Abstract
WormBase (http://www.wormbase.org/) is the central data repository for information about Caenorhabditis elegans and related nematodes. As a model organism database, WormBase extends beyond the genomic sequence, integrating experimental results with extensively annotated views of the genome. The WormBase Consortium continues to expand the biological scope and utility of WormBase with the inclusion of large-scale genomic analyses, through active data and literature curation, through new analysis and visualization tools, and through refinement of the user interface. Over the past year, the nearly complete genomic sequence and comparative analyses of the closely related species Caenorhabditis briggsae have been integrated into WormBase, including gene predictions, ortholog assignments and a new synteny viewer to display the relationships between the two species. Extensive site-wide refinement of the user interface now provides quick access to the most frequently accessed resources and a consistent browsing experience across the site. Unified single-page views now provide complete summaries of commonly accessed entries like genes. These advances continue to increase the utility of WormBase for C.elegans researchers, as well as for those researchers exploring problems in functional and comparative genomics in the context of a powerful genetic system.
DESCRIPTION
Caenorhabditis elegans is a soil nematode whose small size (1 mm), rapid generation time (3 days) and maintenance via clonal or sexual reproduction have all contributed to its widespread use as a genetic model organism. Furthermore, C.elegans is transparent, exhibits an invariant cell lineage and has a relatively simple nervous system. Its compact genome (100 Mbp) and complete genome sequence have extended the benefits of C.elegans to studies in genomics and proteomics (1). Finally, the recent sequencing and analysis of the C.briggsae genome brings to bear powerful techniques of comparative genomics, making the system ideal for studies of genome structure and evolution as well (2).
The WormBase Consortium is a team of researchers whose ultimate aim is to consolidate the growing body of information pertaining to C.elegans and related nematodes into a web-accessible, highly curated resource (3,4). WormBase, however, functions not just as a static data repository, but also as a research tool in its own right, providing a large array of research and analysis tools, making it an effective data mining environment.
This review provides a brief overview of the contents and navigation of WormBase, outlines new data integrated into the resource, explores some of the enhancements to the user interface, discusses tools to facilitate bioinformatic analysis using WormBase and outlines future objectives of the WormBase Consortium.
OVERVIEW OF THE RESOURCE
Contents of the database
The core of WormBase centers on those areas that helped establish C.elegans as a model organism, including: (i) the complete genome sequence (5); (ii) mutant phenotypes, genetic markers and genetic map information; (iii) the developmental lineage of the worm (6,7); (iv) the connectivity of the nervous system (8,9); (v) gene expression described at the level of single cells; and (vi) bibliographic resources including paper abstracts and author contact information. WormBase also contains extensive information from large-scale genomics analyses, including precomputed sequence similarity searches, protein motif analyses, results from systematic RNAi screens (10–12), single nucleotide polymorphisms (SNPs) (13,14), microarray expression studies (15) and the assignment of Gene Ontology (GO) terms to gene products. All these categories continue to be actively curated, with new results added and past results refined or revised as they are experimentally verified. Of significant note, the continuing efforts of the C.elegans Genome Sequencing Consortium resulted in the closure of the last gap in the C.elegans genomic sequence in November 2002. C.elegans is the first and so far only metazoan to have reached sequencing closure of all of its chromosomes.
Searching and browsing WormBase
WormBase users typically enter the site from the main page. From this page, they may search directly for a variety of information, including loci, predicted genes, proteins, clones, alleles and bibliographic entries. Searches that identify multiple results return an intermediary selection screen displaying the object type and identifier. Searches that identify a single result return a specialized report page specific to the data type returned.
Several page elements simplify the navigation of WormBase. First, a general static menu bar at the top of every page links to the most commonly accessed pages, including an index of available searches (Fig. 1A). Second, a search box embedded in the graphical banner enables users to conduct quick searches from any page (Fig. 1B). Finally, on every report page, a navigational bar presents contextually sensitive options (Fig. 1C). For example, on the Gene page, the navigational bar displays options to view the Locus page, the Sequence page and Nearby Genes. Two options appear in this navigational bar for every page view: the ‘Schema’ option displays the underlying data model; the ‘Tree Display’ option shows the contents of the current object filled into a tabular representation of the model.
Figure 1.
Navigational elements of the WormBase interface. (A) A static navigational bar at the top of every page gives quick access to the most commonly accessed features of the site. (B) A new search box in the graphical banner enables basic searches from any page. (C) The navigational bar of Report pages shows contextually sensitive options.
