate the in vivo application in preclinical animal models of these tumors.
Materials and Methods
Materials.
Ultra-pure curcumin (>99% diferuloylmethane) was a kind gift from Sabinsa Corporation. Monomers for polymer nanoparticle synthesis—specifically N-isopropylacrylamide (NIPAAM, 415324), vinylpyrrolidone (VP, V3409) and acrylic acid (AA, 147230)—were obtained from Sigma Aldrich. Reagents for the polymerization step, including NN'-methylenebis-acrylamide (MBA, 146072), ammonium persulfate (APS, 248614) and ferrous sulfate (FeSO4, F8048) were also from Sigma. For each experiment, nanocurcumin (NanoCurc™)16 was dissolved in sterile PBS to prepare a fresh 1 mM stock solution and diluted further in cell culture media to the appropriate free curcumin-equivalent concentrations.
Synthesis of nanocurcumin (NanoCurc™).
The predistilled monomers of NIPAAM, VP and AA are mixed together in a molar ratio of 60:20:20, respectively, hence the acronym “NVA622” for the resulting polymer. Polymerization was performed for 24 hours at 30°C under an inert (nitrogen) atmosphere, using APS and FeSO4 as initiator and activator, respectively. After complete polymerization, the total aqueous solution of polymer was purified using dialysis, and then lyophilized for post loading of curcumin, as described in reference 53. Typically, a 10 ml stock solution of polymeric nanoparticles (100 mg) was slowly mixed with 150 µl of curcumin solution in chloroform (10 mg/ml), and gently stirred for 15–20 minutes on low heating, in order to load curcumin and evaporate chloroform simultaneously. The resulting solution, corresponding to 1.5% (w/w) loading of curcumin in nanoparticles, was then snap frozen on a dry ice/acetone bath and lyophilized. The lyophilized nanocurcumin powder is stored at 4°C until further use.
Cell lines and cultures.
The DAOY and D283Med cell lines were obtained from the American Type Culture Collection (HTB-186™ and HTB-185™), cultured in MEM and Improved MEM Zn++ Option (Richter's Modification) (10373017, Invitrogen) respectively. The ATCC-obtained adherent GBM cell line U87 was cultured in MEM media. Cell culture media were all supplemented with 10% FBS unless otherwise noted. The glioblastoma-derived neurosphere line HSR-GBM1 was propagated in serum-free media as previously described in reference 54. The glioblastoma-derived low passage neurospheres JHH-GBM14 originated from a tumor resected at Johns Hopkins Hospital, and was propagated the same way.
Cell proliferation assay.
Approximately 2 × 103 viable cells were seeded in each well of a 96-well plate and allowed to attach overnight in their respective culturing media. Wells were washed with PBS and replaced with media containing 2% FBS (for DAOY cells) and various concentrations of nanocurcumin. On the day of assay, fresh media was replaced in each assayed well. Cell mass was measured using CellTiter (MTS) assays according to manufacturer's instructions (G3580, Promega). For neurospheres, the overnight attachment step and replacement with fresh media before each reading step were omitted.
Apoptosis assay.
Cells were plated in 12-well plates at 6 × 104 cells per well (and allowed to attach overnight for adherent cells) and treated with void NVA622 nanoparticle or nanocurcumin for 24 hours unless otherwise stated. Apoptotic cell induction was evaluated using the Guava Annexin V assay kit (4500-0455, Millipore) according to the manufacturer's protocol. Quantification of apoptotic and dead cells were done using the GUAVA-PCA flow cytometry system. Each experiment represents a minimum of 2 × 103 gated events from each of three wells. Apoptotic fractions were assigned using the Guava software.
Cell cycle analysis.
Cells were plated in six-well plates at 1.5 × 105 cells per well to attach overnight and treated with void NVA622 nanoparticle or nanocurcumin for 24 hours unless otherwise stated. Cells were then stained with the cell cycle reagent (4500-0220, Millipore) and assessed on the Guava PCA following the manufacturer's protocol. Each experiment represents a minimum of 5 × 103 gated events from each of three wells. Cell cycle fractions were assigned using the Guava software.
Clonogenic assay.
We assessed anchorage-independent tumor growth potential with clonogenic assay using soft agar. Briefly, the base layer consists of serum-supplemented media in 1% agar (18300012, Invitrogen) containing void NVA622 nanoparticle, 5 or 10 µM nanocurcumin in each well of a 6-well plate. The cell layer contains 1 × 104 cells mixed with serum-supplemented media in 0.5% agar on top of the base layer. After 3–4 weeks, the cells were stained with p-Nitro blue tetrazolium chloride (19535, USB Corporation) and the numbers of colonies larger than 50 µM in three high-powered fields per well were determined by use of computer-assisted image analysis with the MCID Elite software. Each experiment was done at least twice in triplicates.
