Abstract
AMPK has been termed the fuel sensor of mammalian cells because it directly responds to the depletion of the fuel molecule ATP. In previous work, we found that AMPK is strongly activated by tumor-like hypoxia and glucose deprivation, independently of the oxygen response system associated with HIF-1. We also observed high levels of AMPK activity in tumor cells in vivo, using different model tumors. These findings suggested the hypothesis that modulation of AMPK activity could have therapeutic value for the treatment of solid tumors. To investigate this hypothesis, we have been conducting a SAR study of potential small-molecule modulators of AMPK activity. Here we report that the chemotherapeutic drug SU11248 (sunitinib) is at least as potent an inhibitor of AMPK as compound C, which is a commonly used experimental direct inhibitor of the enzyme. We also provide a computational model of the binding pose of SU11248 to an AMPKα subunit, which suggests a structural basis for the affinity of the drug for the ATP site of the catalytic domain. The ability of SU11248 to inhibit AMPK has potential clinical significance—there may be populations of SU11248-treated patients in which AMPK activity is inhibited in normal as well as in tumor tissue.
Key words: AMPK, compound C, SU11248, sunitinib, type II inhibitor
Introduction
AMPK is a ubiquitous sensor of energy status in mammalian and other eukaryotic cells—in response to metabolic (energy) stress, AMPK acts to maintain or restore cellular energy homeostasis by inhibiting ATP-consuming processes and stimulating ATP-generating processes (reviewed in refs. 1 and 2). This physiological model of AMPK function has generated interest in manipulating AMPK activity for therapeutic purposes. For example, activation of AMPK might be useful to manage pathologies associated with deregulated energy metabolism such as type II diabetes and metabolic syndrome.3,4 AMPK has also been implicated in the process of tumorigenesis, although whether AMPK activity inhibits or contributes to tumor development seems to depend on context.5,6 In previous research, we found that AMPK is activated by tumor-like low oxygen (hypoxia) and low glucose conditions both in vitro and in experimental tumors.7,8 These findings suggested that AMPK activity contributes to tumor cell adaptation to energy stress, such as that resulting from severe hypoxia and glucose deprivation.9,10 To investigate the potential translational significance of this hypothesis, we have focused on developing direct small-molecule modulators (inhibitors and activators) of AMPKα, the catalytic subunit of the heterotrimeric AMPK holoenzyme.2 While constructing a SAR for potential kinase domain inhibitors of AMPKα, we observed that SU11248 (sunitinib)—a selective PTK inhibitor currently in clinical use11,12—is a direct AMPK inhibitor. SU11248 was included in the SAR study because of its calculated pharmacological properties and chemotype as an ATP-competitive inhibitor,13 both of which conformed to our selection criteria for a first set of potential AMPK inhibitors. SU12248 is also of interest in the context of AMPK inhibition because compound C, which is an established and commonly used experimental direct AMPK inhibitor,14,15 has overlapping PTK inhibitory activity with SU11248.16,17 In this report, we demonstrate that SU11248 is a more potent inhibitor of AMPK activity than compound C, and that SU11248 strongly inhibits cellular AMPK activity. Finally, we provide an energy-minimized molecular model of human AMPKα2 containing SU11248, which suggests how the drug could act as an ATP-competitive inhibitor of the AMPK holoenzyme.
Results
SU11248 is a direct inhibitor of AMPK.
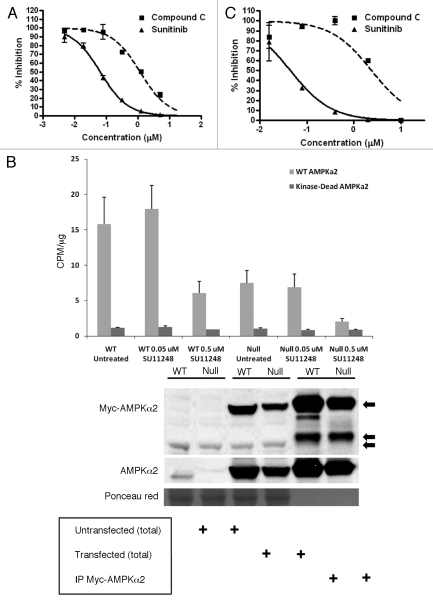
Figure 1A shows an example of dose-response curves from a TR-FRET kinase assay in which purified rat liver AMPK preparations (holoenzymes2) were exposed to compound C and SU11248: both compounds strongly inhibited AMPK activity relative to the control (vehicle-treated AMPK preparations). Sorafenib, a protein kinase inhibitor having overlapping activity with that of SU11248,11 had no activity toward AMPK in this assay, even at a concentration of 40 µM (result not shown). Table 1 presents the IC50 values from multiple independent TR-FRET kinase assays for the inhibition of rat liver AMPK by compound C and SU11248. Figure 1B confirms the specificity of SU11248 for rat AMPK by showing the results of immune complex kinase assays in which epitope (Myc) tagged normal or kinase-dead rat AMPKα2 was immunoprecipitated from lysates of transiently transfected WT and AMPK null MEFS and then exposed to SU11248. Kinase-dead AMPKα2 was used in this assay as a negative control for AMPK activity and as a control for nonspecific protein kinase activity toward the model AMPK substrate. Figure 1B shows that SU11248 strongly inhibited rat AMPKα2 activity at a concentration of 0.5 µM using Myc-tagged AMPK complexes isolated from either WT or AMPK null MEFs, which lack endogenous AMPK activity.7 Relevant values of Myc-tagged AMPK activity (recorded as CPM/µg [95% CI]; see Materials and Methods for details) from Figure 1B are the following: WT Untreated, 15.8 (4.3); WT 0.5 µM SU11248, 6.1 (1.8); Null Untreated, 7.5 (2.0); Null 0.5 µM SU11248, 0.9 (0.07). Figure 1B also shows a representative AMPKα2 immunoblot of total protein and WT Myc-tagged AMPKα2 immunoprecipitates from transfected WT and AMPK null cells. Consistent with the results of the immune complex kinase assays, the level of Myc-tagged AMPKα2 was higher in the WT compared with the AMPK null cells (and substantially higher than the level of endogenous AMPKα2 in the WT cells).