The Genome Browser, a central component of WormBase, provides users with a highly customizable graphical representation of the genome (http://www.wormbase.org/db/seq/gbrowse). Users can enter the Genome Browser through hypertext links from related report pages or by searching from the Genome Browser interface directly using a marker name or position, chromosomal coordinates, oligonucleotide or a description of biological function. Once an area of interest is in view, users may zoom in or out, or slide the display right or left along the genome. Semantic zooming in the Genome Browser displays increasing detail as the magnification is increased (16). Recent enhancements to the browser include the ability to reverse the orientation of a region of a genome, to dump marked-up regions of the genome in GenBank, EMBL, BSML, GAME/XML and other feature formats, to generate restriction maps of a region of interest, to upload private annotations into the browser, and to share third-party annotations with other individuals or groups.
In addition to the Genome Browser, the Gene page also acts as an informational ‘gateway’ to WormBase. This page presents a single consolidated view on the genetic status, physical position, expression, function and literature citations for any given gene. From this page, users may directly navigate to other pages containing more detailed information for any of these attributes.
Specialized search pages enable directed queries on the database. These include a variety of phenotype searches for mutants, RNAi experiments and gene tagging studies, genetic marker searches, and searches across the cell lineage and neuroanatomy of the organism. For searching the nucleotide and protein sequences, WormBase provides the BLAST/BLAT page (http://www.wormbase.org/db/searches/blat). From this page, users may conduct standard similarity searches against both the C.elegans and C.briggsae sequences contained in the database.
WormBase also offers facilities that streamline the retrieval of data en masse. These tools allow batched access of genes, strains and mutants (the ‘Batch Genes’ page; http://www.wormbase.org/db/searches/info_dump), or arbitrary regions of the genome (the ‘Batch Sequences’ page; http://www.wormbase.org/db/searches/advanced/dumper). For example, researchers interested in studying cis-regulatory elements can download a specified length of sequence upstream of every gene via the ‘Batch Sequences’ page. For more advanced data mining, WormBase may be searched using the ACeDB query language or via the Perl scripting language (discussed below).
RECENT ADDITIONS TO THE RESOURCE
Integration of C.briggsae sequence and analysis
Over the past year, WormBase has incorporated the draft C.briggsae genomic sequence into the database (2). These data are available for sequence similarity searching at both the nucleotide and protein levels. Furthermore, WormBase displays C.briggsae gene and protein annotations using the same Gene, Sequence and Protein pages familiar to users of the C.elegans data set.
At the genome level, users can search and navigate the C.briggsae genome using either the standard C.elegans Genome Browser, or a C.briggsae-specific Genome Browser. The standard browser displays the C.briggsae sequence as WABA algorithm alignment features on the C.elegans genome (17). This is ideal for researchers searching for conserved C.elegans regulatory elements or examining the supporting evidence for a gene prediction. When they click on the C.briggsae alignment track, users are taken to the corresponding region of the C.briggsae genome, where the relationship is inverted and the C.elegans genome is shown as an alignment track on C.briggsae. Within the C.briggsae browser, the C.briggsae gene predictions and their supporting data are displayed.
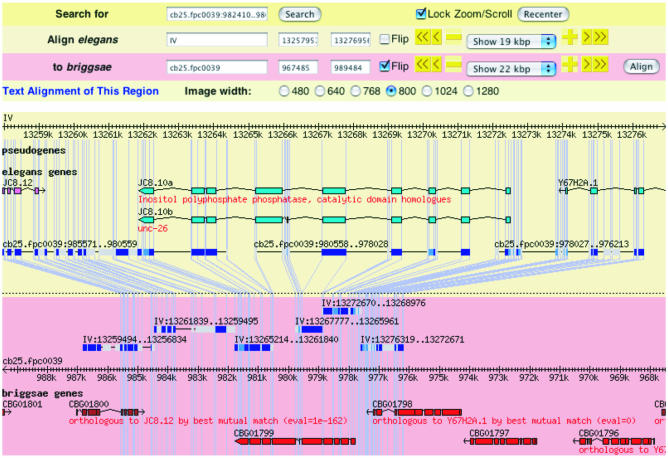
It is also possible to make a direct comparison between the C.elegans and C.briggsae genomes using a new Synteny Browser (Fig. 2). This viewer juxtaposes annotated regions of the two genomes based on WABA alignments, allowing direct assessment of the conservation of gene and gene model structure, the identification of conserved non-coding regions and the presence of gene expansions.
Figure 2.
The Synteny Viewer shows the relationship of syntenic regions between two genomes. Shown here is an alignment between a gene on chromosome IV of C.elegans and the syntenic region in C.briggsae as determined by the WABA algorithm. The cappuccino colored panel shows the C.elegans contig aligned with C.briggsae, including gene models and the actual WABA alignments. The pink panel shows the corresponding region in C.briggsae aligned with C.elegans. The blue lines connect corresponding locations of the two genomes.
