Abstract
Strict one-to-one correspondence between codons and amino acids is thought to be an essential feature of the genetic code. However, here we report that one codon can code for two different amino acids with the choice of the inserted amino acid determined by a specific 3′-UTR structure and location of the dual-function codon within the mRNA. We found that UGA specifies insertion of selenocysteine and cysteine in the ciliate Euplotes crassus, that the dual use of this codon can occur even within the same gene, and that the structural arrangements of Euplotes mRNA preserve location-dependent dual function of UGA when expressed in mammalian cells. Thus, the genetic code supports the use of one codon to code for multiple amino acids.
Although codons can be recoded to specify other amino acids or to have ambiguous meanings (1, 2), and stop codons can be suppressed to insert amino acids (3), insertion of different amino acids into separate positions within nascent polypeptides by the same codeword is believed to be inconsistent with ribosome-based protein synthesis. In ciliated protozoa from the Euplotes genus, cysteine (Cys) is encoded by three codons, UGA, UGU and UGC (4, 5). UGA is a stop signal in the universal genetic code, and this codon can also code for the 21st amino acid, selenocysteine (Sec) (6).
Metabolic labeling with 75Se showed that E. crassus contains multiple selenoproteins (Fig. S1). To identify the codon used for Sec, we sequenced 15,000 E. crassus ESTs (Fig. S2) and the full-length eGPx1 cDNA sequence. The eGPx1 cDNA encodes a 22 kDa protein with a single in-frame UGA codon (Fig. 1A) and a Sec insertion sequence (SECIS) element (7) in its 3′-UTR (Fig. 1B), suggesting that this UGA encodes Sec. Therefore, UGA may be used for both Cys and Sec insertion in Euplotes. Expression of eGPx1 as a fusion protein with GFP in HEK 293 cells revealed specific 75Se incorporation (Fig. 1C). The corresponding full-size protein was also detected by Western blotting (Fig. 1D). Mutation of the core region of the eGPx1 SECIS element prevented 75Se incorporation and protein synthesis (Fig. 1C, D), indicating that SECIS was required for Sec insertion in response to UGA.
Fig. 1. E. crassus selenoproteins encode Sec with UGA codon as directed by SECIS element.
(A) Schematic representation of eGPx1 mRNA. The positions of the start (AUG), Sec (UGA) and stop (UAA) codons, and the SECIS element in the 3′-UTR are shown. (B) eGpx1 SECIS element. Core highlights the mutations made in the SECIS core. (C) Expression of GFP-eGPx1 fusion in HEK 293 cells. Cells were transfected with the pEGFP-C3 vector, pEGFP-eGPx1 construct or construct with mutations in the SECIS core (pEGFP-eGPx1core, see B), labeled with 75Se and analyzed as given in SOM. Selenoprotein patterns were visualized with a PhosphorImager on SDS-PAGE gels. (D) Western blot analysis of samples shown in C with anti-GFP antibodies. Arrows show the positions of GFP and GFP-eGPx1. (E) Expression of GFP-ep22 fusion protein in HEK 293 cells. Cells were transfected with the pEGFP-C3 vector or pEGFP-ep22 construct, labeled with 75Se and analyzed as in C. (F) Western blot analysis of samples shown in E with anti-GFP antibodies. Arrows show the positions of GFP and GFP-ep22. (G) Expression of GFP-eSelW2 fusion protein in HEK 293 cells. Arrow shows the position of the GFP-eSelW2 fusion selenoprotein. Molecular masses of protein standards (in kDa) are shown on the left.
E. crassus genome sequencing and analysis revealed eight selenoprotein genes (Fig. S3-S16) and three tRNAs that recognize UGA codons, including Sec tRNA, mitochondrial Trp tRNA and a novel Cys tRNA (Fig. 2A, S17). A Cys tRNA recognizing UGU and UGC codons was also detected. Four of the eight selenoprotein genes contained multiple UGA codons (Fig. S4). Comparison with known selenoproteins suggested the use of one codon for Sec and (an) additional UGA codon(s) within the same gene for Cys insertion. E. crassus thioredoxin reductases 1 (eTR1) and 2 (eTR2) had seven in-frame UGA codons, with the first six predicted to code for Cys and the last one to code for Sec (Fig. S5, S6 and S18). To examine coding functions of UGA codons, we cloned a novel selenoprotein ep22, selenoprotein W2 (eSelW2) and eTR1 (Fig. S5, S8, S10 and S19), and expressed them in the form of GFP-fusion proteins in HEK 293 cells. Specific 75Se incorporation was observed into GFP-ep22 (Fig. 1E, F) and GFP-eSelW2 (Fig. 1G), which had single UGA codons.
