Abstract
A great deal of progress has recently been made in the discovery and understanding of the genetics of familial dilated cardiomyopathy (FDC). A consensus has emerged that with a new diagnosis of idiopathic dilated cardiomyopathy (IDC), the clinical screening of 1st degree family members will reveal FDC in at least 20-35% of cases. Point mutations in 31 autosomal and 2 X-linked genes representing diverse gene ontogeny have been implicated in causing FDC, but account for only 30-35% of genetic cause. Next generation sequencing (NGS) methods have dramatically decreased sequencing costs, making clinical genetic testing feasible for extensive panels of DCM genes. NGS also provides opportunities to discover additional genetic cause of FDC and IDC. Guidelines for evaluation and testing of FDC and IDC are now available, and when combined with FDC genetic testing and counseling will bring FDC/IDC genetics to the forefront of cardiovascular genetic medicine.
Keywords: cardiomyopathy, genetics, genetic counseling, genetic testing
Introduction
Since our 2005 review of familial dilated cardiomyopathy (FDC) genetics in this Journal (1), a great deal of additional progress has been made. We note other valuable dilated cardiomyopathy (DCM) reviews, consensus documents and guidelines since 2005 (2-17). We review key concepts of genetic research and provide recent updates in FDC genetics. We also note the dramatic innovation in sequencing technologies that are revolutionizing clinical and research genetic studies. Much of this is broadly applicable to all of cardiovascular genetics.
Background: genetic studies, phenotype and genotype
Phenotype studies
In our prior review (1) we cited 19 DCM phenotype studies published between 1981 and 2003, principally focused on estimating the fraction of those patients with idiopathic dilated cardiomyopathy (IDC) who were found to have FDC using family history (FH) or clinical screening of family members. FDC is defined most conservatively as DCM meeting criteria for IDC in at least two closely related family members (1). Large retrospective studies in the 1980's estimated that 2-10% of individuals with IDC had FDC. In the 1990's studies involving larger cohorts of patients with IDC and prospective cardiovascular screening in their close relatives estimated that 20-48% of individuals with IDC could be shown to have FDC (18-20). A consensus has emerged that FDC will be found in at least 20-35% of those with IDC with clinical screening of first-degree family members, where clinical screening includes ECG and echocardiography or some other measure of LV size and function. Notably, a family history without clinical screening is much less sensitive to detect FDC (18).
Genetics studies
In 2005 we listed 19 genes shown to cause nonsyndromic DCM in humans (1). We now list 33 genes, 31 autosomal and 2 X-linked (Table 1) associated with DCM covering significant gene ontogeny (Table 2). Notably, the frequencies of DCM mutations in any one gene are low (<<1% to 6-8%), and a genetic cause is identified in only 30-35% of familial DCM cases (Table 1). In contrast, in HCM genetic cause can be found in 50-75% of familial cases, and in those cases when a mutation is identified, >80% can be found in one of two genes (MYH7, MYBPC3) (13). By inference from HCM (and LQTS, ARVD/C (13, 16)), FDC genetics are inherently more complex.
Table 1.
Genes reported in association with nonsyndromic dilated cardiomyopathy*
| Gene† | Protein | Function‡ | OMIM | Estimated fraction of DCM† |
References |
|---|---|---|---|---|---|
| LMNA | lamin A/C | Structure/stability of inner nuclear membrane; gene expression | 150330 | 0.06 | (21-33) |
| MYH6 | α-myosin heavy chain | Sarcomeric protein; muscle contraction | 160710 | 0.043 | (34, 35) |
| MYH7 | β-myosin heavy chain | Sarcomeric protein; muscle contraction | 160760 | 0.042 | (36-39) |
| MYPN | myopalladin | Sarcomeric protein, z-disc | 608517 | 0.035 | (40) |
| TNNT2 | cardiac troponin T | Sarcomeric protein; muscle contraction | 191045 | 0.029 | (36, 38, 39, 41-44) |
| SCN5A | sodium channel | Controls sodium ion flux | 600163 | 0.026 | (39, 45, 46) |
| MYBPC3 | myosin-binding protein C | Sarcomeric protein; muscle contraction | 600958 | 0.02 | (35, 37) |
| RBM20 | RNA binding protein 20 | RNA binding protein of the spliceosome | 0.019 | ||
| TMPO | thymopoietin | Also LAP2; a lamin-associated nuclear protein | 188380 | 0.011 | (47) |
| LAMA4 | laminin a-4 | Extracellular matrix protein | 600133 | 0.011 | (48) |
| VCL | metavinculin | Sarcomere structure; intercalated discs | 193065 | 0.01 | (38, 49) |
| LDB3 | Cypher/ZASP | Cytoskeletal assembly; clustering of membrane proteins | 605906 | 0.01 | (39, 50) |
| TCAP | titin-cap or telethonin | Z-disc protein that associates with titin; sarcomere assembly | 604488 | 0.01 | (39, 51) |
| PSEN1/2 | presenilin 1 / 2 | Transmembrane proteins, gamma secretase activity | 104311/ 600759 | 0.01 | (52) |
| ACTN2 | α-actinin-2 | Sarcomere structure; anchor for myofibrillar actin | 102573 | 0.009 | (53) |
| CRYAB | alpha B crystalin | Cytoskeletal protein | 123590 | 0.007 | (54) |
| TPM1 | α-tropomyosin | Sarcomeric protein; muscle contraction | 191010 | 0.006 | (35, 55, 56) |
| ABCC9 | SUR2A | Kir6.2 regulatory subunit, inwardly rectifying cardiac KATP channel |
601439 | 0.006 | (57) |
| ACTC | cardiac actin | Sarcomeric protein; muscle contraction | 102540 | 0.005 | (58-63) |
| PDLIM3 | PDZ LIM domain protein 3 | Cytoskeletal protein | 605889 | 0.005 | (64) |
| ILK | integrin-linked kinase | Intracellular serine-threonine kinase; interacts with integrins | 602366 | 0.005 | (48) |
| TNNC1 | cardiac troponin C | Sarcomeric protein; muscle contraction | 191040 | 0.004 | (35, 43) |
| TNNI3 | cardiac troponin I | sarcomeric protein, muscle contraction; also seen as recessive | 191044 | 0.004 | (35, 65, 66) |
| PLN | phospholamban | Sarcoplasmic reticulum Ca++ regulator; inhibits SERCA2 pump | 172405 | 0.004 | (38, 67-70) |
| DES | desmin | DAGC; transduces contractile forces | 125660 | 0.003 | (61, 71, 72) |
| SGCD | δ-sarcoglycan | DAGC; transduces contractile forces | 601411 | 0.003 | (72-74) |
| CSRP3 | muscle LIM protein | Sarcomere stretch sensor/ Z discs | 600824 | 0.003 | (39, 75) |
| TTN | titin | Sarcomere structure/extensible scaffold for other proteins | 188840 | ? | (76, 77) |
| EYA4 | eyes-absent 4 | Transcriptional co-activator | 603550 | ? | (78) |
| ANKRD1 | Ankyrin repeat domain-containing protein 1 | cardiac ankyrin repeat protein (CARP); localized to myopalladin/ titin complex |
609599 | ? | (79) |
| DMD § | dystrophin | DAGC; transduces contractile force | 300377 | ? | (80, 81) |
| TAZ/G4.5 § | tafazzin | Unknown | 300394 | ? | (82, 83) |
See Reference 12 for DCM associated with syndromic disease, and Reference 15 for genes not listed here that routinely cause combined skeletal and cardiac myopathies.
