Abstract
DBTBS (http://dbtbs.hgc.jp) was originally released in 1999 as a reference database of published transcriptional regulation events in Bacillus subtilis, one of the best studied bacteria. It is essentially a compilation of transcription factors with their regulated genes as well as their recognition sequences, which were experimentally characterized and reported in the literature. Here we report its major update, which contains information on 114 transcription factors, including sigma factors, and 633 promoters of 525 genes. The number of references cited in the database has increased from 291 to 378. It also supports a function to find putative transcription factor binding sites within input sequences by using our collection of weight matrices and consensus patterns. Furthermore, though preliminarily, DBTBS now aims to contribute to comparative genomics by showing the presence or absence of potentially orthologous transcription factors and their corresponding cis-elements on the promoters of their potentially orthologously regulated genes in 50 eubacterial genomes.
INTRODUCTION
Bacillus subtilis is one of the most intensively studied bacteria, its genome entirely determined, its essential gene set defined and its systematic functional studies ongoing worldwide (1–3). For further comprehensive understanding of this organism, the existence of reference databases containing the results of previous studies is essential. SubtiList is one successful example (4), but it does not provide users with detailed information of the B.subtilis transcription system. Therefore, we have constructed a database of transcriptional regulation in B.subtilis (DBTBS) that contains transcriptional information specific to this organism (5). In this report, we introduce the recent progress of DBTBS, including the presentation of phylogenetic conservation information of both transcription factors and their recognition elements.
UPDATES AND NEW FEATURES
In Release 3, DBTBS contains information on 114 binding factors and 633 promoters of 525 regulated genes. These binding factors include σ factors [nine σ70-related, one σ54-related and five extracytoplasmic function (ECF) family members]. The promoters include those of six rRNA operons as well as plasmid-encoded promoters. It contains 203 annotated transcriptional start sites and 129 operons. The number of cited references now amounts to 378, which is a ∼30% increase on the previous release. Although we are trying to keep the database as comprehensive as possible, there still remain some references that should be incorporated. For example, information obtained from microarray experiments has not been included fully. Users’ feedback to fix errors or to add more data are welcome.
For each transcription factor, the collected sequences around the binding sites were realigned using MEME (6) as well as by eye. Then, a weight matrix of its sequence specificity was constructed considering pseudocounts if a sufficient number of examples (say, three) are available; if there are too few samples, the consensus pattern (such as ‘TATAAT’) was derived, instead of a weight matrix. The obtained set of weight matrices and consensus patterns is used to support a new function where input sequences are annotated with the hits of this set. The set will be available upon request.
Following requests from several researchers, the position of each binding site in the genome was newly recorded in the database. These data will make it easier to obtain the surrounding sequence at any length. Such data could be used for training or evaluating motif-finding software such as MEME.
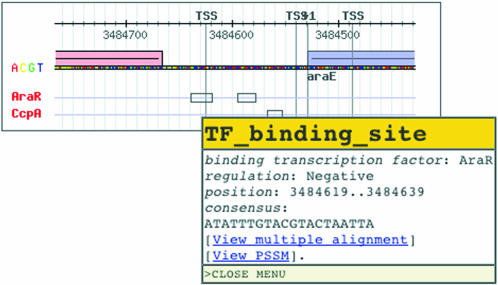
We also renewed the style of the graphical representation of promoters (Fig. 1). One notable feature is that overlapping binding sites can be perceived more easily (core consensus regions are also featured with color). Another is that the sequence information is represented in four colors. When users move the mouse over objects, relevant information such as the gene name and the binding factor is displayed; clicking the binding site will turn the page into a list of known binding sites. The annotation style follows the convention of the sequence ontology project (http://song.sourceforge.net), so that our annotation can easily be integrated into other systems.
Figure 1.
Graphical representation of cis-elements in a promoter. See text for details.