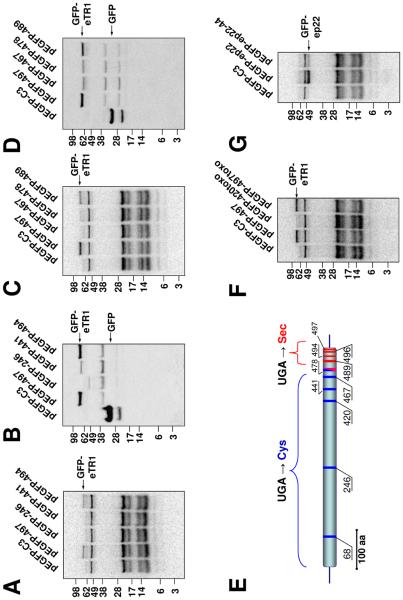
Fig. 2. Sec and Cys insertion in eTR1.
(A) Structures of E. crassus tRNAs. Sec tRNA, Cys tRNAs with UCA and GCA anticodons and a mitochondrial Trp tRNA are shown. Anticodons are highlighted in red (UCA) or blue (GCA). (B) Expression of GFP-eTR1 in HEK 293 cells. Cells were transfected with pEGFP-C3 vector (lane 1), pEGFP-eTR1 (lane 2) or constructs with multiple UGA to UGC mutations in which the number indicates the amino acid residue for which the UGA codon is retained: pEGFP-68 (lane 3), pEGFP-420 (lane 4) and pEGFP-497 (lane 5). Cells were analyzed as described in Figure 1C. Arrow shows the position of the GFP-eTR1 fusion selenoprotein. (C) Western blot analysis of samples shown in B with anti-GFP antibodies. Arrows show the positions of GFP and truncated and full-size GFP-eTR1. Asterisks show the position of truncated GFP-eTR1 fusions in lanes 4 and 5. (D) Partially purified eTR sample analyzed by 2D-PAGE and stained with Coomassie Blue. (E) Visualization of the 75Se-labeled sample shown in D with a PhosphorImager. The spots of eGR are indicated by a blue oval (in D) and of eTR by red ovals (in D and E).
In the case of GFP-eTR1, we initially did not observe 75Se incorporation (Fig. 2B, lane 2). This was likely due to termination at UGAs coding for Cys in Euplotes, which were recognized as stop signals in mammalian cells. We therefore prepared mutant forms of GFP-eTR1, in which six of the seven UGA codons were replaced with UGC, leaving single UGA at positions 68, 420 or 497. Of these, amino acids 68 and 420 corresponded to Cys, and 497 corresponded to Sec in other TRs. We found that 75Se (and therefore, Sec) could be inserted only at position 497 (Fig. 2B, lane 5). Western blotting confirmed the synthesis of truncated proteins when UGA was at positions 68 and 420, and of the full-size protein at position 497 (Fig. 2C). Thus, Sec was only inserted into the classical Sec site in eTR1, whereas other UGA positions were not served by SECIS for Sec insertion and instead supported termination of translation in mammalian cells (in Euplotes, Cys would be inserted).
To confirm Cys insertion at UGA codons other than codon 497 in eTR1, we purified the 75Se-labeled 55 kDa selenoprotein band from E. crassus after a series of chromatographic steps (Fig. 2D,E). LC-MS/MS sequencing revealed peptides corresponding to eTR1 and a more abundant glutathione reductase (eGR) (Fig. S20-S22). This analysis identified eTR1 peptides containing Cys in positions 63, 68, 208, and 270, which are encoded by UGA codons (Fig. S5, S18), whereas peptides containing Sec at these positions were not detected. Thus, UGA differentially codes for Cys and Sec in different positions within the E. crassus eTR1 gene.
To determine whether Cys and/or Sec insertion is associated with UGA position within the gene, we prepared GFP-eTR1 mutants containing single UGA codons in unnatural codon positions: 246, 441, 467, 478, 489, 494 or 496. 75Se-labeling and Western blotting revealed that UGA terminated translation in positions 246, 441 and 467, but inserted Sec in positions 489 and 494 (Fig. 3A-E). Sec was also inserted at position 496 (Fig. S23), whereas position 478 was intermediate, supporting a low level of Sec insertion (Fig. 3C, D). Thus, Sec insertion was restricted to approximately the last 20 codons, whereas the region upstream supported termination by UGA in mammalian cells (and therefore, Cys insertion in E. crassus).
Fig. 3. Position-dependent Sec insertion in eTR1.