Genes ordered by estimates of the fraction of DCM probands carrying mutations from primary and secondary reports; all are autosomal except as indicated; ? denotes inadequate data for estimate.
DAGC, dystrophin-associated glycoprotein complex.
X-linked genes.
Table 2.
DCM Gene Ontology
| Sarcomere | Cytoskeleton |
Nuclear envelope |
| ACTC | DMD | |
| MYH7 | DES | LMNA |
| MYH6 | LDB3 | TMPO |
| MYBPC3 | SGCD |
Gamma secretase activity |
| TNNT2 | PDLIM3 | |
| TNNC1 | VCL | |
| TNNI3 | RYAB | PSEN1 |
| TPM1 | ILK | PSEN2 |
| TTN | LAMA4 |
Sarcoplasmic reticulum |
| Z-disc | Mitochondrial | |
| TCAP | TAZ/G4.5 | PLN |
| CSRP3 | RNA binding |
Transcription factor |
| ACTN2 | RBM20 | |
| MYPN | Ion Channel | EYA4 |
| ANKRD1 | ABCC | |
| SCN5A |
The number of DCM genes will continue to increase with ongoing discovery efforts. Also ‘crossover’ DCM phenotypes of desmosomal genes usually associated with ARVD/C present as DCM with low frequency (84); DCM phenotypes have also been observed for genes principally observed in HCM or the long QT syndrome, as previously reviewed (12, 13). We and others have recently shown that rare variant genetics are at play in some cases of peripartum cardiomyopathy (85-87).
What has not changed over the past 5 years
The core approach to human genetic studies remains the same: the careful and comprehensive phenotyping of subjects and their family members, and then correlating those phenotypes with genetic information. The challenge of this approach is to assure oneself that the genetic variation identified is causative of the phenotype of interest.
Gene mutations
The term ‘mutation’ is most commonly applied in Mendelian disease to one or a short string of variants in coding DNA (Table 3). The most common are missense mutations, but less common types include nonsense, splice site, and short insertion or deletion mutations (Table 3). Synonymous variants do not change the amino acid of that codon, while nonsynonymous variants do change the amino acid of that codon.
Table 3.
Considerations for Molecular Genetic Testing
| A. Types of molecular genetic variants * | |
| Those affecting exonic (coding) sequence. | |
| Missense | Single base variant that changes an amino acid |
| Nonsense | Single base variant that changes an amino acid to a stop codon |
| Insertion/deletion (indel) | Usually one or a few nucleotides inserted or deleted. Unless the indel is in a multiple of three, a frameshift occurs that garbles the usual amino acid sequence. This usually results in an eventual stop codon. |
| Those affecting intronic or splice site sequences | |
| Splice site | Affects exon splicing; one or more exons may be skipped |
| Intronic | By definition intronic sequencing is non-coding. While intronic variation is more common than coding sequence, it has been infrequently associated with disease. |
| B. Testing categories of sequence variations relevant to a phenotype of interest (90) |
|
| C. Criteria used to assess the relevance of a genetic variant for a phenotype of interest | |
| Property | Comment |
| Prior molecular genetic classification, if available |
This may be definitive for variants previously established as disease-causing. |
| Type of variant (see Section A in this table). | A synonymous variant only in unusual circumstances is considered relevant for disease (e.g., a variant that opens a cryptic splice acceptor site). |
| Weight of evidence, in the gene in question, that rare nonsynonymous variants cause DCM. |
**see comment below. |
| This is especially relevant for a novel gene under consideration in a discovery study. Disruption of a functional protein in the tissue of interest could lead to plausible pathophysiology. |
Examples of established genes include those encoding proteins of the contractile apparatus (see Table 1). For discovery studies, evidence of cardiac expression or the presence of the protein product in cardiac tissue may aid in assessing relevance. |
| Rarity in the population | Many Mendelian variants may be ‘private’ or unique to a proband or family |
| Variant segregates with the DCM phenotype, ideally in one or more large families; lacking large families, the variant segregates with DCM in multiple smaller families, or is observed in multiple sporadic DCM cases. |
In genetic DCM (and other multi-locus Mendelian diseases), many variants are ‘private’ so that multiple probands or families with any one specific variant are uncommon. |
| Functional data derived from the variant: cellular or animal models that recapitulate the disease phenotype |
All model systems have inherent limitations and seldom provide definitive studies; however, such functional data increases the certainty that the variant under study is relevant for phenotype of interest. |
These variants do not account for copy number variants (CNV's; also termed structural variants), which are insertions, deletions, duplications or inversions of larger portions of DNA. CNVs range widely in size; from very small (fewer than a hundred nucleotides) to very large (many megabases), and all sizes in between. They may affect both coding and non-coding DNA. Structural variants are not detected by usual sequencing approaches. Systematic evaluation of structural variants has not been undertaken in DCM, and hence their relevance for DCM has not been established.
Some genes (e.g., LMNA, MYH7, TNNT2, see Table 1) have abundant evidence that point mutations can cause DCM. Nevertheless, because of the marked allelic heterogeneity in DCM genes, it is uncommon for any one specific variant to be found in multiple unrelated probands, even in these genes. Whether any of these novel nonsynonymous rare variants can be considered disease-causing by usual molecular genetic diagnostic standards is an open question. Further, because most of the DCM genes (Table 1) have had only a few reported pathologic variants, newly identified rare variants in such genes with less prior DCM sequencing data available are commonly reported as variants of unknown significance (VUS; this table, Section B).
Classifying a variant as a disease-causing mutation
Ascertaining whether any one specific variant is causing the phenotype of interest requires weighing several types of evidence, and achieving a high level of certainty for any one variant is challenging, especially if that variant is novel (1, 88-90) (Table 3). In most cases, the sum of all of the evidence is required to decide if the identified variants are relevant (Table 3). As noted below, with next generation sequencing (NGS) approaches, the unique genetic variants identified in an individual affected with a specific phenotype can be many – hundreds to thousands – creating new challenges.
Phenotypic considerations
The term ‘Mendelian disease’ has been applied to heritable genetic disease, usually familial, with identifiable inheritance patterns (dominant or recessive, and autosomal, X-linked or mitochondrial) (1). Many Mendelian diseases are uncommon to rare, with population frequencies well below 1%. For Mendelian disease demonstrating autosomal dominant inheritance (which is the case in most FDC families) (1), the most powerful evidence that a putative mutation is indeed disease-causing is segregation of the variant of interest with the disease phenotype in at least one large, multigenerational family with multiple affected individuals who carry the variant and multiple unaffected individuals who do not carry the variant (Table 3). Multiple large families available to assess segregation increases the strength of evidence. While this concept is superficially simple, certain features of adult-onset Mendelian disease commonly observed with FDC complicates this approach.