Figure 1.
Direct effects of SU11248 and compound C on rat and human AMPK activity in cell-free assays. (A) Representative TR-FRET kinase assays for purified rat AMPK exposed to various concentrations of compound C (squares) or SU11248 (triangles). Error bars are standard deviations (S.D.) for samples from independent experiments (n = 4); concentrations are indicated on a logarithmic scale. (B) Top: results of immune complex kinase assays in which immunoprecipitates of Myc-tagged rat AMPKα2 (WT and kinase-dead AMPKα2) were untreated or exposed to the indicated concentrations of SU11248 in the presence of the AMARA peptide substrate. Error bars are S.D. for triplicate samples. Bottom: immunoblots of total cellular protein (total) and WT Myc-tagged AMPKα2 immunoprecipitates from transiently transfected WT and AMPK null cells.7 Replicate blots were probed for the relative levels of Myc tag or AMPKα2: top arrow, Myc-tagged AMPKα2; middle arrow, IP antibody heavy chain (−50 kD); bottom arrow, nonspecific protein band in the total protein lysates. The replicate AMPKα2 immunoblot was stained with Ponceau red as a loading control for the total lysates. (C) Results of immune complex kinase assays in which immunoprecipitated human AMPKα1 complexes were exposed to various concentrations of compound C (squares) or SU11248 (triangles) in the presence of the AMARA peptide. Error bars are S.D. for triplicate samples; concentrations are indicated on a logarithmic scale. See Materials and Methods for details.
Table 1.
IC50 values for inhibition of AMPK by SU11248 and compound C in cell-free kinase assays
| aRat AMPK (µM ± S.D.) | bHuman AMPKα1 µM (95% CI) | |
| SU11248 | 0.062 ± 0.029 | 0.045 (0.027 to 0.075) |
| Compound C | 1.28 ± 0.69 | 2.38 (1.28 to 4.44) |
TR-FRET kinase assays, n = 4; p = 0.013.
Immune complex kinase assays, triplicate results (CI, 95% confidence interval).
To investigate the effect of SU11248 and compound C on human AMPK activity, we used PC3 human prostate cancer cells to immunoprecipitate AMPK complexes for cell-free kinase assays. This choice of cell line was based on a recent report in which a panel of human prostate cancer cell lines including PC3 cells was used to investigate the effect of AMPK inhibition on tumor cell proliferation and survival in vitro.18 Figure 1C shows the results of immune complex kinase assays of the effect of compound C and SU11248 on the basal activity of human AMPKα1-containing complexes, which are the most abundant form of AMPK present in these cells (at least 10-fold more abundant than AMPKα2; result not shown). The IC50 values for the inhibition of human AMPKα1 by compound C and SU11248 are also given in Table 1.
In summary, we found that SU11248 is substantially more potent than compound C as an inhibitor of rat AMPK in cell-free kinase assays. In addition, we found that SU11248 is at least as potent as compound C as an inhibitor of human AMPK—strictly, immunoprecipitable AMPKα1-containing AMPK—in cell-free kinase assays. It must be emphasized that compared with AMPK, it is well documented that SU11248 has greater potency as a direct inhibitor of specific receptor PTKs, including VEGF receptors 1–3 and the KIT receptor (e.g., consensus IC50 values of 15–30 nM and 1–10 nM, respectively19).
SU11248 inhibits AMPK activity in cultured cells.
AMPK activity is routinely monitored in cell cultures by using immunoblotting to detect specific phosphorylation of the metabolic enzyme ACC, which defines ubiquitous enzymes for long-chain fatty acid synthesis (ACC1) and oxidation (ACC2)2,20—AMPK uniquely phosphorylates ACC1 on Ser79 in rodent and human cells.7,20 It should be noted that micromolar concentrations of the AMPK inhibitor compound C are typically necessary to achieve complete inhibition of AMPK in cultured cells.15 For this reason, we used micromolar concentrations of compound C and SU11248 to investigate the effects of these compounds on cellular AMPK activity.