(A) Expression of GFP-eTR1 in HEK 293 cells. Cells were transfected with pEGFP-C3 vector, a GFP-eTR1 construct containing a single UGA codon at the natural Sec position 497 (pEGFP-497) or constructs that had UGA at unnatural positions 246 (pEGFP-246), 441 (pEGFP-441) or 494 (pEGFP-494). Cells were analyzed as described in Figure 1C. (B) Western blot analysis of samples shown in A with anti-GFP antibodies. (C) Cells were transfected with pEGFP-C3 vector, a GFP-eTR1 construct containing a single UGA codon at the natural Sec position 497 (pEGFP-497) or constructs that had UGA at unnatural positions 467 (pEGFP-467), 478 (pEGFP-478) or 489 (pEGFP-489). (D) Western blot analysis of samples shown in C with anti-GFP antibodies. (E) Summary of experimental evidence for Sec and Cys insertion in eTR1. The positions of Cys insertion (corresponding to termination in mammalian cells) are shown by blue, and Sec insertion by red lines. Position 478 supported low level Sec insertion. (F) Cells were transfected with pEGFP-C3 vector, a GFP-eTR1 construct containing a single UGA codon at the natural Sec position 497 (pEGFP-497) or constructs containing a 3'-UTR segment of Toxoplasma SelT and UGA at position 420 (pEGFP-420toxo) or 497 (pEGFP-497toxo). (G) Cells were transfected with pEGFP-C3 vector or with ep22 constructs in which UGA corresponded to positions 190 (pEGFP-ep22) or 44 (pEGFP-ep22-44). Arrows show the positions of GFP and full-size GFP-eTR1 or GFP-ep22.
We replaced a segment corresponding to part of the eTR1 3′-UTR, including the entire SECIS element, with the 3′-UTR region of Toxoplasma selenoprotein T (SelT), which also has a SECIS element (8). In this mutant, Sec insertion was detected at position 420, i.e., upstream of codon 478 (Fig. 3F), indicating that replacement of the functional 3′UTR region changed the coding function of UGA. Similarly, Sec could be inserted in the N-terminal region of ep22, in addition to its natural C-terminal penultimate position (Fig. 3G), suggesting a model wherein Sec insertion is dependent on an RNA structure (Fig. S24).
We demonstrate that UGA can designate different amino acids within the same gene, with the choice of the amino acid inserted determined by availability of the functional element within the 3′-UTR and the location of UGA within the gene. Although dual functions of stop codons have previously been described, they support the insertion of single amino acids (e.g., Sec or pyrrolysine) in competition with termination (9) or ambiguous codon function due to dual specificity of a particular tRNA (10). Here, we show that one codon supports specific insertion of multiple amino acids, indicating that evolutionary expansion of genetic code is possible.
One sentence summary.
One codon can naturally evolve to code for two different amino acids, even within one gene, with the choice of the inserted amino acid determined by an RNA structure in the 3′-untranslated region.
Supplementary Material
Acknowledgments
We thank Kasia Hammar for constructing the EST library and Irina Sorokina (Midwest Bio Services) for protein sequencing. Supported by NIH GM061603 and GM065204 to VNG, AI058054 to MLS, NSF 0343813 to LAK, and the Intramural Research Program, NCI, NIH, to DLH. Sequences for tRNA and selenoprotein genes have been deposited in the GenBank database under accession numbers FJ440148-FJ440159
References and notes
- 1.Namy O, Rousset JP, Napthine S, Brierley I. Mol Cell. 2004;13:157. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- 2.Atkins JF, Baranov PV. Nature. 2007;448:1004. doi: 10.1038/4481004a. [DOI] [PubMed] [Google Scholar]
- 3.Anderson JC, et al. Proc Natl Acad Sci USA. 2004;101:7566. doi: 10.1073/pnas.0401517101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osawa S, Jukes TH, Watanabe K, Muto A. Microbiol Rev. 1992;56:229. doi: 10.1128/mr.56.1.229-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer F, et al. Proc Natl Acad Sci USA. 1991;88:3758. [Google Scholar]
- 6.Hatfield DL, Gladyshev VN. Mol Cell Biol. 2002;22:3565. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry MJ, et al. Methods Enzymol. 2002;347:17. doi: 10.1016/s0076-6879(02)47004-8. [DOI] [PubMed] [Google Scholar]
- 8.Novoselov SV, et al. Proc Natl Acad Sci USA. 2007;104:7857. [Google Scholar]
- 9.Srinivasan G, James CM, Krzycki JA. Science. 2002;296:1459. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki T, Ueda T, Watanabe K. Embo J. 1997;16:1122. doi: 10.1093/emboj/16.5.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.