PHYLOGENETIC CONSERVATION STUDY
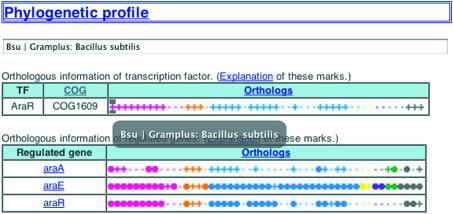
One of the problems of using weight matrices or consensus patterns to identify novel recognition positions of known transcription factors is that it often produces a number of false positives. To overcome this problem, the use of sequence conservation information between closely related species, called phylogenetic footprinting, is widely used. For example, we predicted B.subtilis regulons based on the conservation of upstream sequence segments between B.subtilis, Bacillus halodurans and Bacillus stearothermophilus (7). In this new release, we expand such an analysis into a systematic survey of the conservation of known cis-elements, as well as their binding factors, through available eubacterial genome sequences. For this purpose, we used the orthology information of the COG database (8) and applied the above-mentioned weight matrices/consensus patterns to the upstream sequence of orthologous genes. The diagram summarizing the result is shown in Figure 2. In this diagram, each column represents a bacterium (its name is displayed on the above window when the mouse is put over its position). In the upper table, the ‘.’/’+’ sign shows the absence/presence of an orthologous binding factor. In the lower table, the ‘.’ indicates the absence of the orthologous regulated gene; the ‘+’ means the presence of the ortholog of the regulated gene but the absence of the conserved element; and a filled circle means the presence of both (by clicking the circle, users can see the sequences of all detected sites as putative binding sites). Of course, the determination of presence/absence of orthologs itself is a highly delicate problem depending on, say, the setting of the cut-off values. In this sense, the current version is only a starting point for more comprehensive analyses. However, such kinds of data will undoubtedly stimulate many interesting studies on the evolution of transcriptional networks in eubacteria.
Figure 2.
Summarized representation of the conservation information of a binding factor and its known binding sites. See text for details.
Another direction of future studies using our data is the construction of a comprehensive model of the transcription system in B.subtilis. In such studies, the incorporation of accumulating microarray data will be a challenging task. We believe that DBTBS is a useful resource for both experimental and theoretical researchers of B.subtilis as well as of other eubacteria.
Acknowledgments
ACKNOWLEDGEMENTS
We thank T. Ishii, G. Terai, K. Yoshida and Y. Fujita for their contribution to previous releases; Michiel J. L. de Hoon for checking the contents of DBTBS and critically reading the manuscript. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas ‘Genome Biology’ from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Kunst F., Ogasawara,N., Moszer,I., Albertini,A.M., Alloni,G., Azevedo,V., Bertero,M.G., Bessieres,P., Bolotin,A., Borchert,S. et al. (1997) The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature, 390, 249–256. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi K., Ehrlich,S.D., Albertini,A., Amati,G., Andersen,K.K., Arnaud,M., Asai,K., Ashikaga,S., Aymerch,S., Bessieres,P. et al. (2003) Essential Bacillus subtilis genes. Proc. Natl Acad. Sci. USA, 100, 4678–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogasawara N. (2000) Systematic function analysis of Bacillus subtilis genes. Res. Microbiol., 151, 129–134. [DOI] [PubMed] [Google Scholar]
- 4.Moszer I., Jones,L.M., Moreira,S., Fabry,C. and Danchin,A. (2002) SubtiList: the reference database for the Bacillus subtilis genome. Nucleic Acids Res., 30, 62–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishii T., Yoshida,K., Terai,G., Fujita,Y. and Nakai,K. (2001) DBTBS: a database of Bacillus subtilis promoters and transcription factors. Nucleic Acids Res., 29, 278–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey T.L. and Elkan,C. (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Altman,R., Brutlag,D., Karp,P., Lathrop,R. and Searls,D. (eds), Proceedings of the 2nd International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA, pp. 28–36. [PubMed] [Google Scholar]
- 7.Terai G., Takagi,T. and Nakai,K. (2001) Prediction of co-regulated genes in Bacillus subtilis on the basis of upstream elements conserved across three closely related species. Genome Biol., 2, RESEARCH0048.1–0048.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tatusov R.L., Fedorova,N.D., Jackson,J.J., Jacobs,A.R., Kiryutin,B., Koonin,E.V., Krylov,D.M., Mazumder,R., Mekhedov,S.L., Nikolskaya,A.N. et al. (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics, 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]