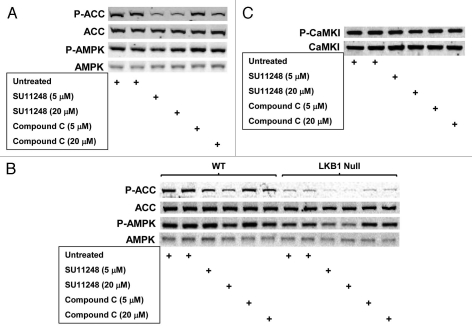
Figure 2A compares the effect of equimolar SU11248 and compound C on basal AMPK activity (detected by immunoblotting for ACC1 Ser79 phosphorylation; P-ACC) in WT MEFs: both compounds inhibited cellular AMPK activity (P-ACC levels), but SU11248 was a more potent AMPK inhibitor than compound C (e.g., 5 µM, 6 h). These findings for the inhibition of cellular AMPK by SU11248 and compound C are consistent with the results shown in Table 1 for the direct inhibition of purified AMPK by these compounds in cell-free assays—together, these two studies indicate that SU11248 directly inhibits AMPK in MEFs. In this connection, Figure 2A also shows that exposure of MEFs to SU11248 did not significantly alter basal phosphorylation of AMPK on Thr172 (P-AMPK), which is the site for AMPK phosphorylation by AMPK kinases (AMPKKs) in the activation loop of the kinase domain of the mouse or rat a subunit.1,2 The insensitivity to SU11248 of basal P-AMPK levels detected using lysates of treated MEFs is consistent with a direct mechanism of AMPK inhibition—there was no evidence of an inhibitory effect of SU11248 on AMPKK activity that could, in principle, attenuate basal AMPK activity. In support of this conclusion, Figure 2B shows an identical experiment using WT MEFs and genetically matched cells lacking expression of LKB1, which is a prominent AMPKK (reviewed in ref. 6). Here, the inhibitory effect of SU11248 on P-ACC levels detected using lysates of the paired WT and LKB1 null cells was essentially identical to the inhibitory effect of the drug on P-ACC levels in the different WT cells7 shown in Figure 2A (e.g., 5 µM, 6 h). The relatively higher P-ACC levels found on immunoblots of WT compared with LKB1 null cells (Fig. 2B)—which both showed the same overall pattern of P-ACC levels in response to SU11248 exposure—can be attributed to alternative AMPKK activity in the LKB1 null cells, such as CaMKKβ.1 Figure 2C shows that these WT cells do express CaMKKβ activity, as detected by immunoblotting for CaMKI Thr177 phosphorylation (P-CaMKI);21–24 however, there was no obvious effect of either SU11248 or compound C on the relative levels of P-CaMKI in these cells. Considering that there are at least two other AMPKKs (CaMKKα, TAK1,1), it is not possible at this time to exclude an effect of the compounds on AMPKK activity in general. Finally, Figure 2B shows that compound C, like SU11248, did not significantly alter basal P-AMPK levels detected using lysates of treated cells.
Figure 2.
Effects of SU11248 and compound C on AMPK-dependent ACC phosphorylation (P-ACC) and activating AMPK phosphorylation (P-AMPK) in WT MEFs, AMPK null MEFS, and LKB1 null MEFs cultured in complete medium. (A) Immunoblots of total protein from WT MEFs harvested following exposure to SU11248 (5 or 20 µM) or compound C (5 or 20 µM) for 6 h. (B) Immunoblots of total protein from WT and LKB1 null MEFs harvested following exposure to SU11248 (5 or 20 µM) or compound C (5 or 20 µM) for 6 h. Replicate blots were probed for the relative levels of P-ACC, total ACC, P-AMPK, or total AMPK. (C) Immunoblots of total protein from WT MEFs from (B) harvested following exposure to SU11248 (5 or 20 µM) or compound C (5 or 20 µM) for 6 h. Replicate blots were probed for the relative levels of P-CaMKI or total CaMKI.
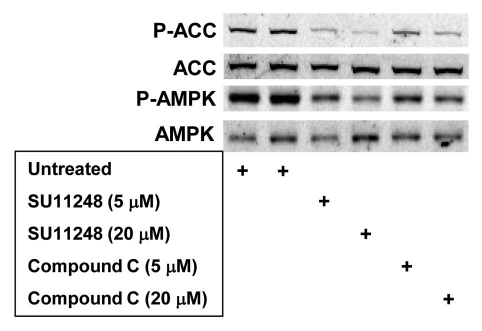
To investigate the effect of SU11248 and compound C on AMPK activity in human tumor cells, we again used PC3 human prostate cancer cells. Figure 3 shows that both SU11248 and compound C strongly inhibited basal AMPK activity (P-ACC levels) in PC3 cells. In this case, there seemed to be a small inhibitory effect of compound treatment on basal AMPK activation (P-AMPK levels) relative to basal activation in untreated cells. However, the pattern of P-ACC levels detected using lysates of PC3 cells exposed to SU11248 or compound C was essentially identical to the pattern detected using lysates of MEFs exposed to these compounds (Fig. 2).
Figure 3.
Effects of SU11248 and compound C on AMPK-dependent ACC phosphorylation (P-ACC) and activating AMPK phosphorylation (P-AMPK) in PC3 human prostate cancer cells cultured in complete medium. Immunoblots of total protein from PC3 cells harvested following exposure to SU11248 (5 or 20 µM) or compound C (5 or 20 µM) for 6 h. Replicate blots in were probed for the relative levels of P-ACC, total ACC, P-AMPK or total AMPK.
In summary, the results shown in Figures 2 and 3 demonstrate that SU11248, which directly inhibits AMPK in cell-free kinase assays (Fig. 1 and Table 1), also inhibits endogenous AMPK activity in cultured mouse and human cell lines.
Modeling of SU11248 bound to the kinase domain of AMPKα2 shows a conformational preference.