One feature is incomplete penetrance, which refers to individuals who carry a mutation but do not manifest any evidence of the disease phenotype. Thus, in gene discovery studies, the absence of a DCM phenotype in someone carrying a putative disease-causing variant can never be considered absolute evidence that the variant is not relevant: the individual in question may simply be manifesting incomplete penetrance. A key corollary for clinicians caring for at-risk family members is that a negative clinical cardiovascular evaluation at any age does not rule out the possibility that the family member may develop later disease. This provides the rationale for the periodic rescreening of at-risk family members who have normal evaluations.
A related concept, ‘age-dependent’ or ‘age-related’ penetrance, is also observed with FDC, where a disease-causing mutation usually manifests a disease phenotype only in the adult years, most commonly in the 4th to 6th decades or later.
Another feature that complicates FDC assessment is variable expressivity, which means that only some aspects of the DCM phenotype are present. For example, only mild left ventricular enlargement (LVE) without systolic dysfunction, or the onset of arrhythmia or conduction system disease with only borderline DCM may be observed. Also, age of onset can vary significantly, with variable severity of disease progression. Thus, within a large FDC family a wide range of clinical findings may be present without fully developed DCM. Reliance on endophenotypes (partial or sub-phenotypes) as an indication of genetic DCM/FDC also has been problematic, in part because subtle clinical changes may result from other more common causes of CV disease, making it difficult to decipher genetic from non-genetic cause.
While usually nonsyndromic, DCM can be included in syndromic disease involving various organ systems, but most commonly skeletal muscle disease (muscular dystrophy) (12).
Genotype considerations
Other criteria to assign causality (Table 3), in addition to segregation of the variant with the phenotype, include its relative rarity in control DNAs (commonly <<1%). The rationale for this is that if it were common in the population, it would be unlikely to cause a rare genetic disease. Nevertheless, how rare is rare (<0.01, <0.005, < 0.001, <0.0001)? Some analyses have suggested that the majority of rare alleles (0.001 – 0.003) may be injurious (91). The caveat with control DNAs is that they should be representative of the race and/or ethnicity of the DCM family, as variants observed to be common (>1%) in one population can be rare in a different population.
Conservation of the amino acid or nucleotide (i.e., lack of variation in the protein structure or specific nucleotide sequence (92, 93) of lower species) is also used to assess variants, with the rationale that an amino acid or a nucleotide position with greater variation in lower species may have increased tolerance to variants at that position and are therefore less likely to be disease-causing. Other features are also relevant (Table 3).
Much of this, vital for discovery efforts, is also relevant for FDC clinical genetics. These fundamental principles of human genetics investigations have not changed, but with NGS the quantity of data to which they are applied has changed dramatically.
Genetic counseling
Text limitations do not permit a reiteration of the components and importance of skilled genetic counseling, especially for difficult, confusing or syndromic cases, supported by geneticist consultations as needed (1). Unlike most cardiologists, genetic counselors are trained to deal with the family as a unit of inquiry rather than the individual patient, an essential quality for genetic medicine. Genetic counselors are also trained to emphasize disease prevention in contrast to the focus on disease treatment taken by most cardiovascular specialists. Both of these qualities are particularly relevant for facilitating genetic risk assessment. The availability of genetic counselors with cardiovascular training or experience can provide the support needed to initiate the practice of cardiovascular genetic medicine. We refer the reader to several citations that deal with these important points (1, 12, 15, 16, 94-96).
What has changed over the past 5 years
Sequencing methods
The most significant change is the dramatic improvement in efficiency and speed of gene sequencing methods. Next generation sequencing (NGS) is the term used to describe several diverse methods that improve sequencing throughput by several magnitudes, resulting in markedly reduced sequencing costs per nucleotide. This has led recently to sequencing the human exome routinely for research applications (97-99). The exome is defined as the protein coding portion (the exons) of the 18,000 – 19,000 genes, estimated at 1-2% of the human genome. NGS is also used to sequence the entire human genome (coding and non-coding regions of DNA), referred to as whole genome sequencing (WGS) (100). Because Mendelian disease typically affects the protein coding portions of the genome, exome sequencing is particularly relevant for rare variant Mendelian disease. As of 2010, typical costs of exome sequencing for research purposes are approximately $2000 per DNA sample. New instruments and new methods for multiplexing DNAs on NGS instruments are being developed that will improve throughput and decrease cost, making <$1000, or even <$500 exome sequences likely in the near future. WGS charges on the open market now range from $10,000 to $20,000; these costs are also expected to decrease dramatically (10- to 20-fold) in the next few years, which will bring even WGS into the realm of clinical genetic testing, as well as within the domain of the NIH research budgets of many cardiovascular genetics studies. NGS, whether for exome or WGS, is dramatically transforming the experimental possibilities – study designs unthinkable even 1-2 years ago can now be proposed and attained (97-99).
Along with this rapidly expanding universe of opportunity from NGS will come monstrous quantities of human DNA sequence data, challenging the hardware and software of informatics platforms, and necessitating novel approaches to data assembly, storage and analysis. Computational budgets for even modest exome projects now (terabytes of data) cost tens of thousands of dollars; larger projects containing hundreds to thousands of terabytes of data will require more robust outlays. These realities will require new ‘pipelines’ to be developed to efficiently analyze these massive data sets and reduce the cost of storage. This will also require new control DNA data sets to be generated, some of which is now underway (101).
Impact of NGS on clinical molecular genetic testing
NGS is directly related to the emergence of clinical genetic testing for FDC. As recently as 2-3 years ago, clinical genetic testing costing thousands of dollars was available only for a few HCM genes. Now panels of dozens of genes at reduced cost, incorporating many or all reported for any of the genetic cardiomyopathies (DCM, HCM, RCM, ARVD/C, and LVNC), are rapidly emerging using NGS methods. While this increase in data comes with a host of limitations and complications in interpretation, FDC testing sensitivity (the probability of finding a genetic cause with the genetic testing) now ranges from 15-25%, making pre-symptomatic testing feasible. Testing laboratories for DCM genes are catalogued at GeneTests (102), an online service hosted by NCBI.
The Genetic Information Nondiscrimination Act (GINA) of 2008
After many years of effort, a new federal law using the eponym of GINA now protects individuals from genetic discrimination in health care or employment. Further information is available at the National Genome Research Institute website (103).
