Abstract
This paper reports the use of natural amino acids, the tripeptide β-strand mimic Hao, and the β-turn mimic δ-linked ornithine to generate water-soluble 54-, 78-, and 102-membered ring macrolactams. These giant macrocycles were efficiently prepared by synthesis of the corresponding protected linear peptides, followed by solution-phase cyclization and deprotection. The protected linear peptide precursors were synthesized on 2-chlorotrityl chloride resin by conventional Fmoc-based solid-phase peptide synthesis. Macrocyclization was typically performed using HCTU and N,N-diisopropylethylamine in DMF at ca. 0.5 mM concentration. The macrocycles were isolated in 13–45% overall yield after HPLC purification and lyophilization. 1D, 2D TOCSY, and 2D ROESY 1H NMR studies of the 54- and 78-membered ring macrolactams establish that these compounds fold to form β-sheet structures in aqueous solutions.
Introduction
Macrocyclization can impart exceptional stability and a variety of biological activities into peptides by enforcing β-sheet structures.1 Gramicidin S, Θ-defensin, and many cyclotides achieve their antimicrobial activities because of their unusual β-sheet macrolactam structures.2 Robinson and coworkers have extensively developed macrocyclic peptides that adopt β-sheet structures to present biologically essential epitopes of structured target proteins. These macrocyclic β-sheets have served as antimicrobial peptides, protease inhibitors, p53—HDM2 inhibitors, and Tat protein—transactivation response region RNA inhibitors.3
Macrocyclization provides an efficient strategy to promote and stabilize β-sheet structures by bringing otherwise remote residues into proximity. Nature helps enforce β-sheet structures or other well-defined structures in cyclotides through cyclization by linkage of the N- and C-termini with a peptide bond.1,4 Cyclization with side-chain disulfide bridges also occurs in natural antibacterial peptides, such as tachyplesins and protegrins.5 Gellman and coworkers have created macrocycles that form stable parallel and antiparallel β-sheets in water through either backbone linkage or side-chain linkage with disulfide bridges.6 Our research group has introduced macrocyclic β-sheets that are cyclized through δ-linked ornithine.7
Synthetic macrocyclic β-sheets can serve as chemical models of protein β-sheets and provide insights into complicated β-sheet interactions in biological systems. The Gellman and Waters groups have demonstrated that macrocyclic β-sheets can act as spectroscopic references for β-sheet structures formed by linear peptides.6a,8 Our research group has used 42-membered ring macrocyclic β-sheets to inhibit the aggregation of amyloidogenic peptides.9 We have also shown that 54-membered ring macrocyclic β-sheets can mimic protein quaternary structure through intermolecular β-sheet interactions.7b-f
Our research group previously introduced the amino acid Hao, a tripeptide β-strand mimic, and δ-linked ornithine (δOrn), a β-turn mimic.10 In our macrocyclic β-sheet peptides, Hao not only serves as a template to present one hydrogen-bonding edge for intramolecular hydrogen bonding but also minimizes edge-to-edge aggregation through intermolecular hydrogen bonding. δ-Linked ornithine serves as a β-turn mimic and helps induce β-hairpin structures. The 42- and 54-membered ring macrocyclic β-sheets 1 and 2 were constructed from these two key components, and most form well-defined non-aggregating β-sheets in water.

In developing these macrocyclic β-sheet peptides, we have become impressed with the reliability of macrocycles in enforcing β-sheet structure and the efficiency of macrocyclization in synthesis. In this paper, we use natural amino acids, Hao, and δ-linked ornithine to generate water-soluble 54-, 78-, and 102-membered ring macrolactams 3a–f, 4a and 4b, 5, and 6. These giant macrocycles were efficiently synthesized by conventional Fmoc-based solid-phase peptide synthesis of the corresponding protected linear peptides, followed by solution-phase cyclization and deprotection. Here, we describe the design, synthesis, and study of these giant macrocycles.
Results and Discussion
1. Synthesis of 54-, 78-, and 102-Membered Ring Macrolactams
Peptide 3a is a 54-membered ring macrolactam containing eight natural amino acids, two Hao units, and two δ-linked ornithines. We chose Lys-Glu pairs for the hydrogen-bonded positions of peptide 3a because of their pairwise statistical and empirical preference for β-sheet formation and to enhance the solubility of peptide 3a in water.11,12 We chose Phe-Leu pairs for the non-hydrogen-bonded positions, because they statistically favor β-sheet formation.12 The Lys-Glu and Phe-Leu pairs provide peptide 3a with two peptide strands Glu-Phe-Hao-Leu-Lys connected by two δOrn turns. The two Hao units in the middle of each peptide strand were designed to minimize aggregation by blocking intermolecular hydrogen bonding among the peptide strands. Although complementary polar and hydrophobic amino acids were selected for initial studies, this design feature was subsequently found to be unnecessary (Section 4, e.g., peptide 3f).
Peptide 3a was synthesized by conventional Fmoc-based solid-phase peptide synthesis on 2-chlorotrityl chloride resin, followed by solution-phase cyclization and deprotection. Scheme 1 summarizes this synthesis.13 Protected linear peptide 7a was synthesized by standard automated Fmoc-based solid-phase peptide synthesis,14 followed by cleavage from the resin with AcOH. A 54% yield of the unpurified protected linear peptide was obtained based on the loading of Boc-Orn(Fmoc)-OH on the resin.