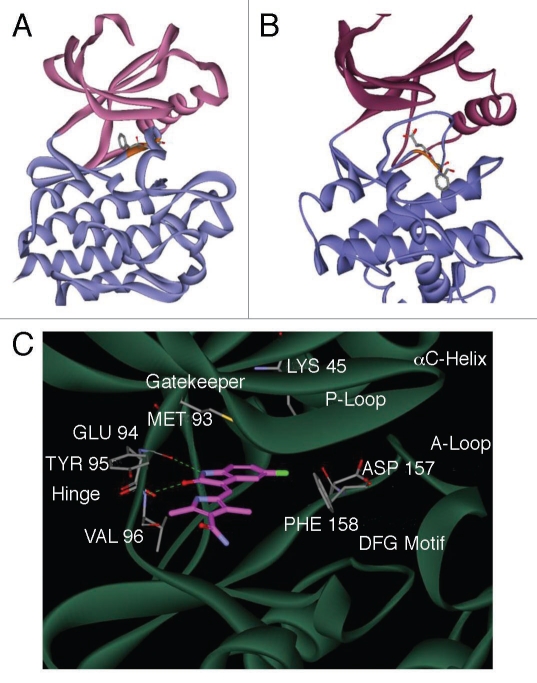
SU11248 was unexpectedly found by X-ray crystallography to have a preference for the ‘out’ conformation of the DFG loop (DFG-out25,26) of the KIT human receptor tyrosine kinase.27 As described in Materials and Methods, we used this structure—the kinase domain of KIT (PDB ID 3G0E) co-crystallized with SU11248 in the ATP binding site—as an experimentally validated model for the binding mode of SU11248 to a protein kinase. The DFG-out conformation of the kinase domain in the KIT:SU11248 structure is also the conformation of the DFG loop in the publicly available crystal structure of the human AMPK α2 subunit (PDB ID 2H6D). The presence of the DFG-out conformation in the crystal structure of AMPKα2 suggested the hypothesis that this conformation of the α2 apoprotein is accessible to binding by SU11248 (Fig. 4). In the DFG-out conformation of the model of AMPKα2:SU11248 (Fig. 4C), the side chain of the DFG loop phenylalanine is in close contact with the hydrophobic surface of SU11248 formed by the 3-methyl group of the 1H-pyrrole and the methylene linker between the indolin-2-one core and the pyrrole. Moreover, the fluorine of SU11248 is in close contact with the β-carbon atom of the DFG loop phenylalanine side chain, forming van der Waals contacts that favor binding. In contrast, in the DFG-in conformation, the DFG loop is moved further toward the allosteric pocket,25,28 resulting in the energetically unfavorable exposure of the hydrophobic surface of SU11248 to solvent. Consistent with these structural considerations, optimization of the binding pose of SU11248 in the kinase domain of AMPKα2 by energy minimization generated a stable model with SU11248 bound to the DFG-out (Fig. 4A) but not the DFG-in conformation (Fig. 4B). Small-molecule ligands that stabilize the DFG-out conformation of a protein kinase—which is considered a catalytically inactive state—are called type II inhibitors.25,26
Figure 4.
An energy-minimized computational model of SU11248 bound to the kinase domain of human AMPKα2. Ribbon diagram models of the inactive DFG-out (A) and active DFG-in (B) conformations of the catalytic domain of AMPKα2, showing the N-terminal (pink) and C-terminal (blue) lobes of the subunit. The conserved DFG loop is depicted (orange) with the side chains for the DFG aspartic acid and phenylalanine residues represented as sticks. A detailed view of the active site of AMPKα2:SU11248 (C), showing key hydrogen bonds between SU11248 (magenta carbons) and protein residues (gray carbons) of the ATP-binding hinge region (dashed green lines).
Discussion
SU11248 is currently approved for treating two human cancers, advanced renal cell carcinoma (RCC) and imatinib-resistant gastrointestinal stromal tumor (GIST), and is also under clinical investigation for other tumor types.11,12,29 Although SU11248 was originally developed as a selective inhibitor of receptor PTKs (e.g., VEGF receptor 1-3, KIT receptor),19 it is considered a multi-targeted protein kinase inhibitor with various other determined or suspected targets.30,31 The major finding reported here is that SU11248 is a potent inhibitor of the serine/threonine protein kinase AMPK—directly, as determined by cell-free (in vitro) assays of purified rat AMPK, immunoprecipitated epitope-tagged rat AMPKα2-containing AMPK, and immunoprecipitated human AMPKα1-containing AMPK (Fig. 1); and in cultures of mouse and human cell lines (Figs. 2 and 3). We also demonstrated that SU11248 is at least as potent an inhibitor of rat and human AMPK as compound C (Figs. 1–3), which is a widely used experimental ATP-competitive AMPK inhibitor.14,15 Our computational model of SU11248 bound to human AMPKα2 (Fig. 4) provides a structural rationale for these experimental findings by predicting that SU11248 can bind to an AMPKα kinase domain as a type II protein kinase inhibitor, thus stabilizing the catalytically inactive DFG-out conformation of the protein.25,26 SU11248 was previously reported to inhibit AMPK in cultures of rat myocytes, although direct inhibition was demonstrated only in cell-free assays using recombinant human AMPKα1:β1:γ2.32 Our experimental findings demonstrating the ability of SU11248 to inhibit human AMPK (Figs. 1 and 3) has potential clinical significance—there may be a substantial population of SU11248-treated patients11,12,29 in which AMPK activity is inhibited in both normal and tumor cells. Considering the physiological model of AMPK function as a regulator of cellular energy (ATP) status,1,2 it is conceivable that SU11248 could inhibit stress-inducible AMPK activity within hypoxic or glucose-deprived solid tumor microenvironments.7
Materials and Methods
Materials.
The following antibodies were obtained from Cell Signaling Technology (Danvers, IL): rabbit polyclonal anti-phospho-acetyl-CoA carboxylase (Ser79) antibody (Cat. No. 3661; P-ACC hereafter); rabbit polyclonal anti-ACC antibody (Cat. No. 3662); and rabbit monoclonal anti-phospho-AMPKα (Thr172) antibody (Cat. No. 2535; P-AMPK hereafter). The following antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA): goat polyclonal-anti-AMPKα2 antibody (Cat. No. sc-19131); rabbit polyclonal-anti-phospho-CaMKI (Thr177) antibody (Cat. No. sc-28438-R); and goat polyclonal-anti-CaMKI antibody (Cat. No. sc-1543). An agarose-bead-conjugated rabbit anti-AMPKα1 antibody was obtained from Bethyl Laboratories, Inc., (Montgomery, TX; Cat. No. S300-507). SU11248 (N-[2-(diethylamino) ethyl]-5-[(Z)-(5-fluoro-1,2-dihydro-2-oxo-3H-indol-3-ylidine) methyl]-2,4-dimethyl-1H-pyrrole-3-carboxamide) and sorafenib (4-[4-[[4-chloro-3-(trifluoromethyl)phenyl]-carbamoylamino]-phenoxy]-N-methyl-pyridine-2-carboxamide) were obtained from LC Laboratories (Woburn, MA; malate salt, Cat. No. S-8803; p-toluenesulfonate salt, Cat. No. S-8502, respectively); compound C ([4-(2-piperidin-1-yl-ethoxy)-phenyl]-3-pyridin-4-yl-pyrrazolo[1,5-a]-pyrimidine) was obtained from Calbiochem (Carlsbad, CA; Cat. No. 171260). The AMARA peptide was obtained from AnaSpec (San Jose, CA). Mammalian expression constructs that constitutively express a Myc-tagged fusion protein of normal AMPKα2 (rat AMPKα2) or kinase-dead AMPKα2 (rat AMPKα2 K45R) were obtained from Dr. Morris Birnbaum (University of Pennsylvania School of Medicine).33,34
Cell culture.
