Abstract
Methionine sulfoxide reductases (Msrs) are enzymes that repair oxidized methionine residues in proteins. This function implicated Msrs in antioxidant defense and the regulation of aging. There are two known Msr types in animals: MsrA specific for the reduction of methionine-S-sulfoxide, and MsrB that catalyzes the reduction of methionine-R-sulfoxide. In a previous study, overexpression of MsrA in the nervous system of Drosophila was found to extend lifespan by 70%. Overexpression of MsrA in yeast also extended lifespan, whereas MsrB overexpression did so only under calorie restriction conditions. The effect of MsrB overexpression on lifespan has not yet been characterized in any animal model systems. Here, the GAL4-UAS binary system was used to drive overexpression of cytosolic Drosophila MsrB and mitochondrial mouse MsrB2 in whole body, fatbody, and the nervous system of flies. In contrast to MsrA, MsrB overexpression had no consistent effect on the lifespan of fruit flies on both corn meal and sugar yeast diets. Physical activity, fecundity, and stress resistance were also similar in MsrB-overexpressing and control flies. Thus, MsrA and MsrB, the two proteins with identical function in antioxidant protein repair, have different effects on aging in fruit flies.
Keywords: aging, lifespan, methionine sulfoxide reductase, Drosophila melanogaster, protein repair
1. Introduction
One of the prominent theories of aging is known as the free radical theory (Harman, 1956; Beckman and Ames, 1998). It postulates that an age-associated accumulation of macromolecules (e.g., proteins, nucleic acids, lipids) oxidatively damaged by reactive oxygen species (ROS) results in senescence. Oxidative damage appears from an imbalance among oxidant production, antioxidant defense, and repair processes. A logical prediction based on this theory is that the overexpression of antioxidant/repair enzymes should increase lifespan. Drosophila melanogaster is a short-lived model-organism that has been widely used to examine the correlation between antioxidant defense and aging; however, a number of conflicting data were obtained.
Strong evidence of lifespan extension in flies overexpressing Cu,Zn-superoxide dismutase (SOD) and Mn-SOD (Parkes et al., 1998; Sun and Tower, 1999; Sun et al., 2002; 2004) or neutral effect of catalase overexpression (Sun et al., 2002; Orr and Sohal, 1992) were found. The simultaneous overexpression of these enzymes in different combinations resulted in life-prolonging effects (Orr and Sohal, 1994; Sun et al., 2004). Additionally, flies lacking either SODs (Kirby et al., 2002; Phillips et al., 1989) or catalase (Orr et al., 1992) had reduced lifespan. However, other studies have shown that overexpression of these antioxidant enzymes in long-lived genetic background had no effect (Orr and Sohal, 2003; Seto et al, 1990) on survivorship. The simultaneous overexpression of Cu,Zn-SOD, Mn-SOD and catalase in different combinations in long-lived flies also did not result in lifespan extension (Orr et al., 2003) and even leaded to a decrease in lifespan by 43% for flies overexpressing Mn-SOD and catalase (Bayne et al., 2005). As the correlation between these enzymes and lifespan was not direct and the observed differences in lifespan of transgenic flies were the function of 1) genetic background, 2) tissue/cell distribution of antioxidants, 3) temporal pattern of expression, and 4) source of transgene and type of expression system it was suggested that antioxidant defense resulting from antioxidant enzymes is not a limiting factor for Drosophila aging (Helfand and Inouye, 2003; Mockett et al., 2001; Sohal et al., 2002). Recent studies showed that overexpression of two enzymes, glucose-6-phosphate dehydrogenase (Legan et al., 2008) and glutamate-cysteine ligase (Orr et al., 2005), that increase low molecular weight antioxidants in cells (NADPH and GSH, respectively), increases mean and maximum lifespan of long-lived Drosophila strain up to 50%. These studies, performed in one genetic background and with different transgene expression patterns, provide insights for understanding processes that favor long lifespan.