SCHEME 1.
Synthesis of Peptide 3a
Protected linear peptide 7a was cyclized to the corresponding protected cyclic peptide 8a by using HCTU15and N,N-diisopropylethylamine (DIEA) in DMF.16 Although we routinely perform macrocyclization at ca. 0.5 mM concentration (e.g., 0.07 mmol peptide in 140 mL of DMF), we have cyclized protected linear peptide 7a at concentrations as high as 10 mM without observing polymerization.17 These macrocyclization conditions do not result in epimerization because the C-terminus of the protected linear peptide comprises an α-amino acid carbamate (Boc-NH-CHR-COOH), rather than an α-amino acid amide (RCO-NH-CHR-COOH). Final deprotection with trifluoroacetic acid (TFA) followed by reverse-phase HPLC purification produced peptide 3a as the TFA salt in 45% overall yield, based on the loading of Boc-Orn(Fmoc)-OH on the resin. HPLC analysis of protected linear peptide 7a and crude deprotected cyclized peptide 3a shows little degradation in purity on cyclization (Figure 1, left).
FIGURE 1.
Analytical HPLC traces in the syntheses of peptides 3a, 4a, and 6. HPLC data were recorded using an Agilent Zorbax SB-C18 column (50 mm × 4.6 mm) with a gradient of 5–100% CH3CN in H2O with 0.1% TFA and a flow of 1.0 mL/min over 20 minutes.
To determine whether we could generate larger macrocyclic peptides, we designed 78- and 102-membered ring macrocyclic homologues of 3a. Peptides 4a and 5 are both 78-membered ring macrolactams: peptide 4a contains two Hao units while peptide 5 contains four Hao units. We designed peptide 4a by directly extending both ends of the upper and lower peptide strands of peptide 3a to give Lys-Leu-Glu-Phe-Hao-Leu-Lys-Phe-Glu. This peptide strand comprises Hao in the middle, flanked by two tetrapeptides. We designed peptide 5 using the upper peptide strand Lys-Leu-Hao-Val-Hao-Phe-Glu and the lower peptide strand Lys-Leu-Hao-Ile-Hao-Phe-Glu. The four Hao units should provide peptide 5 with better protection from edge-to-edge aggregation through intermolecular hydrogen bonding. We chose Val and Ile for the middle positions of the upper and lower β-strands, because these residues favor β-sheet formation. The synthesis of peptides 4a and 5 followed the procedures described above. Cyclization of the corresponding protected linear peptides 9a and 10, followed by deprotection with TFA and reverse-phase HPLC purification, produced peptides 4a and 5 as the TFA salts in 32% and 23% overall yield, respectively. As in the synthesis of the smaller homologue, HPLC analysis of the linear precursor of peptide 4a and its cyclization product, crude peptide 4a, shows little degradation in purity on cyclization (Figure 1, middle).
Peptide 6 is a 102-membered ring macrolactam, an expanded homologue of peptide 5. We designed peptide 6 using a core structure similar to that of peptide 5, with the upper peptide strand Glu-Phe-Lys-Ile-Hao-Phe-Hao-Tyr-Glu-Val-Lys and the lower peptide strand Glu-Phe-Lys-Ile-Hao-Tyr-Hao-Tyr-Glu-Val-Lys. We synthesized peptide 6 using the procedures described above. Cyclization of the corresponding protected linear peptide 11, followed by deprotection with TFA and reverse-phase HPLC purification, produced peptide 6 as the TFA salt in 13% overall yield. Unlike the syntheses of the smaller macrocycles, HPLC analysis of the linear precursor of peptide 6 and its cyclization product, crude peptide 6, shows moderate degradation in purity on cyclization (Figure 1, right).
In all of these syntheses, the protected linear peptides are easily prepared by standard automated Fmoc-based solid-phase peptide synthesis. Macrocyclization of protected linear peptides 7a, 9a, 10, and 11 is surprisingly easy to achieve. The macrocyclization does not appear to cause polymerization and does not require highly dilute conditions in which large volumes of solvent, slow addition, or syringe pumps are used.17 After deprotection with TFA, peptides 3a, 4a, 5, and 6 are easy to purify by reverse-phase HPLC. The yields of these peptides drop from 45% to 13% when the macrocycles increase from a 54-membered ring to a 102-membered ring. The lower yield of peptide 6 may result from the formation of more impurities in the synthesis of the corresponding linear peptide and the further formation of impurities upon macrocyclization. The diminished yields and increasing impurities suggest that the syntheses of macrolactams above 102-membered rings may encounter more difficulties.
2. Design, Synthesis, and Study of a Cyclic Control
To test whether the efficient cyclization of these macrocyclic peptides requires the formation of a preorganized β-sheet structure to template cyclization, we designed and synthesized peptide 12, which cannot form a preorganized β-sheet structure. Peptide 12 is a homologue of peptide 3a in which two amino acids have been deleted from the “bottom” strand. The deletion makes the “bottom” strand shorter than the “top” strand and thus prevents the formation of a hydrogen-bonded β-sheet structure. Linear precursor 13 cyclized cleanly when subjected to the cyclization conditions used for the other macrocycles. Peptide 12 was isolated in 36% yield after macrocyclization, deprotection, and HPLC purification. The successful synthesis of peptide 12 suggests that macrocyclization of these linear peptides is able to proceed without preorganization through β-sheet formation. Although this experiment demonstrates that preorganization is not required for macrocyclization, it does not preclude that preorganization may facilitate macrocyclization.