The generation of wild-type (WT) and AMPKα-/- (AMPK null) mouse embryo fibroblasts (MEFs) immortalized with SV40 large T antigen (TAg) has been described in detail elsewhere.7 WT and LKB1-/-(LKB1 null) MEFs were obtained from Dr. Reuben Shaw (Salk Institute, La Jolla, CA); these cells were immortalized with TAg using pBABE puro SV40 LT (Cat. No. 13970, Addgene, Cambridge, MA). PC3 human prostate cancer cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA). WT and AMPK null cells were cultured in DMEM medium (GIBCO Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (Sigma Chemical Co., St. Louis, MO) and 25 mM HEPES buffer (pH 7.4) in a 5% CO2/air atmosphere at 37°C. PC3 cells were cultured similarly in RPMI 1640 medium (GIBCO Invitrogen) containing 10% FBS and 25 mM HEPES buffer (pH 7.4). For immune complex kinase assays, WT MEFs were transiently transfected with Myc-tagged AMPKα2 or AMPKα2 K45R constructs using TurboFectin 8.0 (Origene, Rockville, MD) at a ratio of 1 µg DNA:4 µl transfection reagent. After incubation at 37°C for 24 h, cells were lysed as described below.
Time-resolved fluorescence/förster resonance energy transfer (TR-FRET) kinase assay.
The TR-FRET kinase assay used for this study has two key reagents: a model AMPK peptide substrate conjugated with a FRET acceptor (ULightTM-Acetyl-CoA Carboxylase [Ser79] Peptide, PerkinElmer, Waltham, MA; acceptor emission at 665 nm); and a phosphorylation-specific antibody conjugated with a lanthanide FRET donor (Europium-anti-phospho-Acetyl CoA Carboxylase [Ser79] Antibody, PerkinElmer; donor excitation at 320/340 nm), which recognizes the model AMPK substrate phosphorylated at a consensus sequence. AMPK purified from rat liver (Millipore Upstate, Danvers, MA; Cat. No. 14-305) was used to screen compounds for inhibition of the enzyme. Briefly, inhibition of AMPK activity by a compound decreases the concentration of the FRET acceptor + donor complex comprised of the AMPK-phosphorylated substrate and the phosphorylation-specific antibody, and thus attenuates the FRET-mediated emission signal at 665 nm: the intensity of the signal is proportional to the level of substrate phosphorylation. The TR-FRET kinase assay was performed using an HTS microplate reader (Analyst HT 96.384, Molecular Devices, Sunnyvale, CA). Only compounds that are not intrinsically fluorescent (here, SU11248 and compound C) are evaluated as inhibitors of kinase activity using this assay.
Assays were performed in a 384-well plate in which each test well first received a 4x solution of 800 µM AMP, 40 µM ATP and 200 nM peptide substrate in a reaction buffer (50 mM HEPES, pH 7.5; 1 mM EGTA, 10 mM MgCl2, 0.01% Tween 20, 2 mM DTT). AMPK protein (2.3 × 10−3 units) in reaction buffer was added, and then appropriate wells received various concentrations of test compounds (SU11248, compound C, sorafenib) or vehicle to a final volume of 10 µl. Reactions were allowed to proceed at room temperature for 30 min, stop solution (5 µl; 40 mM EDTA in LANCE Detection Buffer, PerkinElmer) was added, and 5 min later detection antibody (5 µl) in Lance Detection Buffer was added. Plates were kept at room temperature for 60 min, and then emission signals at 665 nm were recorded using the plate reader in its TR-FRET mode.
Immunoblotting analysis.
Immunoblotting protocols have been described in detail in reference.7 Briefly, cells were placed on ice and the medium was removed. Cells were lysed by adding 200 µl of ice cold high salt lysis buffer (LB1; 50 mM Tris-HCl, pH 7.4, 0.5% NP-40, 250 mM NaCl, 1 mM DTT, 50 mM NaF, 15 mM Na4P2O7, 25 mM β-glycerophosphate, 1 mM Na3VO4, 100 nM okadaic acid, 1x Protease Inhibitor Cocktail III, PIC III, Calbiochem, Carlsbad, CA). After spinning at 9,000 xg for 5 min at 4°C, the protein concentrations of the supernatants were determined by using a bicinchoninic acid assay (Pierce Biotechnology, Rockford, IL). Equal protein samples (typically 10–15 µg) were resolved in 4–12% SDS-polyacrylamide gels (Nu-PAGE Bis-Tris Gels, Invitrogen Corp.,) and electroblotted onto Immobilon-FL membranes (Millipore, Billerica, MA). Blots were blocked in a 1:1 mixture of Odyssey Blocking Buffer (LI-COR Biosciences, Lincoln, NE) and PBS (pH 7.4) at 4°C overnight. For protein detection, blots were incubated overnight at 4°C with a primary antibody typically diluted 1:1,000 in Odyssey Blocking Buffer containing 0.1% Tween-20. Blots were then incubated for 1 h at room temperature containing a species-specific IRDye 800-conjugated IgG antibody (LI-COR Biosciences) diluted in Odyssey Blocking Buffer containing 0.1% Tween-20 and 0.01% SDS (e.g., 1:10,000). Primary antibody binding was detected and visualized by using an Odyssey Infrared Imaging System (LI-COR Biosciences) according to the supplier's instructions.