In Drosophila, concentrations of low molecular weight antioxidants (and their precursors) decline in senescence-associated manner (Mockett et al., 1999; Rebrin et al., 2004). GSH the most abundant low molecular weight antioxidant is considered as a major determinant of the cellular redox state. The reduced GSH/oxidized GSH (GSSG) ratio was decreased in exponential manner in the whole body of Drosophila (by over 70% from 10 to 60 days of age) (Rebrin et al., 2004). In the same time scale, the concentration of free methionine, which also serves as the source for cysteine production via the transsulfuration pathway, was decreased by 50% (Rebrin et al., 2004). Cysteine is a precursor for the GSH biosynthesis (Stipanuk, 2004). Various studies (reviewed in (Stadtman et al., 2005)) revealed an age-related increase in methionine sulfoxide (MetO) levels in different tissues and organisms.
The repair of age-related protein damage has, to some extent, been studied in Drosophila (Chavous et al., 2001; Ruan et al., 2002). A balance between oxidative modifications of proteins and their repair was proposed to play an important role in senescence. Almost all amino acids are susceptible to oxidation by radicals that cause damage to amino acid side chains as well as protein backbones (Levine and Stadtman, 1996; Sohal, 2002; Stadtman, 2008). Thus, Drosophila shows an age-related increase in modified and oxidized proteins (Chavous et al., 2001; Orr and Sohal, 1994; Sohal et al., 1993). To repair oxidized side chains of sulfur-containing amino acids, cysteine and methionine, cells have evolved enzymes specific for oxidized forms of these amino acids. For example, two conserved redox systems, the thioredoxin and glutathione systems, maintain intracellular thiol groups of proteins and low molecular weight thiols in the reduced state in organisms from bacteria to humans (Holmgren, 2008; Lou, 2008).
Methionine sulfoxide reductases (Msrs) occur in most organisms and catalyze the thioredoxin-dependent reduction of free and protein-bound MetO to methionine (Brot and Weissbach, 2000; Kim and Gladyshev, 2007; Moskovitz, 2005). Msrs as well as glutathione and thioredoxin reductases are ubiquitously expressed in cells (Holmgren, 2008; Kim and Gladyshev, 2007; Lou, 2008) and together with their substrates and cofactors form repair systems that protect cells from oxidative stress, and maintain cellular redox homeostasis (Holmgren, 2000; Moskovitz, 2005; Kim and Gladyshev, 2007; Levine et al., 2000).
Msrs are classified with respect to their substrate specificity into two types: MsrA that is specific for the reduction of methionine-S-sulfoxide (Met-SO) in both protein-bound and free forms, and MsrB that catalyzes only the reduction of methionine-R-sulfoxide (Met-RO) primarily in proteins. Msr genes are found in most organisms except some parasites and hypertheromophiles (Kim and Gladyshev, 2007). The number of MsrA and MsrB genes varies in different organisms. One MsrA and one MsrB are present in Escherichia coli, Saccharomyces cerevisiae, Caenorhabditis elegans and D. melanogaster. Mammals have one MsrA and three MsrBs (MsrB1, MsrB2, and MsrB3). Although MsrA and MsrB enzymes have the same function of MetO reduction, they are different not only in substrate specificity, but also in active site composition, protein fold, subcellular localization and evolution (Kim and Gladyshev, 2005, 2006, 2007; Kryukov et al., 2002).