Flow cytometry analysis.
Flow cytometric analysis of CD133 was done with antibodies from Miltenyi Biotec according to manufacturer's instructions using a FACScan machine (BD). In brief, cells were treated for 1 (DAOY, D283Med) or 2 (HSR-GBM1, JHH-GBM14) days with void NVA622 nanoparticle or nanocurcumin and harvested. Cells were blocked with FcR blocking reagent (130-059-901) and incubated with CD133/1 (AC133)-phycoerythrin antibody (130-080-801) in the dark at 4°C. The cells were then washed and resuspended in PBS containing 0.5% BSA and 2 mM EDTA. Cells expressing levels of CD133 higher than those seen in unconjugated CD133 IgG controls (130-090-422) were considered positive.
Quantitative PCR analysis.
RNA was extracted with Qiagen's RNeasy kit. RNA levels were assayed by real-time PCR analysis performed in triplicate with SYBR Green reagent (4364346, Applied Biosystems) according to the manufacturer's instructions on an I-Cycler IQ real-time detection system (Bio-Rad), with all reactions normalized to GAPDH. Primer sequences are in the Supplemental Materials and Methods.
Protein analysis.
For IGF-1R western blot analysis, cells were treated with nanocurcumin in serum-free media for 26 hr and stimulated with 50 ng/ml IGF-1 (I3769, Sigma) for 5 minutes before extracting proteins. In other western analyses, cells were cultured in serum-free media during nanocurcumin treatment. Proteins were extracted in a buffer containing 50 mM Tris, 150 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM PMSF, 10 mM NaF, 2 mM Na3VO4 and complete protease inhibitors (11697498001, Roche). Western blots containing at least 20 µg total protein per lane on a NuPAGE 4–10% Bis-Tris gel (NP0321BOX) were electrophoresed in 1× NuPAGE MOPS SDS running buffer (NP0001) on XCell SureLock mini gel apparatus (all from Invitrogen). Proteins were then transferred to Biorad's Immun-blot PVDF membrane (162-0177) in 1× NuPAGE transfer buffer in X Cell II Blot module (Invitrogen). Membranes were blocked for 1 hour at room temperature in 5% non-fat milk or 5% BSA (for phospho-antibodies) and incubated at 4°C overnight in primary antibodies containing 5% non-fat milk or 5% BSA (for phospho-antibodies). Mouse anti-rabbit GAPDH antibodies were purchased from Research Diagnostics (RDI-TRK5G4-6C5). Akt (9272), STAT3 (9132), Phospho-Akt (Ser 473) (4060), phospho-IGF-IRβ (Tyr 1135/1136) (3024), Phospho-STAT3 (Tyr 705 and Ser 727) antibodies were all purchased from Cell Signaling Technology. IGF-IRβ antibodies were purchased from Santa Cruz (sc-713). Bcl2 antibodies were from Calbiochem (OP60). Secondary antibodies from KPL (peroxidase-conjugated goat anti-mouse [074-1806] or rabbit IgG 074-1506])were diluted 1:5,000 in blocking solution. Blots were developed with enhanced chemiluminescence reagent (NEL103001EA, PerkinElmer).
Statistics.
Cell proliferation, apoptosis, cell cycle, clonogenic and quantitative PCR assays were performed in triplicate, and repeated at least twice unless otherwise noted. As JHH-GBM14 is a low passage line, experiments with these cells were done one time in triplicate (other than the MTS assay, which was performed twice). The software GraphPad Prism was used for all statistical analyses. Mean values ± SEM for representative experiments are shown. Statistical differences were determined by Student's two-tailed t-test. A p value of < 0.05 was considered significant. Bands visualized via western blot were subjected to band densitometry analysis with ImageJ software (National Institutes of Health). The amount of specific signal for each protein was corrected for sample loading and represented as a value compared to vehicle. Normalized intensities from two experiments are depicted as histograms.
Acknowledgements
We thank Karisa Schreck for helpful discussions, and Angelo Vescovi and Francesco DiMeco for their kind gift of HSR-GBM1 cells. NanoCurc™ development and synthesis supported by R01CA113669 (A.M.); R01CA13767 (A.M.); P01CA134292 (A.M.); Flight Attendants Medical Research Institute (FAMRI) (A.M.); and SignPath Pharmaceuticals Inc. (A.M.). Brain tumor assays are supported by R21 AT003893 (C.G.E.).
