Abstract
The rapid pace at which genomic and proteomic data is being generated necessitates the development of tools and resources for managing data that allow integration of information from disparate sources. The Human Protein Reference Database (http://www.hprd.org) is a web-based resource based on open source technologies for protein information about several aspects of human proteins including protein–protein interactions, post-translational modifications, enzyme–substrate relationships and disease associations. This information was derived manually by a critical reading of the published literature by expert biologists and through bioinformatics analyses of the protein sequence. This database will assist in biomedical discoveries by serving as a resource of genomic and proteomic information and providing an integrated view of sequence, structure, function and protein networks in health and disease.
INTRODUCTION
Completion of sequencing of the human genome (1,2) has ushered in an era of characterizing genes and their gene products or proteins in greater detail. High-throughput technologies such as mass spectrometry and the yeast two-hybrid assay are being applied to generate proteomic data on an unprecedented scale (3,4). However, this wealth of data being generated can be fully harnessed only if it can be visualized and understood in the context of the existing information about proteins and their role in biology and disease.
The Human Protein Reference Database (HPRD) is a novel comprehensive protein information resource that depicts various features of proteins such as domain architecture, post-translational modifications, tissue expression, molecular function, subcellular localization, enzyme–substrate relationships and protein–protein interactions (5). This database is completely object oriented and was developed using Zope and Python, both open source technologies. HPRD is web based and is freely available to the academic community at http://www.hprd.org.
REPRESENTING COMPLEX PROTEIN DATA
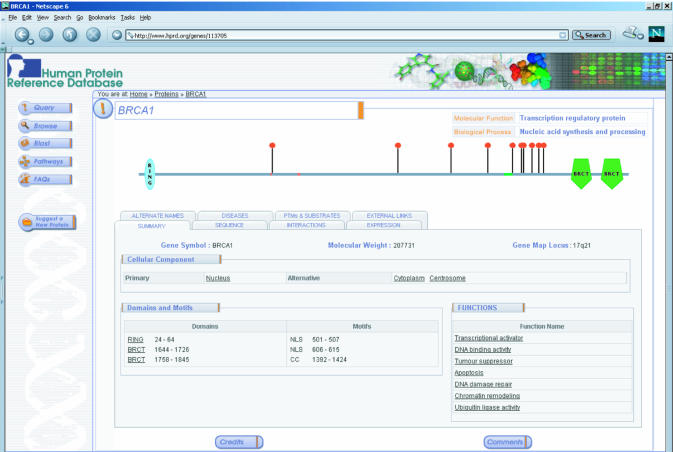
The complexity of protein data is intimately related to their diverse functional roles in various biological processes. Annotation and display of such complex data is quite challenging. Although most types of data can be tackled computationally, a visually appealing graphical interface that is intuitive and easy to use is more likely to be accepted by users. We have therefore designed HPRD to provide as many features graphically as possible along with links to more detailed text-based descriptions in research articles. Figure 1 shows a molecule page of the BRCA1 protein with a graphic showing the various protein domains and motifs along with sites of post-translational modification. Tabs allow simple navigation between different features of the protein.
Figure 1.
A screenshot of the molecule page for BRCA1. The molecule page serves as the entry point for accessing detailed annotation of proteins in HPRD. Molecular function and biological process for each molecule are indicated at the top right with a graphic representation of the protein domains and motifs and PTMs displayed in the middle. Under the summary tab, the HUGO gene symbol, the calculated molecular weight and the gene locus are indicated along with the cellular component, which refers to subcellular localization. The extent of domains and motifs is shown under the summary tab but can also be accessed by mousing over an individual region or motif in the graph. All underlined text is linked to PubMed entries that provide further details or to a database source.