Ongoing issues
Clinical progress
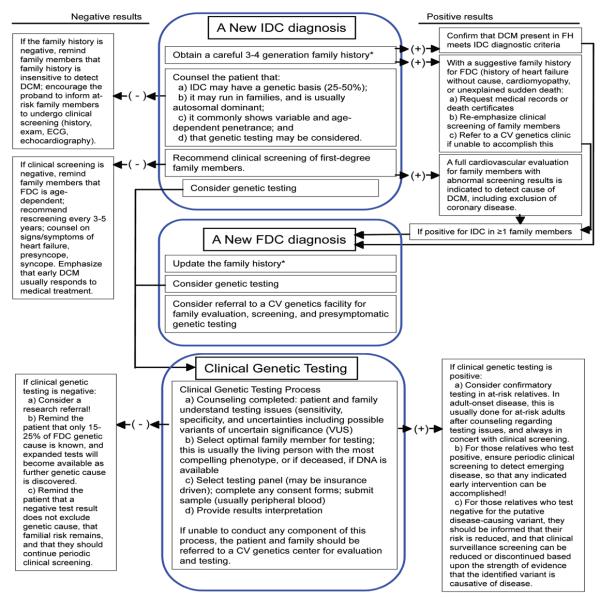
Despite the evidence supporting a genetic basis of IDC/FDC, the implementation of guidelines (13) by practitioners has been tepid. Adherence to such guidelines will require a shift in focus from strictly therapeutic measures for a single patient presenting with advanced disease to the consideration and assessment of DCM risk for an entire family (1, 13, 16) (Figure 1).
Figure 1. Flow diagram of genetic risk assessment for patients newly diagnosed with idiopathic dilated cardiomyopathy (IDC) or familial dilated cardiomyopathy (FDC).
The left- and right-sided boxes provide guidance for negative or positive results, respectively, based on the results of history or testing recommended in the central boxes.
*Always search for history or exam findings consistent with syndromic disease, particularly skeletal muscle symptoms. However, with any suggestion of syndromic disease in the proband or family members, strongly consider referral to a geneticist or CV genetic medicine clinic with genetics collaboration. Some features of early onset conduction system disease or arrhythmia (usually from LMNA rare variants) may be particularly susceptible to genetic testing. Rare variants of unknown significance are not helpful for predictive testing.
The rationale for these recommendations is that most IDC/DCM presents late in its causal pathway (advanced disease, usually with heart failure or sudden cardiac death), but early detection of asymptomatic DCM through screening enables presymptomatic intervention that may prevent or ameliorate the progression to advanced disease (95).
With a new IDC diagnosis, genetic risk evaluation should be initiated, including taking a 3-4 generation family history and recommending that 1st degree family members undergo clinical cardiovascular screening (Figure 1). Clinical genetic testing may also be warranted, including the competent interpretation of genetic results with appropriate counseling (1, 16). All of this may require a referral of patients to centers providing expertise in cardiovascular genetics and guidance on implementation of gene and/or mutation-specific therapies if indicated (13), ideally in centers with geneticists or genetic counselors working in collaboration with cardiologists in cardiovascular genetic medicine clinics (95).
How many genes might be involved in DCM?
Even though rare variants have been identified in >30 genes, we estimate that this accounts for only one-third of genetic cause of FDC. We predict this number will expand significantly. Discovery of additional genetic cause of DCM is still key to further understanding DCM genetics.
Genetic model for DCM
We have only scratched the surface in understanding DCM genetics. We have almost no insight into the causes of the marked variation in age of onset, disease penetrance, or clinical severity observed even for the same mutation within a large extended family, or between families with the same variant. Gene-enviroment interactions may explain some of this, but additional genetic variation may also explain a portion of this variability. Most of the FDC genetic data thus far supports a ‘one gene’ Mendelian model with marked locus (many genes) and marked allelic heterogeneity (many private mutations within any one gene). The impact of multiple mutations in the same individual has been recognized for HCM (104-109) and the long QT syndrome (108, 110), where two or more mutations have been shown to be associated with earlier onset and more severe disease in 3-7% of subjects. However, considerable additional genetic variation may be at play. Such genetic variation could include ‘less common’ common variants (e.g., allele frequencies 0.5 – 5%), additional rare variants (including the bi-allelic models as shown in HCM and ARVD/C (111, 112)), epigenetic factors, gene promoter site variants, or alterations in other genetically-driven regulatory processes such as microRNAs or their target sites. All of these remain to be evaluated for genetic DCM.
Is idiopathic dilated cardiomyopathy (IDC) a genetic disease?
A related issue involves the genetics of IDC, or DCM after all known causes (except genetic) have been ruled out, and its relationship to FDC. This is important because understanding the genetic basis of IDC could have a major public health impact, as non-ischemic DCM makes up a significant proportion of all forms of cardiomyopathy, and IDC is by far the largest component of nonischemic DCM. Here we differentiate a ‘true’ IDC as a patient with IDC who has had their first-degree family members clinically screened (history, exam, echocardiogram, ECG) to rule out FDC versus a ‘presumptive’ IDC – one who is negative for familial disease by a careful 3-4 generation family history but has not had family members screened beyond the FH. Preliminary data from our resequencing studies suggested that the frequency of possibly or likely disease-causing rare variants in a cohort of FDC and IDC probands (>300 in total) was similar (39, 113). However, in those studies the family members of the IDC probands were not systematically screened beyond FH, making it difficult to accurately assess the familial nature of disease in the ‘apparently sporadic’ IDC portion of our cohort. Therefore, whether ‘true’ sporadic IDC differs from FDC in gene composition, penetrance or expressivity remains untested in a large prospective study,
Summary
Recent progress for DCM genetics has been significant, although much remains to be learned. Clinical genetic testing is rapidly emerging, and NGS technology now permits patients to undergo clinical genetic testing for many genes at reduced cost. However, enthusiasm for DCM genetic testing remains tempered in 2011 in large part due to the testing sensitivity of 15-25%, and the plethora of DCM genes that makes the rare variants ‘established as disease-causing’ in any one gene only a very few. The discovery of new DCM genes and other DCM genetic cause, accelerated now by exome sequencing and soon by WGS, will lead to knowledge of the remainder of the genetic makeup of FDC and IDC. The careful and systematic phenotyping of DCM probands and family members, whether sporadic or familial, when combined with the cataloging of many DCM rare variants, will enable DCM genetics to move into the mainstream of cardiovascular genetic medicine.
Acknowledgments
This work was supported by NIH award RO1-HL58626 (Dr. Hershberger).
No relationships with industry of any kind are present.