Immune complex kinase assays.
Cells were lysed as described above for immunoblotting analysis, except that 200 µl (MEFs) or 300 µl (PC3 cells) of ice cold low salt lysis buffer (LB2; 50 mM Tris-HCl, pH 7.4, 1% Triton X, 50 mM NaCl, 1 mM DTT, 50 mM NaF, 15 mM Na4P2O7, 25 mM β-glycerophosphate, 1 mM Na3VO4, 100 nM okadaic acid, 1x PIC III) was used. After determining the protein concentrations of the supernatants, 740–800 µg of total protein were used for each immunoprecipitation. To immunoprecipitate endogenous AMPKα complexes, samples received 10 µl of agarose-bead-conjugated AMPKα1 (controls included lysates with no added antibody, and LB2 + conjugated antibody only). To immunoprecipitate Myc-tagged AMPKα2 constructs, samples received 25 µl of Protein A/G Agarose (Santa Cruz Biotechnology, Santa Cruz, CA). Immunoprecipitations were performed by tumbling samples overnight at 4°C. The beads were washed twice with 1 ml of LB2 and twice with 1 ml of kinase buffer (KB; 60 mM HEPES, pH 7.0, 120 mM NaCl, 5 mM MgCl2, 2 mM EGTA, 1 mM DTT, 10 mM β-glycerophosphate). Then, each sample received 50–55 µl of KB containing 100 µM AMARA peptide,35 300 µM AMP, 200 µM ATP and 4–5 µCi [γ-32P]-ATP (250 µCi, 30 Ci/mmol; PerkinElmer). Doses of SU11248 or compound C in dimethyl sulfoxide (DMSO) solution were added to some of the immunocomplex solutions (controls received DMSO only). Kinase reactions were kept at 30°C for 20 min, and stopped by adding 5–6 µl of 0.5 M EDTA, pH 8. After spinning at 15,000 xg for 5 min at 4°C, 20 µl of each supernatant was spotted in triplicate on a phosphocellulose filter (P30 Filtermat; Cat. No. 1205-406, PerkinElmer). Filters were washed three times for 5 min each time with 0.75% phosphoric acid and then washed in acetone and air dried. Radioactivity retained on the filters was measured by using a 1205 Betaplate reader (Perkin Elmer). Measurements of counts/min (CPM) in the AMARA peptide substrate were normalized to total cellular protein (µg); assay results were expressed as % inhibition of compound-treated relative to untreated AMPKα immunoprecipitate (endogenous human AMPK) or CPM/µg (Myc-tagged rat AMPKα2).
Statistical analysis.
Statistical calculations were performed using a GraphPad Prism 4 package (GraphPad Software, Inc.,) or Microsoft Office Excel 2007. Data (mean ± SD) for independent trials were evaluated using an unpaired t test (p < 0.05 was considered a significant difference). Data for replicate trials were evaluated using the 95% confidence interval (95% CI).36
Molecular modeling.
SU11248 is classified as an ATP-competitive protein kinase inhibitor.13 Therefore, to develop a hypothesis for the binding mode of SU11248 to the ATP binding site of an AMPKα subunit, computational models of human AMPKα2 structures containing SU11248 were generated in which the drug was placed in the ATP binding site, using both the DFG-in and DFG-out conformations of the protein25,26 (Fig. 4). The α2 subunit was chosen for this purpose because the only publicly available X-ray crystal structure of a human AMPKα subunit is that of AMPKα2 (PDB ID 2H6D). It is noteworthy that the catalytic domains of human AMPKα2 and −α1 are highly similar at the amino acid (aa) level (e.g., 90.1% identity by sequence alignment; AMPKα1, UniProt KB accession no. Q13131, aa 27–279; AMPKα2, UniProt KB accession no. P54646, aa 16–268); this overall similarity suggests that small-molecule inhibitors of AMPKα2 would also be capable of inhibiting AMPKα1.
Putative AMPKα2 structures without bound SU11248 (α2 apoproteins) were generated by structural superposition. To model the DFG-out conformation of the α2 apoprotein, X-ray crystal structures of the kinase domain of the human receptor tyrosine kinase KIT in complex with SU11248 (PDB ID 3G0E27), and of the human AMPKα2 subunit were used. The DFG loop is in the out position in the AMPKα2 crystal structure; moreover, the side chain conformation of Phe158 in the structure is predicted to occlude the binding of ATP-competitive inhibitors. The crystal structures were superimposed in the Molecular Operating Environment (MOE, v2008.10; Chemical Computing Group, Montreal, Quebec) using the coordinates of the C-α atoms of the backbones, with the secondary structures accented. The sequence alignment generated from the structural superposition was used to model the DFG-out conformation of the α2 apoprotein. Because a substantial portion of the activation loop (A-loop) in the crystal structure of the α2 subunit was not resolved, the model included the A-loop from the KIT structure (Fig. 4A). To model the DFG-in conformation of the α2 apoprotein, X-ray crystal structures of the kinase domain of microtubule affinity regulating kinase-1 (MARK-1) (PDB ID 2HAK) and AMPKα2 were superimposed using the MOE, as described above. MARK-1 and AMPK α2 have ∼50% sequence identity between the two kinase domains. Residues 157–179 from the MARK-1 structure were used to model the DFG-in conformation, as well as the A-loop residues that are missing in the α2 structure (Fig. 4B). Intermediate models of the putative AMPKα2 apoprotein structures (DFG-out or DFG-in) were generated by using the AMBER99 force field37 for energy minimization and the Generalized Born/Volume Integral (GB/VI) method38 to calculate electrostatic solvation energies. The final models of the AMPKα2 apoproteins were based on the best-scoring intermediate models.