ANNOTATION
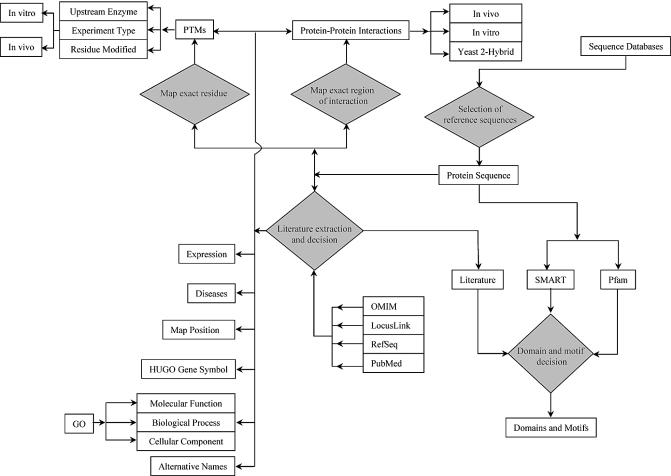
The proteins in HPRD are annotated manually by reading the published literature as well as by bioinformatics analyses of the protein sequences. Figure 2 shows the different steps in the annotation process involving various features of proteins. Interpretive annotation is crucial for classifying types of protein–protein interaction, delineating regions of interaction, type of experiment showing modification of a substrate by an enzyme, domain and motif analysis and subcellular localization. Links to PubMed entries are provided in each case so that the user can access the primary data for more details.
Figure 2.
A schematic outlining the basic steps in the annotation procedure. The annotations are carried out by trained biologists who critically read the published literature. The entries to be annotated are carefully selected from all the existing database entries by BLAST analysis (6) as well as manual inspection to provide the reference sequence. Interpretive annotation steps are represented by orange diamonds in the schematic. For instance, the annotator performs domain and motif analysis using the SMART (7) and Pfam (8) programs as well as by reading the literature. The OMIM database is used for disease annotations (9), and RefSeq and LocusLink (10) for sequences and links to other databases.
AN OBJECT-ORIENTED DATABASE ARCHITECTURE
HPRD is an object-oriented database. We used Zope (http://www.zope.org) for development of HPRD. Zope is a leading open source web application server and is built using the programming language Python (http://www.python.org). Zope was especially suited for developing HPRD because it provides a powerful dynamic site generation system, a clustering system and a robust and transparent object database, which is ideal for storing hierarchical data such as protein interactions, PTMs and domains (11). We used the Zope object database (ZODB), a robust object database that transparently stores persistent objects, to store the data in HPRD. This allowed the programmers to develop a whole application without imposing restrictions for the creation of data structure. We used another Zope-based object called Zcatalog that provides powerful indexing and searching on a Zope database. Zcatalog allows fast and robust searches. Since it catalogs objects and not file handles, all the contents in the database are easily searchable.
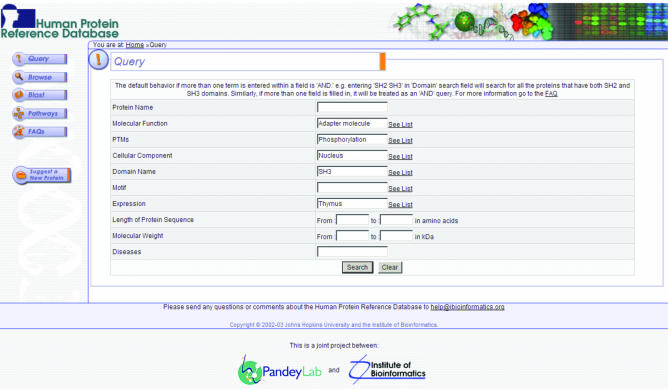
In HPRD, the proteins are accessible by using the query page, by browsing or by using BLAST. The search method is one of the powerful tools in HPRD and the power comes from the ability to search any field in the HPRD. Multiple fields can also be queried simultaneously as shown in Figure 3. Entering a protein name as a query automatically searches the main name as well as alternative names of all entries in HPRD. The browse page allows users to access proteins based on categorization of their function, domains, motifs, post-translational modifications (PTMs) or cellular component.
Figure 3.