Several studies have provided evidence for an important role of Msrs in aging. An age-related decrease in total Msr activity and MsrA levels was observed in rat liver and kidney (Petropoulos et al., 2001). Expression of MsrA and MsrB2 was decreased in senescent human WI-38 fibroblasts (Picot et al., 2004). MsrA null mutants of yeast (Kryukov et al., 2002; Moskovitz et al., 1997) and mice (Moskovitz et al., 2001) were more sensitive to oxidative stress than wild type organisms and had a shortened lifespan (decreased by ~26% in yeast (Koc et al., 2004) and ~40% in mice (Moskovitz et al., 1997)). Moreover, overexpression of MsrA in yeast (Moskovitz et al., 1998) and fruit flies (Ruan et al., 2002) led to protection against oxidative stress. In S. cerevisiae, overexpression of host MsrA extended lifespan by 25% (Koc et al., 2004). Most remarkably, expression of GFP-fused bovine MsrA in the nervous system of Drosophila increased the median lifespan of fruit flies by 70% (Ruan et al., 2002). Moreover, the authors observed that the MsrA-overexpressing transgenic flies had better physical activity, reproduction, and resistance to oxidative stress at the old age. Since MsrB is responsible for the reduction of one diastereomer of MetO (i.e., half of total MetO reduction), this enzyme is expected to have the same role in aging as MsrA. However, thus far, the effects of MsrB overexpression and knockout on aging were studied only in yeast (Koc et al., 2004). The knockout of MsrB gene had little effect on the lifespan. Moreover, the overexpression of MsrB in yeast had no effect on the lifespan under standard growth conditions, but prolonged the lifespan under caloric restriction conditions (Koc et al., 2004).
Since the most dramatic effect of MsrA overexpression on lifespan was observed in Drosophila (Ruan et al., 2002), it was of interest to examine whether MsrB overexpression affects lifespan in this organism. Further considering the findings on the roles of MsrA and MsrB on aging in yeast, it was important to determine the effects of MsrB overexpression under both regular and calorie restriction conditions. In this study, we developed transgenic fruit flies overexpressing mouse mitochondrial MsrB2 or Drosophila cytosolic MsrB, and used them to address these questions.
2. Materials and methods
2.1. Constructs
The ORF of mouse MsrB2 was PCR-amplified from a pET28a-based construct (Kim and Gladyshev, 2004) with 5’-AAAAGAATTCATGGCGCGACTGCTGCGAG-3’ and 5’-ACACCTCGAGTTAGGGCTTGCTGGGTTTGAACT-3’ primers and cloned into EcoRI/XhoI sites of pUAST vector (Brand and Perrimon, 1993) to yield pUAST-mMsrB2. The ORF of D. melanogaster MsrB was subcloned into EcoRI/XhoI sites of pUAST vector from pET28_MsrB construct (Kumar et al., 2002) by using 5’-ACCGGAATTCATGGATAACAAGAGCGAGAAG-3’ and 5’-AAAACTCGAGTCACTGCTGGGCAATG-3’ primers to yield pUAST-dMsrB. Constructs were verified by DNA sequencing.
2.2. Transgenic Drosophila lines
Transgenic flies were obtained using standard techniques for germline cell injection (Rubin and Spradling, 1982) and subsequent crosses. Briefly, white (w1118) embryos were co-injected with pUAST-mMsrB2 or pUAST-dMsrB constructs and the P{Δ2–3} plasmid carrying transposase (Robertson et al., 1988). Microinjection of the recombinant P-element plasmids yielded 8 independent transformants carrying UAS-mMsrB2 and 5 transformants carrying UAS-dMsrB transgenes. P-element insertion was mapped to specific chromosome by genetic segregation analysis. A transgenic male was crossed with virgin females carrying the second-chromosome balancer [w1118; amosTft/CyO] and third-chromosome balancer [w1118; : croc5F59e*/TM3, Sb1]. Phenotypic analysis of the offsprings allowed for generation of the homozygous lines for each MsrB with P-element insertion on X, second or third chromosomes. Two independent homozygous UAS-MsrB-responder lines for each MsrB were used in the study. These MsrB lines were designated as mMsrB22A, mMsrB23A (mouse MsrB2 lines) and dMsrB2A, dMsrB2B (Drosophila MsrB lines). The designations of transgenic lines display the transgene (i.e., mouse MsrB2, mMsrB2) followed by the chromosome of insertion (i.e., 2 or 3), followed by a letter (i.e., A or B lines) indicating the independent insertion on that chromosome. None of these insertions had influence on viability or development of homozygous UAS-MsrBs lines.