3. 1H NMR Structural Studies of 54-, 78-, and 102-Membered Ring Macrolactams
1D, 2D TOCSY, and 2D ROESY 1H NMR studies of the 54-, 78-, and 102-membered ring macrolactams establish the folding of peptides 3a, 4a, and 5.18Interstrand NOEs are a hallmark of well-defined β-sheet structures. Most notably, antiparallel β-sheets exhibit a pattern of strong NOEs between the α-protons in non-hydrogen-bonded positions. δ-Linked ornithine forms a well-defined β-turnlike structure with characteristic NOEs and magnetic anisotropy.10e In a well-folded δOrn turn structure, the pro-S δ-proton (HδS) shows a strong NOE with the α-proton and the two diastereotopic δ-protons exhibit ca. 0.6 ppm magnetic anisotropy (ΔδδOrn) in water.10e Additional indicators of β-sheet folding, such as downfield shifting of the α-protons, appear to be less applicable to these macrolactams, possibly because of magnetic anisotropy of the Hao amino acid.
The 1H NMR spectrum of peptide 3a at 2.0 mM in D2O shows sharp, disperse resonances, indicating peptide 3a to be non-aggregating in water. The ROESY spectrum of peptide 3a exhibits a strong interstrand NOE associated with the twofold symmetrical contact between the α-protons of Leu and Phe and between the α- and pro-S δ-protons of the δOrn turns (Figure 2).19 These NOEs suggest peptide 3a to adopt a well-folded β-sheet structure in aqueous solution. The 600 MHz ROESY spectrum of peptide 3a in H2O/D2O (9:1) shows additional interstrand NH–NH and Hα–NH NOEs, further supporting the complete folding of peptide 3a in water. Figure 3 summarizes these key Hα – Hα , NH–NH, and Hα–NH NOEs.20 Peptide 3a exhibits a ΔδδOrn value of 0.59 ppm, suggesting it to contain well-folded δ Orn turns and thus to adopt a well-defined β-sheet structure. Collectively, both the strong interstrand NOEs and the large ΔδδOrn value of peptide 3a demonstrate that peptide 3a adopts a β-sheet structure in aqueous solution.
FIGURE 2.
Selected expansion of the ROESY spectrum of peptide 3a (2.0 mM in D2O at 500 MHz and 298 K with a 300-ms spin lock time).
FIGURE 3.
Key interstrand Hα–Hα, NH–NH, and Hα–NH NOEs of peptide 3a observed in H2O/D2O (9:1) at 2.0 mM and 298 K.
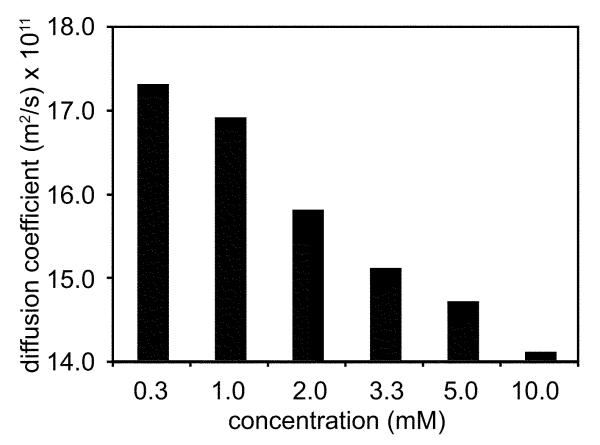
Upon increasing the concentration of peptide 3a to 10 mM, its 1H NMR spectrum broadens slightly, suggesting that self-association occurs at higher concentrations. To investigate the degree of self-association of peptide 3a, we measured its diffusion coefficients at different concentrations using DOSY (diffusion-ordered spectroscopy) experiments.21 Self-association diminishes the diffusion coefficient. Dimerization, for example, results in a smaller diffusion coefficient, about 0.7–0.8 that of the monomer. Peptide 3a exhibits decreasing diffusion coefficients form 17.3 × 10−11 m2/s to 14.1 × 10−11 m2/s upon increasing concentrations up to 10.0 mM in D2O at 298 K (Figure 4). These changes in diffusion coefficients suggest that peptide 3a is largely monomeric at submillimolar and low millimolar concentrations, but self-associates at higher concentrations.
Figure 4.
Diffusion coefficients of peptide 3a in D2O at 298 K.
The 600 MHz 1H NMR spectrum of peptide 4a shows sharp resonances in D2O at 1.0 mM and 298 K. The spectrum is severely broadened at concentrations above 5.0 mM, indicating self-association at higher concentrations. At 1.0 mM, peptide 4a exhibits a diffusion coefficient of 14.4 × 10−11 m2/s in D2O at 298 K, consistent with its larger size than peptide 3a and little self-association. Peptide 4a exhibits a ΔδδOrn value of 0.56 ppm and a strong NOE between the α- and pro-S δ-protons of the δOrn turns, indicating the δOrn turns of peptide 4a are in well-folded β-turnlike structures. Although the ROESY spectrum of peptide 4a exhibits a strong NOE between the α-protons of Leu and Phe, the α-proton resonances of the two types of Leu residues overlap, preventing unambiguous elucidation of key contacts between the Leu and Phe α-protons.