Abbreviations
- Bcl2
B cell lymphoma 2 associated oncogene
- IGF
insulin-like growth factor
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- Gli
glioma-associated oncogene
- MAPK
mitogen-activated protein kinase
- NVA622
N-isopropylacrylamide (NIPAAM), vinylpyrrolidone (VP) and acrylic acid (AA), in a molar ratio of 60:20:20
- Ptch
patched homolog
- PI3K
phosphoinositide-3-kinase
- STAT
signal transducer and activator of transcription
Conflicts of Interest
NanoCurc™ is a registered trademark of SignPath Pharmaceuticals Inc., Quakerstown, PA. Dr. Maitra is a member of the scientific advisory board of SignPath Pharma Inc. and any conflicts of interest under this arrangement are handled in accordance with the Johns Hopkins University Office of Policy Coordination (OPC) guidelines.
Supplementary Material
References
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. WHO classification of tumours of the central nervous system. Lyon: IARC; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bengmark S, Mesa MD, Gil A. Plant-derived health: the effects of turmeric and curcuminoids. Nutr Hosp. 2009;24:273–281. [PubMed] [Google Scholar]
- 3.Hatcher H, Planalp R, Cho J, Torti FM, Torti SV. Curcumin: from ancient medicine to current clinical trials. Cell Mol Life Sci. 2008;65:1631–1652. doi: 10.1007/s00018-008-7452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aggarwal BB, Sundaram C, Malani N, Ichikawa H. Curcumin: the Indian solid gold. Adv Exp Med Biol. 2007;595:1–75. doi: 10.1007/978-0-387-46401-5_1. [DOI] [PubMed] [Google Scholar]
- 5.Aggarwal BB, Kumar A, Bharti AC. Anticancer potential of curcumin: preclinical and clinical studies. Anticancer Res. 2003;23:363–398. [PubMed] [Google Scholar]
- 6.Kunnumakkara AB, Anand P, Aggarwal BB. Curcumin inhibits proliferation, invasion, angiogenesis and metastasis of different cancers through interaction with multiple cell signaling proteins. Cancer Lett. 2008;269:199–225. doi: 10.1016/j.canlet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Maheshwari RK, Singh AK, Gaddipati J, Srimal RC. Multiple biological activities of curcumin: a short review. Life Sci. 2006;78:2081–2087. doi: 10.1016/j.lfs.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Ravindran J, Prasad S, Aggarwal BB. Curcumin and cancer cells: how many ways can curry kill tumor cells selectively? AAPS J. 2009;11:495–510. doi: 10.1208/s12248-009-9128-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atkinson GP, Nozell SE, Benveniste ET. NFkappaB and STAT3 signaling in glioma: targets for future therapies. Expert Rev Neurother. 2010;10:575–586. doi: 10.1586/ern.10.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt AL, Brunetto AL, Schwartsmann G, Roesler R, Abujamra AL. Recent therapeutic advances for treating medulloblastoma: focus on new molecular targets. CNS Neurol Disord Drug Targets. 2010;9:335–348. doi: 10.2174/187152710791292602. [DOI] [PubMed] [Google Scholar]
- 11.Woo MS, Jung SH, Kim SY, Hyun JW, Ko KH, Kim WK, et al. Curcumin suppresses phorbol ester-induced matrix metalloproteinase-9 expression by inhibiting the PKC to MAPK signaling pathways in human astroglioma cells. Biochem Biophys Res Commun. 2005;335:1017–1025. doi: 10.1016/j.bbrc.2005.07.174. [DOI] [PubMed] [Google Scholar]
- 12.Choi BH, Kim CG, Bae YS, Lim Y, Lee YH, Shin SY. p21 Waf1/Cip1 expression by curcumin in U-87MG human glioma cells: role of early growth response-1 expression. Cancer Res. 2008;68:1369–1377. doi: 10.1158/0008-5472.CAN-07-5222. [DOI] [PubMed] [Google Scholar]
- 13.Aoki H, Takada Y, Kondo S, Sawaya R, Aggarwal BB, Kondo Y. Evidence that curcumin suppresses the growth of malignant gliomas in vitro and in vivo through induction of autophagy: role of Akt and extracellular signal-regulated kinase signaling pathways. Mol Pharmacol. 2007;72:29–39. doi: 10.1124/mol.106.033167. [DOI] [PubMed] [Google Scholar]
- 14.Bangaru ML, Chen S, Woodliff J, Kansra S. Curcumin (diferuloylmethane) induces apoptosis and blocks migration of human medulloblastoma cells. Anticancer Res. 2010;30:499–504. [PubMed] [Google Scholar]