Two GAL4-activator lines used in this study, da-GAL4 [w*; ; P{w+mW.hs = GAL4-da.G32}UH1] (stock #5460), elav-GAL4 [w*, P{w+mW.hs = GawB}elavc155] (stock #458) and second-chromosome balancer w1118; amosTft/CyO (stock #424) were obtained from the Bloomington Drosophila Stock Center at Indiana University. The tubP-GAL4 [w*; P{w+mC=tubP-GAL4}/CyO] was kindly provided by J.P. Vincent (National Institute for Medical Research, London, U.K.). The third chromosome balancer [w1118; ; croc5F59e*/TM3, Sb1] (Häcker et al., 1995) and the fatbody-GAL4 driver [y*w*; P(w+mW.hs = GawB)FB P(w+m*UAS-GFP 1010T2)] (Grönke et al., 2003) was a kind gift from H. Jäckle and R.P. Kühnlein correspondingly (Max-Planck-Institut für Biophysikalische Chemie, Göttingen, Germany).
2.3. Fly culture media
Two food types were used in the lifespan experiments. The first was corn meal food (85.7 g corn meal “Aunt Jemina”; The Quaker Oats Company, Chicago, IL), 50 ml golden A unsulfured molasses (Groeb Farms Inc, Onsted, MI), 71.4 g Torula yeast (MP Biomedicals, Solon, OH), 2.86 g p-hydroxybenzoic acid methyl ester (Sigma), 6.4 g agar (MoorAgar Inc, Loomis, CA) and 5.7 ml propionic acid (Sigma) per liter water). The second type was sugar yeast (SY) food. It included 20 g agar, 3 g p-hydroxybenzoic acid methyl ester, 3 ml propionic acid, and variable amounts of sugar and yeast per liter of water. 50 g/L dextrose was present in the dietary restriction sugar yeast food (DR SY) and 100 g in normal food (N SY). Autolysed yeast powder (T.P. Drewitt, London, UK) was added in quantity 50 g per liter of water DR SY food and 100 g in N SY food (Chapman and Partridge, 1996).
2.4. Genetic crosses and Drosophila husbandry
The GAL4-UAS binary system (Brand and Perrimon, 1993) was used to drive overexpression of mouse mitochondrial MsrB2 and fruit fly cytosolic MsrB in Drosophila. To obtain heterozygous flies expressing mouse MsrB2 or Drosophila MsrB, homozygous mMsrB22A, mMsrB23A, dMsrB2A, and dMsrB2B virgin males were crossed to the corresponding virgin females containing the GAL4-driver. F1 offspring from crosses between GAL4 drivers and white flies (GAL4-activator/w1118) and between white flies and homozygous MsrB lines (w1118/UAS-MsrBs) were obtained for control concurrently. Experimental flies were reared on corn meal food from vials seeded with less than 70 eggs. Newly emerged flies were transferred to fresh corn meal food and allowed to mate for 1–2 days. 3-day-old mated flies were collected using ether, sorted by sex, and used for lifespan studies, paraquat resistance test, and starvation test. Unless otherwise stated, flies used in the experiment were held on the corn meal food and transferred to fresh vials without anesthesia every three days. Embryonic, larval, pupal, and adult animals were kept in temperature-controlled chamber at 25 °C with 12 h light/dark cycle and approximately 60% humidity.