To model the binding mode of SU11248 to AMPKα2, the crystal structure of KIT in complex with SU11248 (KIT:SU11248) was superimposed on the AMPKα2 apoprotein models to orient the coordinates of bound SU11248 in the ATP binding site. The KIT:SU11248 structure did not include the fully solvated triethylamine fragment of SU11248; therefore, energy optimization of the AMPKα2 models containing SU11248 (AMPKα2:SU11248) were computed without this part of the molecule. The AMPKα2:SU11248 structures were energy minimized using the AMBER99 force field with the generalized Born solvation model; Gasteiger partial charges were used39 for the bound drug. Multiple rounds of energy minimization were performed by assigning and gradually reducing restraints on the heavy atom positions with force constants of 100, 10 and 1 kcal/mol, followed by a final round of unrestrained minimization. The root mean square of the gradient of 0.05 was used as the convergence criteria for the energy minimizations. While energy minimization resulted in a stable AMPKα2:SU11248 complex for the DFG-out conformation, the resulting complex for the DFG-in conformation was unstable.
Acknowledgements
This research was supported by NIH grant CA 73807 (K.R.L.) and CA 132529 (P.J.E.).
Abbreviations
- AMPK
5′-AMP-activated protein kinase
- ACC
acetyl-CoA carboxylase
- CaMKKβ
calcium/calmodulin-dependent protein kinase kinase β
- HIF-1
hypoxia-inducible factor-1
- IC50
inhibition constant at 50% of control value
- PTK
protein tyrosine kinase
- PDB ID
research collaboratory for structural bioinformatics protein data bank identification number
- SAR
structure-activity relationship
- TR-FRET
time-resolved fluorescence/förster resonance energy transfer
- TAK1
TGFβ-activated kinase 1
- VEGF
vascular endothelial growth factor
Footnotes
Previously published online: www.landesbioscience.com/journals/cbt/article/12162
References
- 1.Bright NJ, Thornton C, Carling D. The regulation and function of mammalian AMPK-related kinases. Acta Physiol (Oxf) 2009;196:15–26. doi: 10.1111/j.1748-1716.2009.01971.x. [DOI] [PubMed] [Google Scholar]
- 2.Fogarty S, Hardie DG. Development of protein kinase activators: AMPK as a target in metabolic disorders and cancer. Biochim Biophys Acta. 2009;1804:581–591. doi: 10.1016/j.bbapap.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 3.Lage R, Dieguez C, Vidal-Puig A, Lopez M. AMPK: a metabolic gauge regulating whole-body energy homeostasis. Trends Mol Med. 2008;14:539–549. doi: 10.1016/j.molmed.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Shaw RJ. LKB1 and AMP-activated protein kinase control of mTOR signalling and growth. Acta Physiol (Oxf) 2009;196:65–80. doi: 10.1111/j.1748-1716.2009.01972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fay JR, Steele V, Crowell JA. Energy homeostasis and cancer prevention: the AMP-activated protein kinase. Cancer Prev Res (Phila Pa) 2009;2:301–309. doi: 10.1158/1940-6207.CAPR-08-0166. [DOI] [PubMed] [Google Scholar]
- 6.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laderoute KR, Amin K, Calaoagan JM, Knapp M, Le T, Orduna J, et al. 5′-AMP-activated protein kinase (AMPK) is induced by low-oxygen and glucose deprivation conditions found in solid-tumor microenvironments. Mol Cell Biol. 2006;26:5336–5347. doi: 10.1128/MCB.00166-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papandreou I, Lim AL, Laderoute K, Denko NC. Hypoxia signals autophagy in tumor cells via AMPK activity, independent of HIF-1, BNIP3 and BNIP3L. Cell Death Differ. 2008;15:1572–1581. doi: 10.1038/cdd.2008.84. [DOI] [PubMed] [Google Scholar]
- 9.Cairns R, Papandreou I, Denko N. Overcoming physiologic barriers to cancer treatment by molecularly targeting the tumor microenvironment. Mol Cancer Res. 2006;4:61–70. doi: 10.1158/1541-7786.MCR-06-0002. [DOI] [PubMed] [Google Scholar]
- 10.Bristow RG, Hill RP. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer. 2008;8:180–192. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- 11.Gridelli C, Maione P, Del Gaizo F, Colantuoni G, Guerriero C, Ferrara C, et al. Sorafenib and sunitinib in the treatment of advanced non-small cell lung cancer. Oncologist. 2007;12:191–200. doi: 10.1634/theoncologist.12-2-191. [DOI] [PubMed] [Google Scholar]
- 12.Polyzos A. Activity of SU11248, a multitargeted inhibitor of vascular endothelial growth factor receptor and platelet-derived growth factor receptor, in patients with metastatic renal cell carcinoma and various other solid tumors. J Steroid Biochem Mol Biol. 2008;108:261–266. doi: 10.1016/j.jsbmb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Mendel DB, Laird AD, Xin X, Louie SG, Christensen JG, Li G, et al. In vivo antitumor activity of SU11248, a novel tyrosine kinase inhibitor targeting vascular endothelial growth factor and platelet-derived growth factor receptors: determination of a pharmacokinetic/pharmacodynamic relationship. Clin Cancer Res. 2003;9:327–337. [PubMed] [Google Scholar]