To address this ambiguity, we synthesized peptide 4b, a close homologue of peptide 4a in which the Leu-Phe pairs at positions 2–15 and 10–7 were replaced with Val-Tyr pairs. The ROESY spectrum of peptide 4b in D2O at 288 K shows strong interstrand NOEs between the Val and Tyr α-protons and between the Leu and Phe α-protons (Figure 5). These interstrand NOEs unambiguously demonstrate the β-sheet structure of peptide 4b. Peptide 4b exhibits a strong NOE between the α- and pro-S δ-protons of the δOrn turns and a ΔδδOrn value of 0.52 ppm, suggesting near complete folding of the δOrn turns. The interstrand NOEs and well-defined β-turn structures substantiate that peptide 4b adopts a well-folded β-sheet in aqueous solution.
FIGURE 5.
Selected expansion of the ROESY spectrum of peptide 4b (1.0 mM in D2O at 500 MHz and 288 K with a 200-ms spin lock time).
Peptide 5 shows severe broadening of the 1H NMR spectrum in D2O at submillimolar concentrations, suggesting severe self-association and precluding 1H NMR structural studies. For this reason, we studied peptide 5 in methanol (CD3OD) rather than in water. Although the 1H NMR spectrum of peptide 5 in CD3OD shows slight broadening, the resonances of peptide 5 can be assigned. The ROESY spectrum in CD3OD exhibits strong NOEs, indicating peptide 5 to adopt a β-sheet structure in methanol (Figure 6). Peptide 5 shows a ΔδδOrn value of 0.70 ppm in methanol, suggesting it to contain well-folded δOrn turns and thus to adopt a well-folded β-sheet structure. The greater ΔδδOrn value of peptide 5 in methanol may result from a more structured conformation through stronger hydrogen bonding in a solvent less competitive than water.22
FIGURE 6.
Key NOEs of peptide 5 observed in CD3OD at 1.0 mM and 298 K.
Peptide 6 exhibits severely broadened 1H NMR resonances both in water and in methanol, precluding further 1H NMR structural investigation in these solvents. The severe broadening of the 1H NMR spectra of peptides 5 and 6 leads to an interesting question: whether macrolactams containing more Hao units have greater propensity to self-associate in water. Both peptides 5 and 6 contain four Hao units, while peptides 3 and 4 only contain two. Although the Hao units help block aggregation through intermolecular edge-to-edge hydrogen bonding, they also impart more hydrophobic aromatic surfaces. The greater hydrophobic surface area may lead to more self-association through face-to-face hydrophobic interactions.
4. Effect of the Sequence on Folding of 54-Membered Ring Macrocyclic β-Sheets 3
To examine the effect of side-chain interactions in non-hydrogen-bonded positions on folding, we designed and synthesized peptides 3b–e in which the Phe-Leu side-chain pairs were respectively replaced with Leu-Leu, Phe-Phe, Tyr-Trp, and Phe-Tyr (Table 1). Peptides 3b–e show ΔδδOrn values of 0.26, 0.18, 0.58, and 0.35 ppm, respectively (Figure 7). The ΔδδOrn values suggest that peptide 3d forms a well-folded β-sheet in aqueous solution, while peptides 3b, 3c, and 3e form partially folded β-sheets. These studies demonstrate that the side-chain pairs in the non-hydrogen-bonded positions play a role in the folding of peptides 3 and that Phe-Leu and Tyr-Trp pairs lead to better folding.
TABLE 1.
Amino Acids at Positions R1-R8 of Peptides 3
| peptide | R1 | R2 | R3 | R4 | R5 | R6 | R7 | R8 |
|---|---|---|---|---|---|---|---|---|
| 3a | Glu | Phe | Leu | Lys | Glu | Phe | Leu | Lys |
| 3b | Glu | Leu | Leu | Lys | Glu | Leu | Leu | Lys |
| 3c | Glu | Phe | Phe | Lys | Glu | Phe | Phe | Lys |
| 3d | Glu | Tyr | Trp | Lys | Glu | Tyr | Trp | Lys |
| 3e | Glu | Phe | Tyr | Lys | Glu | Phe | Tyr | Lys |
| 3f | Val | Phe | Leu | Lys | Val | Phe | Leu | Lys |
FIGURE 7.
ΔδδOrn values of peptides 3 in D2O at 298 K.

To investigate whether the Lys-Glu pairs in the hydrogen-bonded positions are necessary to achieve the complete folding of peptides 3, we designed and synthesized peptide 3f, a close analogue of peptide 3a in which the two Glu residues are replaced with two Val residues. Peptide 3f exhibits a ΔδδOrn value of 0.63 ppm in D2O, suggesting it to be completely folded in aqueous solution. The complete folding of peptide 3f indicates that salt bridges between Lys and Glu are not necessary to stabilize the folded β-sheet structures. Consistent with this observation, the uncorrected pH of peptides 3 and 4 in D2O ranges from 1.9 to 2.8, and acidifying peptides 3a and 4b with 10 mM DCl has little effect upon their ΔδδOrn values.