As with the C.elegans data set, all C.briggsae sequencing data and annotations are available for download from the WormBase FTP site (ftp.wormbase.org).
Refinement of C.elegans gene models
Of major interest to end-users are the integrity and accuracy of gene models. WormBase uses a variety of methods to address difficulties with gene predictions. First, data from GenBank/EMBL submissions and the ongoing C.elegans ORFeome project continue to be updated and integrated into WormBase. The ORFeome project seeks to experimentally confirm the transcription and splicing patterns of predicted genes using systematic amplification by RT–PCR (18,19). PCR primer pairs and positively amplified products from the most recent version of this project, version 1.1, are displayed on the Genome Browser, allowing users to quickly assess whether a given gene model has been experimentally verified. Second, gene models continue to be revised from user submissions and literature curation. Finally, comparative analysis between C.briggsae and C.elegans is having a substantial impact on gene predictions in C.elegans by identifying previously missed genes and suggesting many changes to the internal structure of gene models (2).
Curation and annotation
Curation efforts are focused on providing concise functional annotations for every gene. Data are drawn from published papers, abstracts from C.elegans meetings, analysis of large-scale data sets and from direct submissions from the research community. Annotations appear at the top of every Gene Summary page, incorporating the following fields, when available: homology/orthology, biochemical and cellular function, genetic identity, mutant phenotype, RNAi results, genetic pathway information and expression data. To provide a controlled vocabulary of gene function, we continue to update gene ontology terms for every locus, as well as developing cell and phenotype ontologies. Large scale data sets curated in the past year include a genome-wide analysis of operons (20) and the inclusion of non-C.elegans ESTs. All curated data includes appropriate citations and detailed descriptions of their origin. To ensure that annotations are as current and accurate as possible, we encourage the research community to submit corrections and unpublished observations for inclusion in the database.
ENHANCEMENTS TO THE USER INTERFACE
New physical and genetic map displays
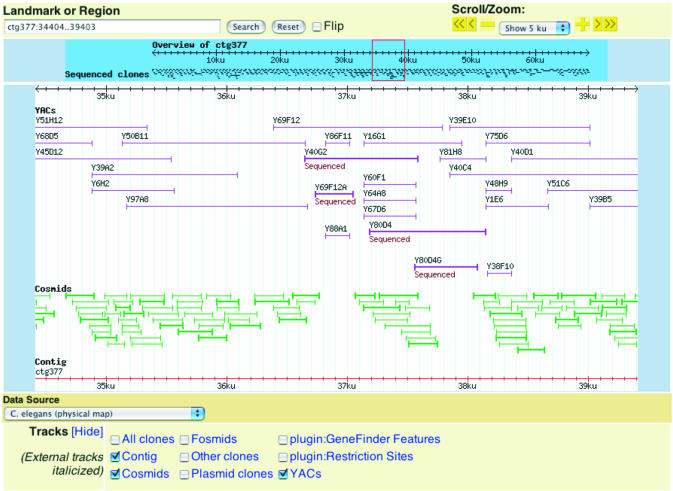
Several tools assist in the use of physical and genetic maps. The clone-based physical map is used extensively by the community as an adjunct to cloning and transformation rescue experiments. A new physical map display assists with this type of experiment (Fig. 3). This display more closely matches the look and feel of the WormBase interface, replacing an ACeDB-generated image. Built using the same program that generates the Genome Browser, the physical map inherits many of the features and capabilities of that tool, including scrolling, zooming and flexible landmark searches. Users may choose to display all clones, or may select specific clone types such as YACs, fosmids or cosmids.
Figure 3.
A new Physical Map displays clones generated during the sequencing of C.elegans. Built with the Genome Browser, the Physical Map inherits the same flexibility of user configuration and navigation as that tool.
A new genetic map display adds the ability to view multiple genetic maps simultaneously and display the relationship between the genetic map and the physical map. This facility will become increasingly useful as labs begin performing genetic mapping on C.briggsae.