- 14.Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108:1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraley ME, Rubino RS, Hoffman WF, Hambaugh SR, Arrington KL, Hungate RW, et al. Optimization of a pyrazolo[1,5-a]pyrimidine class of KDR kinase inhibitors: improvements in physical properties enhance cellular activity and pharmacokinetics. Bioorg Med Chem Lett. 2002;12:3537–3541. doi: 10.1016/s0960-894x(02)00827-2. [DOI] [PubMed] [Google Scholar]
- 17.McGaughey GB, Culberson JC, Feuston BP, Kreatsoulas C, Maiorov V, Shpungin J. Scoring of KDR kinase inhibitors: using interaction energy as a guide for ranking. Mol Divers. 2006;10:341–347. doi: 10.1007/s11030-006-9037-1. [DOI] [PubMed] [Google Scholar]
- 18.Park HU, Suy S, Danner M, Dailey V, Zhang Y, Li H, et al. AMP-activated protein kinase promotes human prostate cancer cell growth and survival. Mol Cancer Ther. 2009;8:733–741. doi: 10.1158/1535-7163.MCT-08-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roskoski R., Jr Sunitinib: a VEGF and PDGF receptor protein kinase and angiogenesis inhibitor. Biochem Biophys Res Commun. 2007;356:323–328. doi: 10.1016/j.bbrc.2007.02.156. [DOI] [PubMed] [Google Scholar]
- 20.Hardie DG, Pan DA. Regulation of fatty acid synthesis and oxidation by the AMP-activated protein kinase. Biochem Soc Trans. 2002;30:1064–1070. doi: 10.1042/bst0301064. [DOI] [PubMed] [Google Scholar]
- 21.Hawley SA, Pan DA, Mustard KJ, Ross L, Bain J, Edelman AM, et al. Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2005;2:9–19. doi: 10.1016/j.cmet.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 22.Hurley RL, Anderson KA, Franzone JM, Kemp BE, Means AR, Witters LA. The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J Biol Chem. 2005;280:29060–29066. doi: 10.1074/jbc.M503824200. [DOI] [PubMed] [Google Scholar]
- 23.Woods A, Dickerson K, Heath R, Hong SP, Momcilovic M, Johnstone SR, et al. Ca2+/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab. 2005;2:21–33. doi: 10.1016/j.cmet.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Hurley RL, Barre LK, Wood SD, Anderson KA, Kemp BE, Means AR, et al. Regulation of AMP-activated protein kinase by multisite phosphorylation in response to agents that elevate cellular cAMP. J Biol Chem. 2006;281:36662–36672. doi: 10.1074/jbc.M606676200. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat Chem Biol. 2006;2:358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gajiwala KS, Wu JC, Christensen J, Deshmukh GD, Diehl W, DiNitto JP, et al. KIT kinase mutants show unique mechanisms of drug resistance to imatinib and sunitinib in gastrointestinal stromal tumor patients. Proc Natl Acad Sci USA. 2009;106:1542–1547. doi: 10.1073/pnas.0812413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okram B, Nagle A, Adrian FJ, Lee C, Ren P, Wang X, et al. A general strategy for creating “inactive-conformation” abl inhibitors. Chem Biol. 2006;13:779–786. doi: 10.1016/j.chembiol.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 29.Gan HK, Seruga B, Knox JJ. Sunitinib in solid tumors. Expert Opin Investig Drugs. 2009;18:821–834. doi: 10.1517/13543780902980171. [DOI] [PubMed] [Google Scholar]
- 30.Faivre S, Demetri G, Sargent W, Raymond E. Molecular basis for sunitinib efficacy and future clinical development. Nat Rev Drug Discov. 2007;6:734–745. doi: 10.1038/nrd2380. [DOI] [PubMed] [Google Scholar]
- 31.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 32.Hasinoff BB, Patel D, O'Hara KA. Mechanisms of myocyte cytotoxicity induced by the multiple receptor tyrosine kinase inhibitor sunitinib. Mol Pharmacol. 2008;74:1722–1728. doi: 10.1124/mol.108.050104. [DOI] [PubMed] [Google Scholar]
- 33.Dyck JR, Gao G, Widmer J, Stapleton D, Fernandez CS, Kemp BE, et al. Regulation of 5′-AMP-activated protein kinase activity by the noncatalytic beta and gamma subunits. J Biol Chem. 1996;271:17798–17803. doi: 10.1074/jbc.271.30.17798. [DOI] [PubMed] [Google Scholar]
- 34.Mu J, Brozinick JT, Jr, Valladares O, Bucan M, Birnbaum MJ. A role for AMP-activated protein kinase in contraction- and hypoxia-regulated glucose transport in skeletal muscle. Mol Cell. 2001;7:1085–1094. doi: 10.1016/s1097-2765(01)00251-9. [DOI] [PubMed] [Google Scholar]
- 35.Dale S, Wilson WA, Edelman AM, Hardie DG. Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett. 1995;361:191–195. doi: 10.1016/0014-5793(95)00172-6. [DOI] [PubMed] [Google Scholar]
- 36.Cumming G, Fidler F, Vaux DL. Error bars in experimental biology. J Cell Biol. 2007;177:7–11. doi: 10.1083/jcb.200611141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem. 2000;21:1049–1074. [Google Scholar]
- 38.Labute P. The generalized Born/volume integral implicit solvent model: estimation of the free energy of hydration using London dispersion instead of atomic surface area. J Comput Chem. 2008;29:1693–1698. doi: 10.1002/jcc.20933. [DOI] [PubMed] [Google Scholar]
- 39.Gasteiger J, Marsili M. Iterative partial equalization of orbital electronegativity—A rapid access to atomic charges. Tetrahedron. 1980;36:3219–3228. [Google Scholar]