5. Design, Synthesis, and Study of Acyclic Controls
To verify that macrocyclization promotes β-sheet formation, we designed peptides 14 and 15 as acyclic controls. Peptide 14 is a homologue of the “upper” peptide strand of peptide 3a, while peptide 15 is a homologue of both the “upper” and “lower” peptide strands linked by a single δOrn β-turn mimic. 1H NMR studies of peptide 14 in D2O do not show an NOE between the Leu and Phe α-protons, suggesting that peptide 14 does not dimerize to form β-sheetlike structures in aqueous solution. Consistent with a monomeric structure, peptide 14 exhibits a diffusion coefficient of 25.7 × 10−11 m2/s in D2O at 2.2 mM and 298 K. Peptide 15 also does not show NOEs between the Leu and Phe α-protons and only exhibits a ΔδδOrn value of 0.24 ppm in D2O at 1.9 mM and 298 K, suggesting the absence of significant β-sheetlike structure and only minimal folding in aqueous solution. The diffusion coefficient of peptide 15 under these conditions is 19.0 × 10−11 m2/s, which is comparable to those measured for peptides 3 and appropriately less than that of peptide 14. The absence of significant β-sheetlike dimerization of peptide 14 and folding of peptide 15 confirms that macrocyclization is necessary for creating β-sheet structure in peptide 3a.

Conclusion
These studies demonstrate that macrocyclization to synthesize giant macrolactams is surprisingly efficient. Macrocyclization does not require extremely dilute conditions to avoid polymerization in cyclizing the corresponding protected linear peptides. Although the present studies are focused on β-sheet structures, the efficient cyclization does not appear to require preorganization through β-sheet formation.
The water-soluble 54-, 78-, and 102-membered ring macrocyclic peptides prepared in these studies are among the largest synthetic macrolactams. These giant macrolactams are easy to synthesize and purify. Macrocyclization requires a few hundreds of milliliters of DMF to produce tens of milligrams of macrocyclic peptides as fluffy white solids after purification and lyophilization. This synthesis provides sufficient quantity of samples for characterization and further studies. Macrocyclization is an effective means to promote and stabilize β-sheet structures, and 1H NMR structural studies show that macrolactams 3 and 4 adopt β-sheet structures in aqueous solution.
We envision giant macrolactams to be a fruitful area for further research. With incorporation of biologically relevant amino acid sequences, macrocyclization may impart peptide-based macrolactams with unique three-dimensional structures and potential biological activities such as those of cyclotides. Moreover, large peptide-based macrolactam structures may be suitable for supramolecular chemistry and nanotechnology. It is not clear whether there is an upper limit to the size of the macrolactam structures that can be prepared, and the synthesis of even larger macrolactams may be envisioned.
Experimental Section
Representative Synthesis of Macrocyclic Peptide 3a.13
Step A
2-Chlorotrityl chloride resin (300 mg, 1.55 mmol/g) was added to a Bio-Rad Poly-Prep chromatography column (10 mL, 0.8 × 4.0 cm). The resin was washed with dry CH2Cl2 and suspended in 10 mL of dry CH2Cl2 to swell the resin. A solution of Boc-Orn(Fmoc)-OH (0.33 equiv, 70 mg, 0.15 mmol), 2,4,6-collidine (12 equiv, 0.24 mL, 220 mg) and 2.4 mL of dry CH2Cl2 was added directly to the resin, and the mixture was gently agitated for 4 h. The solution was then drained and the resin was washed with dry CH2Cl2 (5 times). A mixture of CH2Cl2/MeOH/DIEA (17:2:1, 10 mL) was added, and the suspension was gently agitated for 1 h to cap the unreacted 2-chlorotrityl chloride sites. The capping step was repeated to ensure completion. The resin was then washed with dry CH2Cl2 followed by DMF. The resin was dried by passing nitrogen through the vessel. The resin loading was determined to be 0.40 mmol/g (78% based on Boc-Orn(Fmoc)-OH) by UV analysis of the Fmoc cleavage product.13
Step B
The PS-2-chlorotrityl-Orn(Fmoc)-Boc generated from step A was washed with DMF (3 × 5 mL) and submitted to cycles of Fmoc-based solid-phase peptide synthesis on an automated peptide synthesizer using amino acid building blocks: Fmoc*-Hao-OH, Fmoc-Lys(Boc)-OH, Fmoc-Glu(tBu)-OH, Fmoc-Phe-OH, and Fmoc-Leu-OH. The linear peptide was elongated from the C-terminus to the N-terminus. Each coupling consisted of: (1) Fmoc-deprotection with 20% piperidine in DMF for 3 min; (2) washing with DMF (3 times); (3) coupling of the amino acid (0.5 mmol, 4 equiv) in the presence of HCTU; (4) washing with DMF (6 times). Each amino acid coupling step took 20 min for natural amino acids and 1 h for Hao. Because of the sluggish coupling of Fmoc*–Hao–OH into the growing peptide, the Hao coupling reaction was carried out twice without Fmoc* deprotection in between. After the last amino acid was coupled onto the growing peptide, the terminal Fmoc group was removed with 20% piperidine in DMF.