Restructured reports
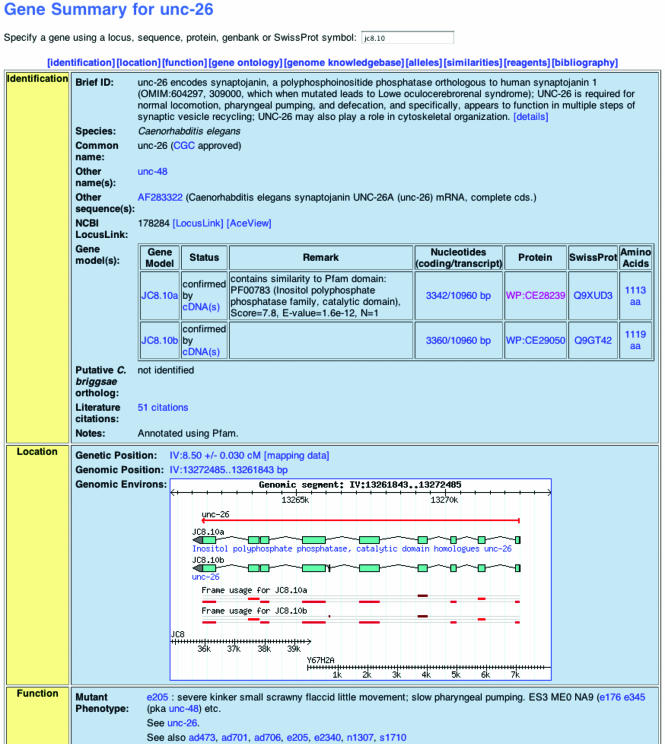
To provide easier access to a broad array of data, a number of pages have been completely redesigned. This is most clearly evident in the new Gene page, which consolidates information previously scattered amongst several other pages (Fig. 4). This consolidation of data allows the most useful information about a gene to be summarized in a single well-organized page, while placing the detailed supporting information in linked pages. For example, the Gene page summarizes the genetic position of a gene, but the mapping data that support that position are available on a separate linked page.
Figure 4.
The revised Gene Report page, which consolidates into a single page information that was previously available across several pages. Many reports in the WormBase site have seen a similar consolidation of information.
RESOURCES FOR THE BIOINFORMATICIST
WormBase offers many tools for those researchers interested in data mining and programmatic analysis of the resource. These include multiple access methods, precomputed data sets, stable releases of the data and the ability to run WormBase locally. First, WormBase provides multiple methods of access. The primary access point to WormBase is via its web interface. This interface itself contains tools useful for the bioinformaticist, such as structured XML dumps of any page, and the ‘Batch’ web forms, which provide quick access to information on groups of genes or fast dumps of sequences. Users may also interact with the underlying ACeDB database directly using the Perl scripting language and the ACePerl module (stein.cshl.org/aceperl/) or through the ACeDB query language AQL. Second, many precomputed data sets, such as spliced, unspliced and translated sequences of predicted and confirmed genes, tRNAs, C.briggsae sequence and analyses, gene predictions, and alignments, are available through the WormBase FTP site (ftp.wormbase.org). Third, WormBase now generates a stable release of the genome and its associated annotations biannually, facilitating consistency across genomic analyses. The first such release, WS100, is available at ws100.wormbase.org, and the next freeze will be available at ws110.wormbase.org. Finally, users may install and run their own local version of WormBase. The latest stable release of the software that drives WormBase can be downloaded from the WormBase ftp site or obtained by anonymous CVS access. The underlying ACeDB database and database builds are available at www.acedb.org and www.sanger.ac.uk/Projects/C_elegans.
FUTURE DIRECTIONS
WormBase is a dynamic resource. A number of new data sets are under curation, literature curation efforts are continuing and enhancements to the user interface are in development. With the completion of the C.briggsae sequence and the sequencing of other related nematodes on the horizon, we are working to expand the tools available for exploring the relationships between these genomes. This includes tools for defining and analyzing orthology and paralogy, new ways to visualize and understand the extent of synteny, and viewers for multiple genome alignments.
We continue to refine the user interface of WormBase to simplify the retrieval and display of diverse data types. A more pliable interface that allows users to select the types of data that they most frequently access as well as its display and formatting is currently under development. A long-range goal is to provide users with a ‘shopping cart’ or ‘workbench’ for continuity between visits and quick access to frequently accessed data and searches. As a growing number of users seeks access to larger amounts of data, we continue to develop methods to enable biologists with minimal programming experience to access and analyze data en masse.
On a closing note, we welcome the submission of data sets, corrections to existing data, and your comments, questions and suggestions (wormbase-help@wormbase.org).
Acknowledgments
ACKNOWLEDGEMENTS
WormBase is supported by grant P41-HG02223 from the US National Human Genome Research Institute and the British Medical Research Council. P.W.S. is an Investigator with the Howard Hughes Medical Institute.