- 15.Dhandapani KM, Mahesh VB, Brann DW. Curcumin suppresses growth and chemoresistance of human glioblastoma cells via AP-1 and NFkappaB transcription factors. J Neurochem. 2007;102:522–538. doi: 10.1111/j.1471-4159.2007.04633.x. [DOI] [PubMed] [Google Scholar]
- 16.Bisht S, Mizuma M, Feldmann G, Ottenhof NA, Hong SM, Pramanik D, et al. Systemic administration of polymeric nanoparticle-encapsulated curcumin (NanoCurc) blocks tumor growth and metastases in preclinical models of pancreatic cancer. Mol Cancer Ther. 2010;9:2255–2264. doi: 10.1158/1535-7163.MCT-10-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang MC, Yang-Yen HF, Yen JJ, Lin JK. Curcumin induces apoptosis in immortalized NIH 3T3 and malignant cancer cell lines. Nutr Cancer. 1996;26:111–120. doi: 10.1080/01635589609514468. [DOI] [PubMed] [Google Scholar]
- 18.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 19.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 20.Lenkiewicz M, Li N, Singh SK. Culture and isolation of brain tumor initiating cells. Curr Protoc Stem Cell Biol. 2009;3:3. doi: 10.1002/9780470151808.sc0303s11. [DOI] [PubMed] [Google Scholar]
- 21.Fan X, Khaki L, Zhu TS, Soules ME, Talsma CE, Gul N, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28:5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beier D, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ, et al. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 23.Joo KM, Kim SY, Jin X, Song SY, Kong DS, Lee JI, et al. Clinical and biological implications of CD133-positive and CD133-negative cells in glioblastomas. Lab Invest. 2008;88:808–815. doi: 10.1038/labinvest.2008.57. [DOI] [PubMed] [Google Scholar]
- 24.Chen R, Nishimura MC, Bumbaca SM, Kharbanda S, Forrest WF, Kasman IM, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17:362–375. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 25.Bar EE, Lin A, Mahairaki V, Matsui W, Eberhart CG. Hypoxia increases the expression of stem-cell markers and promotes clonogenicity in glioblastoma neurospheres. Am J Pathol. 2010;177:1491–1502. doi: 10.2353/ajpath.2010.091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan X, Eberhart CG. Medulloblastoma stem cells. J Clin Oncol. 2008;26:2821–2827. doi: 10.1200/JCO.2007.15.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Del Valle L, Enam S, Lassak A, Wang JY, Croul S, Khalili K, et al. Insulin-like growth factor I receptor activity in human medulloblastomas. Clin Cancer Res. 2002;8:1822–1830. [PubMed] [Google Scholar]
- 28.Sandberg AC, Engberg C, Lake M, von Holst H, Sara VR. The expression of insulin-like growth factor I and insulin-like growth factor II genes in the human fetal and adult brain and in glioma. Neurosci Lett. 1988;93:114–119. doi: 10.1016/0304-3940(88)90022-5. [DOI] [PubMed] [Google Scholar]
- 29.Glick RP, Unterman TG, Blaydes L, Hollis R. Insulin-like growth factors in central nervous system tumors. Ann NY Acad Sci. 1993;692:223–229. doi: 10.1111/j.1749-6632.1993.tb26220.x. [DOI] [PubMed] [Google Scholar]
- 30.Glick RP, Gettleman R, Patel K, Lakshman R, Tsibris JC. Insulin and insulin-like growth factor I in brain tumors: binding and in vitro effects. Neurosurgery. 1989;24:791–797. doi: 10.1227/00006123-198906000-00001. [DOI] [PubMed] [Google Scholar]
- 31.Yadav A, Kalita A, Dhillon S, Banerjee K. JAK/STAT3 pathway is involved in survival of neurons in response to insulin-like growth factor and negatively regulated by suppressor of cytokine signaling-3. J Biol Chem. 2005;280:31830–31840. doi: 10.1074/jbc.M501316200. [DOI] [PubMed] [Google Scholar]
- 32.Zong CS, Chan J, Levy DE, Horvath C, Sadowski HB, Wang LH. Mechanism of STAT3 activation by insulin-like growth factor I receptor. J Biol Chem. 2000;275:15099–15105. doi: 10.1074/jbc.M000089200. [DOI] [PubMed] [Google Scholar]
- 33.de la Iglesia N, Puram SV, Bonni A. STAT3 regulation of glioblastoma pathogenesis. Curr Mol Med. 2009;9:580–590. doi: 10.2174/156652409788488739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherry MM, Reeves A, Wu JK, Cochran BH. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27:2383–2392. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dahmane N, Sanchez P, Gitton Y, Palma V, Sun T, Beyna M, et al. The Sonic Hedgehog-Gli pathway regulates dorsal brain growth and tumorigenesis. Development. 2001;128:5201–5212. doi: 10.1242/dev.128.24.5201. [DOI] [PubMed] [Google Scholar]