Step C
The resin with the linear peptide was transferred from the reaction vessel of the peptide synthesizer to a Bio-Rad Poly-Prep chromatography column and washed with DMF (3 × 5 mL) followed by CH2Cl2 (3 × 5 mL). A mixture of AcOH/CH2Cl2/MeOH (5:4:1, 20 mL) was added to the resin, and the suspension was agitated for 1 h. This cleavage solution was collected into a 250-mL round-bottom flask and the resin was washed with CH2Cl2 (3 × 10 mL). The combined solutions were concentrated by rotary evaporation under reduced pressure. Hexanes (ca. 100 mL) was added to the flask and then removed by rotary evaporation to azeotropically remove residual AcOH. The resulting yellowish oil was dissolved CH2Cl2 (ca. 5 mL), and the solution was diluted with hexanes (ca. 100 mL) and concentrated to dryness. The addition of CH2Cl2 and hexanes, followed by concentration, was repeated two additional times to completely remove residual AcOH. The residue was dried under vacuum to give 153 mg of crude protected linear peptide 7a as a white solid (54% crude yield based on resin loading).
Step D
Crude protected linear peptide 7a (153 mg, 0.068 mmol) was dissolved in DMF (70 mL). The peptide solution was added in drops via a 250-mL pressure-equalizing addition funnel to a 250-mL round-bottom flask containing a magnetic stirring bar, HCTU (111 mg, 0.27 mmol, 4 equiv), and DIEA (94 μL, 0.54 mmol, 8 equiv) in DMF (70 mL). (The final peptide concentration was 0.5 mM.) The reaction mixture was then stirred under nitrogen for 24 h. DMF was then removed by rotary evaporation under reduced pressure to give crude protected cyclic peptide 8a as a yellowish waxy solid. The crude product was used in final deprotection step E without further purification.
Step E
Crude protected cyclic peptide 8a was dissolved in TFA/triisopropylsilane/H2O (18:1:1, 20 mL) in a 50-mL round-bottom flask equipped with a nitrogen-inlet adaptor. The solution was magnetically stirred for 2 h. The reaction mixture was then concentrated by rotary evaporation under reduced pressure to give crude peptide 3a as a yellowish oil. Crude peptide 3a was purified by reverse-phase HPLC (gradient elution with 20–50% CH3CN over 60 min). The pure fractions were lyophilized to give 110 mg of peptide 3a (45% yield, based on resin loading): 1H NMR, (500 MHz, D2O, 298 K) δ 8.24 (appar. dd, J = 7.6, 2.7 Hz, 2 H), 7.99 (d, J = 2.7 Hz, 2 H), 7.23 (d, J = 9.2 Hz, 2H), 6.92 (d, J = 7.2 Hz, 4H), 6.82 (br s, 4H), 6.55 (br s, 2H), 4.75 (dd, J = 8.1, 4.7 Hz, 2H), 4.60 (dd, J = 9.6, 5.9 Hz, 2H), 4.41 (t, J = 7.0 Hz, 2H), 4.36 (d, J = 11.6 Hz, 2H), 4.26 (t, J = 6.8 Hz, 2H), 3.98 (s, 6H), 3.65 (t, J = 11.9 Hz, 2H), 3.13–3.02 (m, 6H), 2.72 (t, J = 7.6 Hz, 4H), 2.53 (t, J = 7.4 Hz, 4H), 2.29–2.26 (m, 2H), 2.10–2.02 (m, 2H), 1.87–1.82 (m, 2H), 1.76–1.56 (m, 14H), 1.51–1.40 (m, 6H), 1.32–1.24 (m, 2H), 1.24–1.15 (m, 2H), 0.85 (d, J = 6.5 Hz, 6H), 0.76 (d, J = 6.4 Hz, 6H); ESIMS m/z for C82H118N20O22 [M + 2H]2+ calcd 867.4, found 867.4.
Supplementary Material
Acknowledgment
We thank the NIH and NSF for grant support, Jing Zheng and Andrew Lam for developing procedures for preparing Fmoc-Hao-OH, and Dr. Philip R. Dennison for assistance with NMR studies.