Abbreviations
- ARVD/C
arrhythmogenic right ventricular dysplasia
- DCM
dilated cardiomyopathy
- IDC
idiopathic dilated cardiomyopathy
- FDC
familial dilated cardiomyopathy
- HCM
hypertrophic cardiomyopathy
- LQTS
long QT syndrome
- NGS
next generation sequencing
- WGS
whole genome sequencing
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burkett EL, Hershberger RE. Clinical and genetic issues in familial dilated cardiomyopathy. J Am Coll Cardiol. 2005;45:969–81. doi: 10.1016/j.jacc.2004.11.066. [DOI] [PubMed] [Google Scholar]
- 2.Morita H, Seidman J, Seidman CE. Genetic causes of human heart failure. J Clin Invest. 2005;115:518–26. doi: 10.1172/JCI200524351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maron BJ, Towbin JA, Thiene G, et al. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation. 2006;113:1807–16. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- 4.Richard P, Villard E, Charron P, Isnard R. The genetic bases of cardiomyopathies. J Am Coll Cardiol. 2006;48(supple A):A79–89. [Google Scholar]
- 5.Ashrafian H, Watkins H. Reviews of translational medicine and genomics in cardiovascular disease: new disease taxonomy and therapeutic implications cardiomyopathies: therapeutics based on molecular phenotype. J Am Coll Cardiol. 2007;49:1251–64. doi: 10.1016/j.jacc.2006.10.073. [DOI] [PubMed] [Google Scholar]
- 6.van Spaendonck-Zwarts KY, van den Berg MP, van Tintelen JP. DNA analysis in inherited cardiomyopathies: current status and clinical relevance. Pacing Clin Electrophysiol. 2008;31(Suppl 1):S46–9. doi: 10.1111/j.1540-8159.2008.00956.x. [DOI] [PubMed] [Google Scholar]
- 7.Taylor MR, Carniel E, Mestroni L. Cardiomyopathy, familial dilated. Orphanet J Rare Dis. 2006;1:27. doi: 10.1186/1750-1172-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fatkin D. Guidelines for the diagnosis and management of familial dilated cardiomyopathy. Heart Lung Circ. 2007;16:19–21. doi: 10.1016/j.hlc.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 9.Karkkainen S, Peuhkurinen K. Genetics of dilated cardiomyopathy. Ann Med. 2007;39:91–107. doi: 10.1080/07853890601145821. [DOI] [PubMed] [Google Scholar]
- 10.Robin NH, Tabereaux PB, Benza R, Korf BR. Genetic testing in cardiovascular disease. J Am Coll Cardiol. 2007;50:727–37. doi: 10.1016/j.jacc.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 11.Judge DP, Johnson NM. Genetic evaluation of familial cardiomyopathy. J Cardiovasc Trans Res. 2008;1:144–54. doi: 10.1007/s12265-008-9025-1. [DOI] [PubMed] [Google Scholar]
- 12.Hershberger RE, Cowan J, Morales A, Siegfried JD. Progress with genetic cardiomyopathies: screening, counseling, and testing in dilated, hypertrophic, and arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circ Heart Fail. 2009;2:253–61. doi: 10.1161/CIRCHEARTFAILURE.108.817346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hershberger RE, Lindenfeld J, Mestroni L, Seidman CE, Taylor MR, Towbin JA. Genetic evaluation of cardiomyopathy--a Heart Failure Society of America practice guideline. J Card Fail. 2009;15:83–97. doi: 10.1016/j.cardfail.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Jefferies JL, Towbin JA. Dilated cardiomyopathy. Lancet. 2010;375:752–62. doi: 10.1016/S0140-6736(09)62023-7. [DOI] [PubMed] [Google Scholar]
- 15.Dellefave L, McNally EM. The genetics of dilated cardiomyopathy. Curr Opin Cardiol. 2010 doi: 10.1097/HCO.0b013e328337ba52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hershberger RE, Morales A, Siegfried JD. Clinical and genetic issues in dilated cardiomyopathy: A review for genetics professionals. Genetics in Medicine. 2010;12:655–67. doi: 10.1097/GIM.0b013e3181f2481f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fatkin D, Otway R, Richmond Z. Genetics of dilated cardiomyopathy. Heart Fail Clin. 2010;6:129–40. doi: 10.1016/j.hfc.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Michels VV, Moll PP, Miller FA, et al. The frequency of familial dilated cardiomyopathy in a series of patients with idiopathic dilated cardiomyopathy. N Engl J Med. 1992;326:77–82. doi: 10.1056/NEJM199201093260201. [DOI] [PubMed] [Google Scholar]
- 19.Grunig E, Tasman JA, Kucherer H, Franz W, Kubler W, Katus HA. Frequency and phenotypes of familial dilated cardiomyopathy [see comments] J Am Coll Cardiol. 1998;31:186–94. doi: 10.1016/s0735-1097(97)00434-8. [DOI] [PubMed] [Google Scholar]
- 20.Baig MK, Goldman JH, Caforio AP, Coonar AS, Keeling PJ, McKenna WJ. Familial dilated cardiomyopathy: cardiac abnormalities are common in asymptomatic relatives and may represent early disease. J Am Coll Cardiol. 1998;31:195–201. doi: 10.1016/s0735-1097(97)00433-6. [DOI] [PubMed] [Google Scholar]
- 21.Fatkin D, MacRae C, Sasaki T, et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med. 1999;341:1715–24. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 22.Brodsky G, Muntoni F, Miocic S, Sinagra G, Sewry C, Mestroni L. Lamin A/C gene mutation associated with dilated cardiomyopathy with variable skeletal muscle involvement. Circ. 2000;101:473–476. doi: 10.1161/01.cir.101.5.473. [DOI] [PubMed] [Google Scholar]
- 23.Becane HM, Bonne G, Varnous S, et al. High incidence of sudden death with conduction system and myocardial disease due to lamins A and C gene mutation. Pacing Clin Electrophysiol. 2000;23:1661–6. doi: 10.1046/j.1460-9592.2000.01661.x. [DOI] [PubMed] [Google Scholar]
- 24.Jakobs PM, Hanson E, Crispell KA, et al. Novel lamin A/C mutations in two families with dilated cardiomyopathy and conduction system disease. J Card Fail. 2001;7:249–256. doi: 10.1054/jcaf.2001.26339. [DOI] [PubMed] [Google Scholar]
- 25.Arbustini E, Pilotto A, Repetto A, et al. Autosomal dominant dilated cardiomyopathy with atrioventricular block: a lamin A/C defect-related disease. J Am Coll Cardiol. 2002;39:981–90. doi: 10.1016/s0735-1097(02)01724-2. [DOI] [PubMed] [Google Scholar]
- 26.Hershberger RE, Hanson E, Jakobs PM, et al. A novel lamin A/C mutation in a family with dilated cardiomyopathy, prominent conduction system disease, and need for permanent pacemaker implantation. Am Heart J. 2002;144:1081–6. doi: 10.1067/mhj.2002.126737. [DOI] [PubMed] [Google Scholar]
- 27.Taylor MR, Fain PR, Sinagra G, et al. Natural history of dilated cardiomyopathy due to lamin A/C gene mutations. J Am Coll Cardiol. 2003;41:771–80. doi: 10.1016/s0735-1097(02)02954-6. [DOI] [PubMed] [Google Scholar]