2.5. Lifespan study
Newly eclosed adult animals were collected within 24 h and kept on corn meal food at density of 50 animals per vial. In a typical lifespan trial, 3-day-old mated flies were placed in cages. The cage was made from 1 L plastic container with a grommet for changing of fresh food vials and a slit for removal of dead flies by aspiration. 35 males and 35 females were kept in each cage for testing lifespan on sugar yeast or corn meal food. In one independent experiment, 70 mated males per cage were kept on dietary restriction SY food. Three replica cages were used for each lifespan simultaneously; survivor curves are presented the average of those independent replicas. Fresh food was supplied into cages and dead flies were removed by aspiration and counted every 3 days. Animals were maintained in temperature-controlled incubator at 25°C with 12 h light/dark cycle and 50–60% humidity. The control and experimental group trials were always performed concurrently.
2.6. Paraquat resistance test
Male flies overexpressing mouse MsrB2 or Drosophila MsrB in nervous system were prepared as described above (section 2.4) and cultured in half pint bottles on corn meal food at a density of 150 animals per bottle. 17-day-old flies were sorted in groups of 10 or 20 animals with etherization 3 days before the test. The test vials were set up with sponge soaked with 2 ml of 20 mM or 30 mM freshly prepared paraquat (Sigma) and 5% sucrose (Sigma) solution and covered with a 3MM filter paper circle. On the day of the test, flies were transferred without etherization to the test vials (for the experiment with 20 mM paraquat) or to vials with 1% agar and kept starved for 6 h (for the experiment with 30 mM paraquat). The starved flies were transferred to the test vials without etherization and the number of dead animals was counted at indicated time points. For the test with 20 mM paraquat, 8 replicates of 10 animals were used. For the test with 30 mM paraquat, 18 replicates of 20 animals were performed for each cross except for control cross elav-GAL4/w1118 where 9 replicates were examined. All data are reported as the means ±S.D.
2.7. Starvation resistance test
Male flies were prepared by standard techniques (Section 2.4) and maintained on corn meal food for 20 days prior to the test. Groups of 20 flies were transferred without etherization to vials with 6 ml 1% agar and kept in 12 h light/dark cycle at 25°C. Total number of flies tested for each genotype was 140 animals. Dead animals were counted every 8 h. All data are reported as the means ±S.D.
2.8. Reproduction
Age-specific changes in pupa production were determined from counts of pupal progeny produced by 5 females in 24 h. Females were placed in vials along with 5 males. 10 replicates of each vial were tested. Flies were transferred to vials with fresh food without anesthetization every 2–3 days. Females laid eggs for 24 h, then animals were removed and the number of pupa was counted after eclosion. All data are reported as the means ±S.D.
2.9. Enzyme activity assays
Homogenates of flies were prepared using the same procedure for enzyme activity assays, HPLC-based assay and Western blot analyses. Flies were homogenized in liquid nitrogen, resuspended in of PBS buffer containing protease inhibitor mixture (Roche), incubated on ice for 20 min and centrifuged at 16,000 × g for 20 min at 4 °C. Total protein concentration in supernatant was measured by the Bradford method using bovine serum albumin as a standard. 35 six-day-old males were resuspended in 350 µl of PBS buffer for activity measurements. MsrB activity was assayed in the supernatant by a standard procedure (Kumar et al., 2002). Briefly, the reaction mixture (100 µl) contained 20 mM dithiothreitol, 200 µM dabsyl-Met-RO, and 200 µg of total protein in PBS, pH 7.4. The reaction was carried out at 37°C for 30 min and stopped by adding 200 µl of acetonitrile. The reaction product, dabsyl-Met, was analyzed by an HPLC procedure using a C18 column (Zorbax Eclipse XDB, USA). Measurements for each sample were performed in triplicate. All data are reported as the means ±S.D.