Footnotes
Supporting Information Available: Full citation for reference 3f. NMR, ESIMS, HPLC data, and details of synthesis of peptides 3, 4, 5, 6, 12, 14, 15. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.a Cascales L, Craik DJ. Org. Biomol. Chem. 2010;8:5035–5047. doi: 10.1039/c0ob00139b. [DOI] [PubMed] [Google Scholar]; b Jagadish K, Camarero JA. Pept. Sci. 2010;94:611–616. doi: 10.1002/bip.21433. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Gerlach SL, Rathinakumar R, Chakravarty G, Göransson U, Wimley WC, Darwin SP, Mondal D. Pept. Sci. 2010;94:617–625. doi: 10.1002/bip.21435. [DOI] [PubMed] [Google Scholar]; d Mylne JS, Wang CK, van der Weerden NL, Craik DJ. Pept. Sci. 2010;94:635–646. doi: 10.1002/bip.21419. [DOI] [PubMed] [Google Scholar]; e Colgrave ML, Korsinczky MJL, Clark RJ, Foley F, Craik DJ. Pept. Sci. 2010;94:665–672. doi: 10.1002/bip.21415. [DOI] [PubMed] [Google Scholar]
- 2.a Kondejewski LH, Farmer SW, Wishart DS, Kay CM, Hancock REW, Hodges RS. J. Biol. Chem. 1996;271:25261–25268. doi: 10.1074/jbc.271.41.25261. [DOI] [PubMed] [Google Scholar]; b Gibbs AC, Kondejewski LH, Gronwald W, Nip AM, Hodges RS, Sykes BD, Wishart DS. Nat. Struct. Biol. 1998;5:284–288. doi: 10.1038/nsb0498-284. [DOI] [PubMed] [Google Scholar]; c Gibbs AC, Bjorndahl TC, Hodges RS, Wishart DS. J. Am. Chem. Soc. 2002;124:1203–1213. doi: 10.1021/ja011005e. [DOI] [PubMed] [Google Scholar]; d Gallo SA, Wang W, Rawat SS, Jung G, Waring AJ, Cole AM, Lu H, Yan X, Daly NL, Craik DJ, Jiang S, Lehrer RI, Blumenthal R. J. Biol. Chem. 2006;281:18787–18792. doi: 10.1074/jbc.M602422200. [DOI] [PubMed] [Google Scholar]; e Tran D, Tran P, Roberts K, Osapay G, Schaal J, Ouellette A, Selsted ME. Antimicrob. Agents Chemother. 2008;52:944–953. doi: 10.1128/AAC.01090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a Späth J, Stuart F, Jiang L, Robinson JA. Helv. Chim. Acta. 1998;81:1726–1738. [Google Scholar]; b Athanassiou Z, Patora K, Dias RLA, Moehle K, Robinson JA, Varani G. Biochemistry. 2006;46:741–751. doi: 10.1021/bi0619371. [DOI] [PubMed] [Google Scholar]; c Fasan R, Dias RLA, Moehle K, Zerbe O, Obrecht D, Mittl PRE, Grütter MG, Robinson JA. ChemBioChem. 2006;7:515–526. doi: 10.1002/cbic.200500452. [DOI] [PubMed] [Google Scholar]; d Robinson JA. Acc. Chem. Res. 2008;41:1278–1288. doi: 10.1021/ar700259k. [DOI] [PubMed] [Google Scholar]; e Robinson JA, et al. Science. 2010;327:1010–1013. [Google Scholar]
- 4.Conlan BF, Gillon AD, Craik DJ, Anderson MA. Pept. Sci. 2010;94:573–583. doi: 10.1002/bip.21422. [DOI] [PubMed] [Google Scholar]
- 5.a Tam JP, Lu Y-A, Yang J-L. Biochem. Biophys. Res. Commun. 2000;267:783–790. doi: 10.1006/bbrc.1999.2035. [DOI] [PubMed] [Google Scholar]; b Tam JP, Lu Y-A, Yang J-L. J. Biol. Chem. 2002;277:50450–50456. doi: 10.1074/jbc.M208429200. [DOI] [PubMed] [Google Scholar]; c Gottler LM, de la Salud Bea R, Shelburne CE, Ramamoorthy A, Marsh ENG. Biochemistry. 2008;47:9243–9250. doi: 10.1021/bi801045n. [DOI] [PubMed] [Google Scholar]
- 6.a Syud FA, Espinosa JF, Gellman SH. J. Am. Chem. Soc. 1999;121:11577–11578. [Google Scholar]; b Stanger HE, Syud FA, Espinosa JF, Giriat I, Muir T, Gellman SH. Proc. Natl. Acad. Sci. 2001;98:12015–12020. doi: 10.1073/pnas.211536998. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Espinosa JF, Syud FA, Gellman SH. Protein Sci. 2002;11:1492–1505. doi: 10.1110/ps.4140102. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Freire F, Gellman SH. J. Am. Chem. Soc. 2009;131:7970–7972. doi: 10.1021/ja902210f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a Woods RJ, Brower JO, Castellanos E, Hashemzadeh M, Khakshoor O, Russu WA, J. S. Nowick. J. Am. Chem. Soc. 2007;129:2548–2558. doi: 10.1021/ja0667965. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Khakshoor O, Demeler B, J. S. Nowick. J. Am. Chem. Soc. 2007;129:5558–5569. doi: 10.1021/ja068511u. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Khakshoor O, J. S. Nowick. Curr. Opin. Chem. Biol. 2008;12:722–729. doi: 10.1016/j.cbpa.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; d J. S. Nowick. Acc. Chem. Res. 2008;41:1319–1330. doi: 10.1021/ar800064f. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Khakshoor O, J. S. Nowick. Org. Lett. 2009;11:3000–3003. doi: 10.1021/ol901015a. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Khakshoor O, Lin AJ, Korman TP, Sawaya MR, Tsai S-C, Eisenberg D, J. S. Nowick. J. Am. Chem. Soc. 2010;132:11622–11628. doi: 10.1021/ja103438w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tatko CD, M. L. Waters. J. Am. Chem. Soc. 2002;124:9372–9373. doi: 10.1021/ja0262481. [DOI] [PubMed] [Google Scholar]