- 36.Fan X, Matsui W, Khaki L, Stearns D, Chun J, Li YM, et al. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 2006;66:7445–7452. doi: 10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- 37.Shahi MH, Lorente A, Castresana JS. Hedgehog signalling in medulloblastoma, glioblastoma and neuroblastoma. Oncol Rep. 2008;19:681–688. [PubMed] [Google Scholar]
- 38.Bar EE, Chaudhry A, Lin A, Fan X, Schreck K, Matsui W, et al. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells. 2007;25:2524–2533. doi: 10.1634/stemcells.2007-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bar EE, Chaudhry A, Farah MH, Eberhart CG. Hedgehog signaling promotes medulloblastoma survival via Bc/II. Am J Pathol. 2007;170:347–355. doi: 10.2353/ajpath.2007.060066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Shu W, Chen W, Wu Q, Liu H, Cui G. Curcumin, both histone deacetylase and p300/CBP-specific inhibitor, represses the activity of nuclear factorkappaB and Notch 1 in Raji cells. Basic Clin Pharmacol Toxicol. 2007;101:427–433. doi: 10.1111/j.1742-7843.2007.00142.x. [DOI] [PubMed] [Google Scholar]
- 41.Wang Z, Zhang Y, Banerjee S, Li Y, Sarkar FH. Notch-1 downregulation by curcumin is associated with the inhibition of cell growth and the induction of apoptosis in pancreatic cancer cells. Cancer. 2006;106:2503–2513. doi: 10.1002/cncr.21904. [DOI] [PubMed] [Google Scholar]
- 42.Elamin MH, Shinwari Z, Hendrayani SF, Al-Hindi H, Al-Shail E, Khafaga Y, et al. Curcumin inhibits the Sonic Hedgehog signaling pathway and triggers apoptosis in medulloblastoma cells. Mol Carcinog. 2010;49:302–314. doi: 10.1002/mc.20604. [DOI] [PubMed] [Google Scholar]
- 43.Fong D, Yeh A, Naftalovich R, Choi TH, Chan MM. Curcumin inhibits the side population (SP) phenotype of the rat C6 glioma cell line: towards targeting of cancer stem cells with phytochemicals. Cancer Lett. 2010;293:65–72. doi: 10.1016/j.canlet.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia Y, Jin L, Zhang B, Xue H, Li Q, Xu Y. The potentiation of curcumin on insulin-like growth factor-1 action in MCF-7 human breast carcinoma cells. Life Sci. 2007;80:2161–2169. doi: 10.1016/j.lfs.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 45.Patti R, Reddy CD, Geoerger B, Grotzer MA, Raghunath M, Sutton LN, et al. Autocrine secreted insulin-like growth factor-I stimulates MAP kinase-dependent mitogenic effects in human primitive neuroectodermal tumor/medulloblastoma. Int J Oncol. 2000;16:577–584. doi: 10.3892/ijo.16.3.577. [DOI] [PubMed] [Google Scholar]
- 46.Wang JY, Del Valle L, Gordon J, Rubini M, Romano G, Croul S, et al. Activation of the IGF-IR system contributes to malignant growth of human and mouse medulloblastomas. Oncogene. 2001;20:3857–3868. doi: 10.1038/sj.onc.1204532. [DOI] [PubMed] [Google Scholar]
- 47.Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294:2542–2545. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- 48.Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW, et al. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–826. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- 49.Zhong Z, Wen Z, Darnell J., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 50.Holland EC, Celestino J, Dai C, Schaefer L, Sawaya RE, Fuller GN. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet. 2000;25:55–57. doi: 10.1038/75596. [DOI] [PubMed] [Google Scholar]
- 51.Uhrbom L, Dai C, Celestino JC, Rosenblum MK, Fuller GN, Holland EC. Ink4a-Arf loss cooperates with KRas activation in astrocytes and neural progenitors to generate glioblastomas of various morphologies depending on activated Akt. Cancer Res. 2002;62:5551–5558. [PubMed] [Google Scholar]
- 52.Clement V, Sanchez P, de Tribolet N, Radovanovic I. Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal and tumorigenicity. Curr Biol. 2007;17:165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bisht S, Feldmann G, Soni S, Ravi R, Karikar C, Maitra A. Polymeric nanoparticle-encapsulated curcumin (“nanocurcumin”): a novel strategy for human cancer therapy. J Nanobiotechnology. 2007;5:3. doi: 10.1186/1477-3155-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.