A screenshot of the query page in HPRD. A user can search multiple annotation fields to retrieve protein entries based on protein name, molecular function, PTMs, cellular component, domains, motifs, tissue expression, length, molecular weight and diseases. The query system allows Boolean searches and the query terms are provided as pop-up lists for query fields other than protein name and diseases. Wild card searches using * are permitted by the query engine. Querying multiple search fields is processed as an ‘AND’ type of query by default. For example, a search for an adapter molecule, phosphorylation, nucleus, SH3 domain and thymus, as shown in the figure will retrieve a single entry for the Fyn binding protein, FYB, which satisfies all these parameters.
DATA STANDARDIZATION
The use of controlled vocabulary facilitates annotation efforts and promotes standardization and interoperability across different platforms and databases. Several annotation projects have already adopted the Gene Ontology (GO) Consortium’s standardization framework in their annotations (12). In order to be compatible with such efforts, the vocabulary used in HPRD is compliant with GO vocabulary, describing protein functions based on molecular function, biological process and cellular component. In addition to the use of controlled vocabulary, standards that govern data format and transfer have made tremendous progress in unifying data across the world wide web. One such standard involves the file format in which the data are stored. The use of eXtensible Markup Language (XML) as a standard for implementation of gene expression microarray data has already been adopted by the microarray community (13) and will soon be adopted for proteomic data as well (14). The major advantage of XML is that it lends itself well to importing from, and exporting to, various database systems while preserving the hierarchical nature of the data. The data contained in HPRD are available as XML files in addition to other flat file formats. To standardize nomenclature of gene names, the Human Genome Organization (HUGO) has put forth officially approved gene symbols for genes in humans (15). HPRD provides HUGO-approved gene symbols linked to all the proteins, which should allow easy linking to other databases because these gene symbols are non-redundant.
VISUALIZATION OF INTERACTION NETWORKS
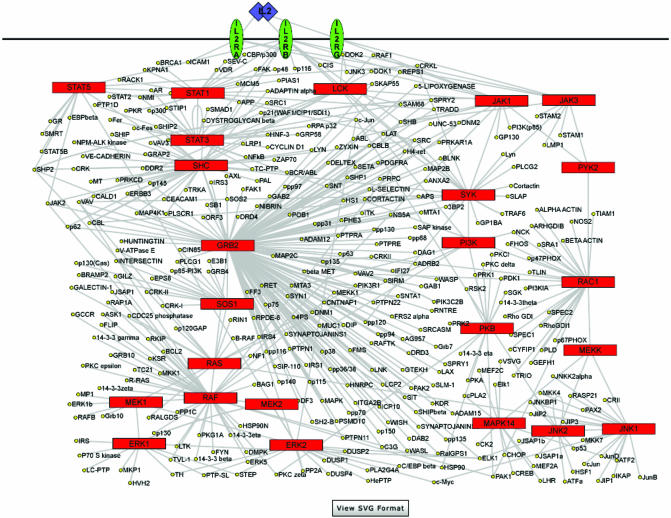
The complexity and intensity of the protein data are difficult to present without proper visualization methods. Of course, ultimately one would like to have an integrated view of genomic as well as proteomic networks as has been demonstrated in the case of certain metabolic pathways in yeast (16). Currently, there are protein interaction network pathway diagrams for nine signal transduction pathways in HPRD and this number is expected to grow as more proteins are annotated (see Fig. 4 for the interleukin-2 receptor pathway diagram). These pathway diagrams were generated using Pajek, which is a large network analysis program (17). These networks are made available both in jpeg format and in Scalable Vector Graphics (SVG) format by clicking on ‘view SVG format’ button. SVG is a language for describing 2D graphics in XML and allows additional functionalities such as zooming in without loss of resolution, search capability and the ability to link to the molecule page of any protein in the network by clicking on its name.
Figure 4.
Visualizing protein interaction networks in HPRD. The interleukin-2 receptor pathway is shown with red boxes indicating the major signaling molecules in the pathway and yellow circles representing the interaction partners. The pathways are displayed as jpeg images by default with the option of viewing them as SVG images by clicking on the ‘view SVG format’ button at the bottom. With an appropriate SVG viewer plug-in, a user is able to visualize the network with additional functionalities such as zoom operations, a search function within the pathway, a hand tool for maneuvering the figure and links to individual molecule pages by clicking on the name of any protein.