- 9.Zheng J, Liu C, Sawaya MR, Vadla B, Khan S, Woods RJ, Eisenberg D, Goux WJ, J. S. Nowick. J. Am. Chem. Soc. 2011;133:3144–3157. doi: 10.1021/ja110545h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a Nowick JS, Tsai JH, Bui Q-CD, S. Maitra. J. Am. Chem. Soc. 1999;121:8409–8410. [Google Scholar]; b Nowick JS, Chung DM, Maitra K, Maitra S, Stigers KD, Y. Sun. J. Am. Chem. Soc. 2000;122:7654–7661. [Google Scholar]; c Nowick JS, Lam KS, Khasanova TV, Kemnitzer WE, Maitra S, Mee HT, R. Liu. J. Am. Chem. Soc. 2002;124:4972–4973. doi: 10.1021/ja025699i. [DOI] [PubMed] [Google Scholar]; d Nowick JS, D. M. Chung. Angew. Chem., Int. Ed. 2003;42:1765–1768. doi: 10.1002/anie.200250750. [DOI] [PubMed] [Google Scholar]; e Nowick JS, J. O. Brower. J. Am. Chem. Soc. 2003;125:876–877. doi: 10.1021/ja028938a. [DOI] [PubMed] [Google Scholar]; f Chung DM, J. S. Nowick. J. Am. Chem. Soc. 2004;126:3062–3063. doi: 10.1021/ja031632z. [DOI] [PubMed] [Google Scholar]; g Chung DM, Dou Y, Baldi P, J. S. Nowick. J. Am. Chem. Soc. 2005;127:9998–9999. doi: 10.1021/ja052351p. [DOI] [PubMed] [Google Scholar]; h J. S. Nowick. Org. Biomol. Chem. 2006;4:3869–3885. doi: 10.1039/b608953b. [DOI] [PubMed] [Google Scholar]
- 11.a Smith CK, L. Regan. Science. 1995;270:980–982. doi: 10.1126/science.270.5238.980. [DOI] [PubMed] [Google Scholar]; b Smith CK, L. Regan. Acc. Chem. Res. 1997;30:153–161. [Google Scholar]
- 12.Wouters MA, P. M. G. Curmi. Proteins: Struct, Funct., Genet. 1995;22:119–131. doi: 10.1002/prot.340220205. [DOI] [PubMed] [Google Scholar]
- 13.Most standard peptide synthesis procedures followed those in the “Synthesis Notes” section of the 2009/2010 Novabiochem catalog.
- 14.Either Fmoc*-Hao-OH or Fmoc-Hao-OH was used to introduce the Hao residue. Fmoc*-Hao-OH has been described previously in reference 10b. Fmoc* refers to the 2,7-di-tert-butyl-Fmoc group. This more soluble analogue of the Fmoc group is described in Stigers KD, Koutroulis MR, Chung DM, J. S. Nowick. J. Org. Chem. 2000;65:3858–3860. doi: 10.1021/jo9915832. For details of the preparation of Fmoc-Hao-OH, see the supporting information.
- 15.a Montalbetti CAGN, V. Falque. Tetrahedron. 2005;61:10827–10852. [Google Scholar]; b Hood CA, Fuentes G, Patel H, Page K, Menakuru M, J. H. Park. J. Pept. Sci. 2008;14:97–101. doi: 10.1002/psc.921. [DOI] [PubMed] [Google Scholar]; c Valeur E, M. Bradley. Chem. Soc. Rev. 2009;38:606–631. doi: 10.1039/b701677h. [DOI] [PubMed] [Google Scholar]
- 16.In the cyclization step, most macrocyclic peptides were synthesized using HCTU/DIEA/DMF. Cyclization using HCTU/DIEA/DMF produces yellowish impurities that remain after deprotection with TFA. When these colored impurities proved difficult to remove by HPLC purification, cyclization was alternatively performed with HBTU/HOBt/N-methylmorpholine/DMF.15
- 17.HPLC analysis of cyclic peptide 8a shows comparable purities of the crude products from cyclization at 0.5 and 10 mM concentrations, indicating that macrocyclization causes no or little polymerization. For details, see the Supporting Information, Figure S1. If polymers or oligomers were to form under these conditions, the HPLC traces from cyclization at 10 mM would show substantially greater impurity than that at 0.5 mM.
- 18.1H NMR studies were performed in D2O with DSA as an internal standard. DSA is described in Nowick JS, Khakshoor O, Hashemzadeh M, J. O. Brower. Org. Lett. 2003;5:3511–3513. doi: 10.1021/ol035347w. Resonances were assigned by 1D, 2D TOCSY, and 2D ROESY 1H NMR studies. 1H NMR studies of peptide 3a were also performed in H2O/D2O (9:1) with water suppression to observe amide resonances and allow sequence-specific assignment of the resonances.
- 19.NOEs between the two Hao units cannot be observed due to symmetry.
- 20.The hydrazide N-H resonances are broadened by chemical exchange with H2O, and the NOEs involving these protons can not be observed under conditions of the ROESY experiments.
- 21.a Yao S, Howlett GJ, R. S. Norton. J. Biomol. NMR. 2000;16:109–119. doi: 10.1023/a:1008382624724. [DOI] [PubMed] [Google Scholar]; b Cohen Y, Avram L, L. Frish. Angew. Chem., Int. Ed. 2005;44:520–554. doi: 10.1002/anie.200300637. [DOI] [PubMed] [Google Scholar]
- 22.Previous studies have shown that a well-folded δOrn turn exhibits a Δδδ Orn value of 1.3 ppm in a chloroform solution. For details, see reference 10c.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.