- 28.Sebillon P, Bouchier C, Bidot LD, et al. Expanding the phenotype of LMNA mutations in dilated cardiomyopathy and functional consequences of these mutations. J Med Genet. 2003;40:560–7. doi: 10.1136/jmg.40.8.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacLeod HM, Culley MR, Huber JM, McNally EM. Lamin A/C truncation in dilated cardiomyopathy with conduction disease. BMC Med Genet. 2003;4:4. doi: 10.1186/1471-2350-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sylvius N, Bilinska ZT, Veinot JP, et al. In vivo and in vitro examination of the functional significances of novel lamin gene mutations in heart failure patients. J Med Genet. 2005;42:639–47. doi: 10.1136/jmg.2004.023283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pethig K, Genschel J, Peters T, et al. LMNA mutations in cardiac transplant recipients. Cardiology. 2005;103:57–62. doi: 10.1159/000082048. [DOI] [PubMed] [Google Scholar]
- 32.Karkkainen S, Reissell E, Helio T, et al. Novel mutations in the lamin A/C gene in heart transplant recipients with end stage dilated cardiomyopathy. Heart. 2006;92:524–6. doi: 10.1136/hrt.2004.056721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parks SB, Kushner JD, Nauman D, et al. Lamin A/C mutation analysis in a cohort of 324 unrelated patients with idiopathic or familial dilated cardiomyopathy. Am Heart J. 2008;156:161–9. doi: 10.1016/j.ahj.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carniel E, Taylor MR, Sinagra G, et al. Alpha-myosin heavy chain: a sarcomeric gene associated with dilated and hypertrophic phenotypes of cardiomyopathy. Circulation. 2005;112:54–9. doi: 10.1161/CIRCULATIONAHA.104.507699. [DOI] [PubMed] [Google Scholar]
- 35.Hershberger R, Norton N, Morales A, Li D, Siegfried J, Gonzalez-Quintana J. Coding sequence rare variants identified in MYBPC3, MYH6, TPM1, TNNC1 And TNNI3 from 312 patients with familial or idiopathic dilated cardiomyopathy. Circ Cardiovasc Genet. 2010;3:155–161. doi: 10.1161/CIRCGENETICS.109.912345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamisago M, Sharma SD, DePalma SR, et al. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N Engl J Med. 2000;343:1688–1696. doi: 10.1056/NEJM200012073432304. [DOI] [PubMed] [Google Scholar]
- 37.Daehmlow S, Erdmann J, Knueppel T, et al. Novel mutations in sarcomeric protein genes in dilated cardiomyopathy. Biochem Biophys Res Commun. 2002;298:116–20. doi: 10.1016/s0006-291x(02)02374-4. [DOI] [PubMed] [Google Scholar]
- 38.Villard E, Duboscq-Bidot L, Charron P, et al. Mutation screening in dilated cardiomyopathy: prominent role of the beta myosin heavy chain gene. Eur Heart J. 2005;26:794–803. doi: 10.1093/eurheartj/ehi193. [DOI] [PubMed] [Google Scholar]
- 39.Hershberger RE, Parks SB, Kushner JD, et al. Coding sequence mutations identified in MYH7, TNNT2, SCN5A, CSRP3, LBD3, and TCAP from 313 patients with familial or idiopathic dilated cardiomyopathy. Clin Translational Science. 2008;1:21–26. doi: 10.1111/j.1752-8062.2008.00017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duboscq-Bidot L, Xu P, Charron P, et al. Mutations in the Z-band protein myopalladin gene and idiopathic dilated cardiomyopathy. Cardiovasc Res. 2008;77:118–25. doi: 10.1093/cvr/cvm015. [DOI] [PubMed] [Google Scholar]
- 41.Hanson E, Jakobs P, Keegan H, et al. Cardiac troponin T lysine-210 deletion in a family with dilated cardiomyopathy. J Card Fail. 2002;8:28–32. doi: 10.1054/jcaf.2002.31157. [DOI] [PubMed] [Google Scholar]
- 42.Li D, Czernuszewicz GZ, Gonzalez O, et al. Novel cardiac troponin T mutation as a cause of familial dilated cardiomyopathy. Circulation. 2001;104:2188–93. doi: 10.1161/hc4301.098285. [DOI] [PubMed] [Google Scholar]
- 43.Mogensen J, Murphy RT, Shaw T, et al. Severe disease expression of cardiac troponin C and T mutations in patients with idiopathic dilated cardiomyopathy. J Am Coll Cardiol. 2004;44:2033–40. doi: 10.1016/j.jacc.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 44.Moller DV, Andersen PS, Hedley P, et al. The role of sarcomere gene mutations in patients with idiopathic dilated cardiomyopathy. Eur J Hum Genet. 2009;17:1241–9. doi: 10.1038/ejhg.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McNair WP, Ku L, Taylor MR, et al. SCN5A mutation associated with dilated cardiomyopathy, conduction disorder, and arrhythmia. Circulation. 2004;110:2163–7. doi: 10.1161/01.CIR.0000144458.58660.BB. [DOI] [PubMed] [Google Scholar]
- 46.Olson TM, Michels VV, Ballew JD, et al. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA. 2005;293:447–54. doi: 10.1001/jama.293.4.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor MR, Slavov D, Gajewski A, et al. Thymopoietin (lamina-associated polypeptide 2) gene mutation associated with dilated cardiomyopathy. Hum Mutat. 2005;26:566–74. doi: 10.1002/humu.20250. [DOI] [PubMed] [Google Scholar]
- 48.Knoll R, Postel R, Wang J, et al. Laminin-alpha4 and integrin-linked kinase mutations cause human cardiomyopathy via simultaneous defects in cardiomyocytes and endothelial cells. Circulation. 2007;116:515–25. doi: 10.1161/CIRCULATIONAHA.107.689984. [DOI] [PubMed] [Google Scholar]
- 49.Olson TM, Illenberger S, Kishimoto NY, Huttelmaier S, Keating MT, Jockusch BM. Metavinculin mutations alter actin interaction in dilated cardiomyopathy. Circulation. 2002;105:431–7. doi: 10.1161/hc0402.102930. [DOI] [PubMed] [Google Scholar]
- 50.Vatta M, Mohapatra B, Jimenez S, et al. Mutations in Cypher/ZASP in patients with dilated cardiomyopathy and left ventricular non-compaction. J Am Coll Cardiol. 2003;42:2014–27. doi: 10.1016/j.jacc.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 51.Hayashi T, Arimura T, Itoh-Satoh M, et al. Tcap gene mutations in hypertrophic cardiomyopathy and dilated cardiomyopathy. J Am Coll Cardiol. 2004;44:2192–201. doi: 10.1016/j.jacc.2004.08.058. [DOI] [PubMed] [Google Scholar]
- 52.Li D, Parks SB, Kushner JD, et al. Mutations of presenilin genes in dilated cardiomyopathy and heart failure. Am J Hum Genet. 2006;79:1030–9. doi: 10.1086/509900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohapatra B, Jimenez S, Lin JH, et al. Mutations in the muscle LIM protein and alpha-actinin-2 genes in dilated cardiomyopathy and endocardial fibroelastosis. Mol Genet Metab. 2003;80:207–15. doi: 10.1016/s1096-7192(03)00142-2. [DOI] [PubMed] [Google Scholar]
- 54.Inagaki N, Hayashi T, Arimura T, et al. Alpha B-crystallin mutation in dilated cardiomyopathy. Biochem Biophys Res Commun. 2006;342:379–86. doi: 10.1016/j.bbrc.2006.01.154. [DOI] [PubMed] [Google Scholar]