REFERENCES
- 1.Riddle D.L., Blumenthal,T., Meyer,B.J. and Priess,J.R. (1997) C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- 2.Stein L.D., Bao,Z., Blasiar,D., Blumenthal,T., Brent,M., Chen,N.S., Chinwalla,A., Clarke,L., Clee,C., Coghlan,A. et al. (2003) The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS Biol., 1, e45. DOI: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris T.W., Lee,R., Schwarz,E., Bradnam,K., Lawson,D., Chen,W., Blasiar,D., Kenny,E., Cunningham,F., Kishore,R. et al. (2003) WormBase: a cross-species database for comparative genomics. Nucleic Acids Res., 31, 133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stein L., Sternberg,P., Durbin,R., Thierry-Mieg,J. and Spieth,J. (2001) WormBase: network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res., 29, 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 6.Sulston J.E. and Horvitz,H.R. (1977) Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol., 56, 110–156. [DOI] [PubMed] [Google Scholar]
- 7.Sulston J.E., Schierenberg,E., White,J.G. and Thomson,J.N. (1983) The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol., 100, 64–119. [DOI] [PubMed] [Google Scholar]
- 8.Chen N., Lee,R.Y., Altun,Z.F., Boulin,T., Sternberg,P.W. and Stein,L.D. (2003) In Kotter,R. (ed.), Neuroscience Databases: A Practical Guide. Kluwer Academic Publishers, Boston, MA, pp. 1–17. [Google Scholar]
- 9.White J.G., Southgate,E., Thomson,J.N. and Brenner,S. (1986) The structure of the nervous system of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond., 314, 1–340. [DOI] [PubMed] [Google Scholar]
- 10.Gonczy P., Echeverri,C., Oegema,K., Coulson,A., Jones,S.J., Copley,R.R., Duperon,J., Oegema,J., Brehm,M., Cassin,E. et al. (2000) Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature, 408, 331–336. [DOI] [PubMed] [Google Scholar]
- 11.Fraser A.G., Kamath,R.S., Zipperlen,P., Martinez-Campos,M., Sohrmann,M. and Ahringer,J. (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature, 408, 325–330. [DOI] [PubMed] [Google Scholar]
- 12.Kamath R.S., Fraser,A.G., Dong,Y., Poulin,G., Durbin,R., Gotta,M., Kanapin,A., Le Bot,N., Moreno,S., Sohrmann,M. et al. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature, 421, 231–237. [DOI] [PubMed] [Google Scholar]
- 13.Swan K.A., Curtis,D.E., McKusick,K.B., Voinov,A.V., Mapa,F.A. and Cancilla,M.R. (2002) High-throughput gene mapping in Caenorhabditis elegans. Genome Res., 12, 1100–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wicks S.R., Yeh,R.T., Gish,W.R., Waterston,R.H. and Plasterk,R.H. (2001) Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nature Genet., 28, 160–164. [DOI] [PubMed] [Google Scholar]
- 15.Kim S.K., Lund,J., Kiraly,M., Duke,K., Jiang,M., Stuart,J.M., Eizinger,A., Wylie,B.N. and Davidson,G.S. (2001) A gene expression map for Caenorhabditis elegans. Science, 293, 2087–2092. [DOI] [PubMed] [Google Scholar]
- 16.Stein L.D., Mungall,C., Shu,S., Caudy,M., Mangone,M., Day,A., Nickerson,E., Stajich,J.E., Harris,T.W., Arva,A. et al. (2002) The generic genome browser: a building block for a model organism system database. Genome Res., 12, 1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent W.J. and Zahler,A.M. (2000) Conservation, regulation, synteny and introns in a large-scale C. briggsae–C. elegans genomic alignment. Genome Res., 10, 1115–1125. [DOI] [PubMed] [Google Scholar]
- 18.Reboul J., Vaglio,P., Rual,J.F., Lamesch,P., Martinez,M., Armstrong,C.M., Li,S., Jacotot,L., Bertin,N., Janky,R. et al. (2003) C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nature Genet., 34, 35–41. [DOI] [PubMed] [Google Scholar]
- 19.Reboul J., Vaglio,P., Tzellas,N., Thierry-Mieg,N., Moore,T., Jackson,C., Shin-i,T., Kohara,Y., Thierry-Mieg,D., Thierry-Mieg,J. et al. (2001) Open-reading-frame sequence tags (OSTs) support the existence of at least 17 300 genes in C. elegans. Nature Genet., 27, 332–336. [DOI] [PubMed] [Google Scholar]
- 20.Blumenthal T., Evans,D., Link,C.D., Guffanti,A., Lawson,D., Thierry-Mieg,J., Thierry-Mieg,D., Chiu,W.L., Duke,K., Kiraly,M. et al. (2002) A global analysis of Caenorhabditis elegans operons. Nature, 417, 851–854. [DOI] [PubMed] [Google Scholar]