FUTURE DEVELOPMENTS
Federated databases that integrate information from various high-throughput experiments are essential to process the massive amount of available information into knowledge. In this respect, we are developing HPRD further such that mass spectrometric database searching algorithms can use certain annotated features of proteins such as PTMs and processing events. This will allow better identification of proteins and their isoforms including PTMs by taking advantage of the known information buried in the literature. Centralized efforts in genome and proteome annotation need to be supplemented by annotation by the entire biomedical community. Such a concerted effort will not only help enrich the databases but also minimize the errors that abound in databases. The Distributed Annotation System (DAS) provides a mechanism for multiple servers/providers to provide annotations for a common sequence database (18). We are developing specifications by which any third party’s annotations can be viewed along with the data contained in HPRD. Finally, it must be noted that HPRD continues to evolve in terms of the number of entries as well as in the depth of annotation for each entry and in the types of information displayed for each protein. With the active involvement of the biomedical community, we wish to make HPRD an evolving knowledge base of human proteins that will provide an integrated view of human proteins and networks.
Acknowledgments
ACKNOWLEDGEMENTS
Akhilesh Pandey is a Sidney Kimmel Scholar of the Sidney Kimmel Foundation for Cancer Research. He serves as Chief Scientific Advisor to the Institute of Bioinformatics. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies.
REFERENCES
- 1.The International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 2.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 3.Mann M. and Pandey,A. (2001) Use of mass spectrometry-derived data to annotate nucleotide and protein sequence databases. Trends Biochem. Sci., 26, 54–61. [DOI] [PubMed] [Google Scholar]
- 4.Tucker C.L., Gera,J.F. and Uetz,P. (2001) Towards an understanding of complex protein networks. Trends Cell Biol., 11, 102–116. [DOI] [PubMed] [Google Scholar]
- 5.Peri S., Navarro,J.D., Amanchy,R., Kristiansen,T.Z., Jonnalagadda,C.K., Surendranath,V., Niranjan,V., Muthusamy,B., Gandhi,T.K.B., Gronborg,M. et.al. (2003) Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res., 13, 2363–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 7.Schultz J., Milpetz,F., Bork,P. and Ponting,C.P. (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl Acad. Sci. USA, 95, 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamosh A., Scott,A.F., Amberger,J., Bocchini,C., Valle,D. and McKusick,V.A. (2002) Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res., 30, 52–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Navarro J.D., Niranjan,V., Peri,S., Jonnalagadda,C.K. and Pandey,A. (2003) From biological databases to platforms for biomedical discovery. Trends Biotechnol., 21, 263–268. [DOI] [PubMed] [Google Scholar]
- 12.Gene Ontology Consortium (2001) Creating the gene ontology resource: design and implementation. Genome Res., 11, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spellman P.T., Miller,M., Stewart,J., Troup,C., Sarkans,U., Chervitz,S., Bernhart,D., Sherlock,G., Ball,C., Lepage,M. et al. (2002) Design and implementation of microarray gene expression markup language (MAGE-ML). Genome Biol., 3, RESEARCH0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orchard S., Hermjakob,H. and Apweiler,R. (2003) The proteomics standards initiative. Proteomics, 3, 1374–1376. [DOI] [PubMed] [Google Scholar]
- 15.Wain H.M., Bruford,E.A., Lovering,R.C., Lush,M.J., Wright,M.W. and Povey,S. (2002) Guidelines for human gene nomenclature. Genomics, 79, 464–470. [DOI] [PubMed] [Google Scholar]
- 16.Ideker T., Thorsson,V., Ranish,J.A., Christmas,R., Buhler,J., Eng,J.K., Bumgarner,R., Goodlett,D.R., Aebersold,R. and Hood,L. (2001) Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science, 292, 929–934. [DOI] [PubMed] [Google Scholar]
- 17.Batagelj V. and Mrvar,A. (1998) Pajek—program for large network analysis. Connection, 21, 47–57. [Google Scholar]
- 18.Dowell R.D., Jokerst,R.M., Day,A., Eddy,S.R. and Stein,L. (2001) The Distributed Annotation System. BMC Bioinformatics, 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]