- 55.Olson TM, Kishimoto NY, Whitby FG, Michels VV. Mutations that alter the surface charge of alpha-tropomyosin are associated with dilated cardiomyopathy. J Mol Cell Cardiol. 2001;33:723–32. doi: 10.1006/jmcc.2000.1339. [DOI] [PubMed] [Google Scholar]
- 56.Lakdawala NK, Dellefave L, Redwood CS, et al. Familial dilated cardiomyopathy caused by an alpha-tropomyosin mutation: the distinctive natural history of sarcomeric dilated cardiomyopathy. J Am Coll Cardiol. 2010;55:320–9. doi: 10.1016/j.jacc.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bienengraeber M, Olson TM, Selivanov VA, et al. ABCC9 mutations identified in human dilated cardiomyopathy disrupt catalytic KATP channel gating. Nat Genet. 2004;36:382–7. doi: 10.1038/ng1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olson TM, Michels VV, Thibodeau SN, Tai YS, Keating MT. Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science. 1998;280:750–752. doi: 10.1126/science.280.5364.750. [DOI] [PubMed] [Google Scholar]
- 59.Mayosa B, Khogali S, Zhang B, Watkins H. Cardiac and skeletal actin gene mutations are not a common cause of dilated cardiomyopathy. J Med Genet. 1999;36:796–797. doi: 10.1136/jmg.36.10.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takai E, Akita H, Shiga N, et al. Mutational analysis of the cardiac actin gene in familial and sporadic dilated cardiomyopathy. Am J Med Genet. 1999;86:325–327. doi: 10.1002/(sici)1096-8628(19991008)86:4<325::aid-ajmg5>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 61.Tesson F, Sylvius N, Pilotto A, et al. Epidemiology of desmin and cardiac actin gene mutations in a european population of dilated cardiomyopathy [In Process Citation] Eur Heart J. 2000;21:1872–6. doi: 10.1053/euhj.2000.2245. [DOI] [PubMed] [Google Scholar]
- 62.Zolty R, Brodsky G, Perryman B, Bristow M, Mestroni L. Epidemiology of cardiac actin gene mutations in dilated cardiomyopathy. J Cardiac Failure. 1999;5(Supple 1):23. [Google Scholar]
- 63.Taylor MR, Slavov D, Ku L, et al. Prevalence of desmin mutations in dilated cardiomyopathy. Circulation. 2007;115:1244–51. doi: 10.1161/CIRCULATIONAHA.106.646778. [DOI] [PubMed] [Google Scholar]
- 64.Arola AM, Sanchez X, Murphy RT, et al. Mutations in PDLIM3 and MYOZ1 encoding myocyte Z line proteins are infrequently found in idiopathic dilated cardiomyopathy. Mol Genet Metab. 2007;90:435–40. doi: 10.1016/j.ymgme.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 65.Murphy RT, Mogensen J, Shaw A, Kubo T, Hughes S, McKenna WJ. Novel mutation in cardiac troponin I in recessive idiopathic dilated cardiomyopathy. Lancet. 2004;363:371–2. doi: 10.1016/S0140-6736(04)15468-8. [DOI] [PubMed] [Google Scholar]
- 66.Carballo S, Robinson P, Otway R, et al. Identification and functional characterization of cardiac troponin I as a novel disease gene in autosomal dominant dilated cardiomyopathy. Circ Res. 2009;105:375–82. doi: 10.1161/CIRCRESAHA.109.196055. [DOI] [PubMed] [Google Scholar]
- 67.Schmitt JP, Kamisago M, Asahi M, et al. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science. 2003;299:1410–3. doi: 10.1126/science.1081578. [DOI] [PubMed] [Google Scholar]
- 68.Haghighi K, Kolokathis F, Pater L, et al. Human phospholamban null results in lethal dilated cardiomyopathy revealing a critical difference between mouse and human. J Clin Invest. 2003;111:869–76. doi: 10.1172/JCI17892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DeWitt MM, MacLeod HM, Soliven B, McNally EM. Phospholamban R14 deletion results in late-onset, mild, hereditary dilated cardiomyopathy. J Am Coll Cardiol. 2006;48:1396–8. doi: 10.1016/j.jacc.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 70.Medin M, Hermida-Prieto M, Monserrat L, et al. Mutational screening of phospholamban gene in hypertrophic and idiopathic dilated cardiomyopathy and functional study of the PLN -42 C>G mutation. Eur J Heart Fail. 2007;9:37–43. doi: 10.1016/j.ejheart.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 71.Li D, Tapscoft T, Gonzalez O, et al. Desmin mutation responsible for idiopathic dilated cardiomyopathy. Circ. 1999;100:461–464. doi: 10.1161/01.cir.100.5.461. [DOI] [PubMed] [Google Scholar]
- 72.Karkkainen S, Miettinen R, Tuomainen P, et al. A novel mutation, Arg71Thr, in the delta-sarcoglycan gene is associated with dilated cardiomyopathy. J Mol Med. 2003;15:15. doi: 10.1007/s00109-003-0480-5. [DOI] [PubMed] [Google Scholar]
- 73.Tsubata S, Bowles KR, Vatta M, et al. Mutations in the human delta-sarcoglycan gene in familial and sporadic dilated cardiomyopathy. J. Clin. Invest. 2000;106:655–662. doi: 10.1172/JCI9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sylvius N, Duboscq-Bidot L, Bouchier C, et al. Mutational analysis of the beta- and delta-sarcoglycan genes in a large number of patients with familial and sporadic dilated cardiomyopathy. Am J Med Genet. 2003;120A:8–12. doi: 10.1002/ajmg.a.20003. [DOI] [PubMed] [Google Scholar]
- 75.Knoll R, Hoshijima M, Hoffman HM, et al. The Cardiac Mechanical Stretch Sensor Machinery Involves a Z Disc Complex that Is Defective in a Subset of Human Dilated Cardiomyopathy. Cell. 2002;111:943–55. doi: 10.1016/s0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- 76.Gerull B, Gramlich M, Atherton J, et al. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat Genet. 2002;14:14. doi: 10.1038/ng815. [DOI] [PubMed] [Google Scholar]
- 77.Gerull B, Atherton J, Geupel A, et al. Identification of a novel frameshift mutation in the giant muscle filament titin in a large Australian family with dilated cardiomyopathy. J Mol Med. 2006;84:478–83. doi: 10.1007/s00109-006-0060-6. [DOI] [PubMed] [Google Scholar]
- 78.Schonberger J, Wang L, Shin JT, et al. Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat Genet. 2005;37:418–22. doi: 10.1038/ng1527. [DOI] [PubMed] [Google Scholar]
- 79.Duboscq-Bidot L, Charron P, Ruppert V, et al. Mutations in the ANKRD1 gene encoding CARP are responsible for human dilated cardiomyopathy. Eur Heart J. 2009;30:2128–36. doi: 10.1093/eurheartj/ehp225. [DOI] [PubMed] [Google Scholar]
- 80.Towbin JA, Hejtmancik JF, Brink P, et al. X-linked dilated cardiomyopathy. Molecular genetic evidence of linkage to the Duchenne muscular dystrophy (dystrophin) gene at the Xp21 locus. Circulation. 1993;87:1854–65. doi: 10.1161/01.cir.87.6.1854. [DOI] [PubMed] [Google Scholar]
- 81.Muntoni F, Cau M, Ganau A, et al. Brief report: Deletion of the dystrophin muscle-promoter region associated with x-linked dilated cardiomyopathy. N Engl J Med. 1993;329:921–925. doi: 10.1056/NEJM199309233291304. [DOI] [PubMed] [Google Scholar]
- 82.D'Adamo P, Fassone L, Gedeon A, et al. The x-linked gene G4.5 is responsible for different infantile dilated cardiomyopathies. Am J Hum Genet. 1997;61:862–867. doi: 10.1086/514886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bione S, D'Adamo P, Maestrini E, Gedeon A, Bolhuis P, Toniolo D. A novel X-linked gene, G4.5, is responsible for Barth syndrome. Nat Genet. 1996;12:385–389. doi: 10.1038/ng0496-385. [DOI] [PubMed] [Google Scholar]
- 84.Elliott P, O'Mahony C, Syrris P, et al. Prevalence of desmosomal protein gene mutations in patients with dilated cardiomyopathy. Circ Cardiovasc Genet. 2010;3:314–22. doi: 10.1161/CIRCGENETICS.110.937805. [DOI] [PubMed] [Google Scholar]
- 85.Morales A, Painter T, Li R, et al. Rare variant mutations in pregnancy-associated or peripartum cardiomyopathy. Circulation. 2010;121:2176–82. doi: 10.1161/CIRCULATIONAHA.109.931220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van Spaendonck-Zwarts KY, van Tintelen JP, van Veldhuisen DJ, et al. Peripartum cardiomyopathy as a part of familial dilated cardiomyopathy. Circulation. 2010;121:2169–75. doi: 10.1161/CIRCULATIONAHA.109.929646. [DOI] [PubMed] [Google Scholar]
- 87.Anderson JL, Horne BD. Birthing the genetics of peripartum cardiomyopathy. Circulation. 2010;121:2157–9. doi: 10.1161/CIRCULATIONAHA.110.956169. [DOI] [PubMed] [Google Scholar]
- 88.Ho CY, MacRae CA. Defining the pathogenicity of DNA sequence variation. Circ Cardiovasc Genet. 2009;2:95–7. doi: 10.1161/CIRCGENETICS.109.864793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu Q, Dewey S, Nguyen S, Gomes AV. Malignant and benign mutations in familial cardiomyopathies: insights into mutations linked to complex cardiovascular phenotypes. J Mol Cell Cardiol. 2010;48:899–909. doi: 10.1016/j.yjmcc.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 90.Richards CS, Bale S, Bellissimo DB, et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 91.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–39. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–13. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Goode DL, Cooper GM, Schmutz J, et al. Evolutionary constraint facilitates interpretation of genetic variation in resequenced human genomes. Genome Res. 2010;20:301–10. doi: 10.1101/gr.102210.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hanson E, Hershberger RE. Genetic counseling and screening issues in familial dilated cardiomyopathy. J Genet Counseling. 2001;10:397–415. doi: 10.1023/A:1016641504606. [DOI] [PubMed] [Google Scholar]
- 95.Hershberger RE. Cardiovascular Genetic Medicine: Evolving Concepts, Rationale and Implementation. J Cardiovasc Trans Res. 2008;1:137–143. doi: 10.1007/s12265-008-9031-3. [DOI] [PubMed] [Google Scholar]
- 96.Judge DP. Use of genetics in the clinical evaluation of cardiomyopathy. JAMA. 2009;302:2471–6. doi: 10.1001/jama.2009.1787. [DOI] [PubMed] [Google Scholar]
- 97.Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–6. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hedges D, Burges D, Powell E, et al. Exome sequencing of a multigenerational human pedigree. PLoS One. 2009;4:e8232. doi: 10.1371/journal.pone.0008232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ng SB, Buckingham KJ, Lee C, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–5. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lupski JR, Reid JG, Gonzaga-Jauregui C, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362:1181–91. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Siva N. 1000 Genomes project. Nat Biotechnol. 2008;26:256. doi: 10.1038/nbt0308-256b. [DOI] [PubMed] [Google Scholar]
- 102.Hershberger RE, Kushner JK, Parks SP. [2007 July 10];Dilated Cardiomyopathy Overview. GeneReviews at GeneTests: Medical Genetics Information Resource (database online) 2008 initial posting, July 27, 2007:[Available from: http://www.genetests.org. [Google Scholar]
- 103.Genetic information nondiscrimination act (GINA) of 2008 [cited 2010 December 17];A brief history of GINA, with information for researchers and health care professionals. 2010 Available from: http://www.genome.gov/24519851. [Google Scholar]
- 104.Van Driest SL, Vasile VC, Ommen SR, et al. Myosin binding protein C mutations and compound heterozygosity in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:1903–10. doi: 10.1016/j.jacc.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 105.Richard P, Charron P, Carrier L, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–32. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 106.Ingles J, Doolan A, Chiu C, Seidman J, Seidman C, Semsarian C. Compound and double mutations in patients with hypertrophic cardiomyopathy: implications for genetic testing and counselling. J Med Genet. 2005;42:e59. doi: 10.1136/jmg.2005.033886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Girolami F, Ho CY, Semsarian C, et al. Clinical features and outcome of hypertrophic cardiomyopathy associated with triple sarcomere protein gene mutations. J Am Coll Cardiol. 2010;55:1444–53. doi: 10.1016/j.jacc.2009.11.062. [DOI] [PubMed] [Google Scholar]
- 108.Kelly M, Semsarian C. Multiple mutations in genetic cardiovascular disease: a marker of disease severity? Circ Cardiovasc Genet. 2009;2:182–90. doi: 10.1161/CIRCGENETICS.108.836478. [DOI] [PubMed] [Google Scholar]
- 109.Hershberger RE. A glimpse into multigene rare variant genetics: triple mutations in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2010;55:1454–5. doi: 10.1016/j.jacc.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tester DJ, Will ML, Haglund CM, Ackerman MJ. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm. 2005;2:507–17. doi: 10.1016/j.hrthm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 111.Bauce B, Nava A, Beffagna G, et al. Multiple mutations in desmosomal proteins encoding genes in arrhythmogenic right ventricular cardiomyopathy/dysplasia. Heart Rhythm. 2010;7:22–9. doi: 10.1016/j.hrthm.2009.09.070. [DOI] [PubMed] [Google Scholar]
- 112.Xu T, Yang Z, Vatta M, et al. Compound and digenic heterozygosity contributes to arrhythmogenic right ventricular cardiomyopathy. J Am Coll Cardiol. 2010;55:587–97. doi: 10.1016/j.jacc.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hershberger RE, Norton N, Morales A, Li D, Siegfried JD, Gonzalez-Quintana J. Coding sequence rare variants identified in MYBPC3, MYH6, TPM1, TNNC1, and TNNI3 from 312 patients with familial or idiopathic dilated cardiomyopathy. Circ Cardiovasc Genet. 2010;3:155–61. doi: 10.1161/CIRCGENETICS.109.912345. [DOI] [PMC free article] [PubMed] [Google Scholar]