2.10. Analysis of Met/MetO content in proteins and free amino acid forms
Homogenates of 70 female flies in 800 µl of PBS buffer containing protease inhibitors were prepared as in Section 2.9. For each type of MsrB-overexpressing fly (mouse MsrB2 or Drosophila MsrB) two independent preparations were made. Proteins (2 mg) were precipitated from the homogenates by 5% trichloroacetic acid (Sigma) on ice for 10 min. Samples were centrifuged at 16,000 × g for 20 min at 4°C, supernatants were used for measurements of Met content immediately (or stored at −80 °C), and pellets were hydrolyzed with p-toluenensulfonic acid. p-Toluenensulfonic acid (Sigma, 98.5% purity) was prepared as described (Bayer et al., 1985. Pellets were treated with 0.5 ml of 3 M p- toluenensulfonic acid under anaerobic conditions at 110 °C for 22 h, acidic samples were neutralized with equal volumes of 2M NaOH and stored at −20 °C. The analysis of Met/MetO content in protein-free and hydrolyzed samples by HPLC was carried out as described (Lee et al., 2008). o-Phthalaldehyde derivatizatied of amino acids were detected using a Zorbax Eclipse XDB-C8 column (for of Met content) or Zorbax Eclipse XDB-C18 column (for MetO content). Measurements for each sample was performed in triplicate. All data are reported as the means ±S.D.
2.11. Western blotting
6-day-old flies were prepared as in section 2.9. 100 µg of total protein was electrophoresed on NuPAGE® Novex 10% Bis-Tris gels, transferred onto PVDF membranes, and immunoblotted with antibodies specific for mouse MsrB2 (Kim and Gladyshev, 2005), Drosophila MsrB (Kumar et al., 2002) or Drosophila β-actin (ab8224, Abcam). Dilution 1:1000 was used for anti-mouse MsrB2 antibodies, 1:2000 for anti-Drosophila MsrB antibodies and 1:1000 for anti-β-actin. Immunoblot signals were visualized using ECL detection system (Sigma).
2.12. Statistic analysis
The significance of the difference between the survivor curves was determined separately for each 3 replicates of transgenic flies and their corresponding control (GAL4-activator/w1118, UAS-MsrB/w1118) using SAS software v. 9.1 (SAS Institute Inc., Cary, NC). To compare the survivor curves of the MsrB-expressing and control flies the nonparametrical estimates of the survivor functions by Kaplan-Meier method were applied using the procedure LIFETEST provided by SAS. The PROC LIFETEST calculated statistics for testing the null hypothesis that the survivor functions are the same for testing and control flies; statistical parameters (χ2 and p-value) of log-rank test were used for estimation of accuracy of the null hypothesis. To get the mean and maximum (90% mortality) lifespans for each set of replicates (transgenic and control flies) the procedure MEANS provided by SAS software was used.
3. Results
3.1. Overexpression of fruit fly MsrB and mouse MsrB2 in Drosophila
We used the GAL4-UAS binary system to drive overexpression of mouse mitochondrial MsrB2 and fruit fly cytosolic MsrB in Drosophila as described in Section 2.4. Two independent homozygous UAS-MsrB-responder lines for each MsrB were used, designated mMsrB22A and mMsrB23A for mouse MsrB2 and dMsrB2A and dMsrB2B for Drosophila MsrB. To test the efficacy of UAS-MsrB transgenes for whole body/ectopic expression, the responder lines were mated to four different GAL4-activator lines (drivers). Two drivers with constitutive broad range GAL4 expression, tubP-GAL4 and da-GAL4, of different strength were selected because 1) flies express endogenous MsrB ubiquitously; and 2) we were interested in examining how MsrB expression level influences lifespan. The GAL4-fatbody driver was chosen as Drosophila fatbody includes many metabolic activities of the vertebrate liver such as storage and metabolism of fat, glycogen and proteins (Söndergaard, 1993). Mammalian liver shows high MsrB expression and activity (Kim and Gladyshev, 2007). The elav-GAL4 activator line that mediates GAL4 expression in embryo, larvae and adult neurons was selected as the expression of bovine MsrA in Drosophila nervous system markedly extended lifespan of fruit flies in a previous study (Ruan et al., 2002). We used elav-GAL4 driver with the same genotype as that in the study that examined MsrA overexpression (Ruan et al., 2002 and Section 2.2).
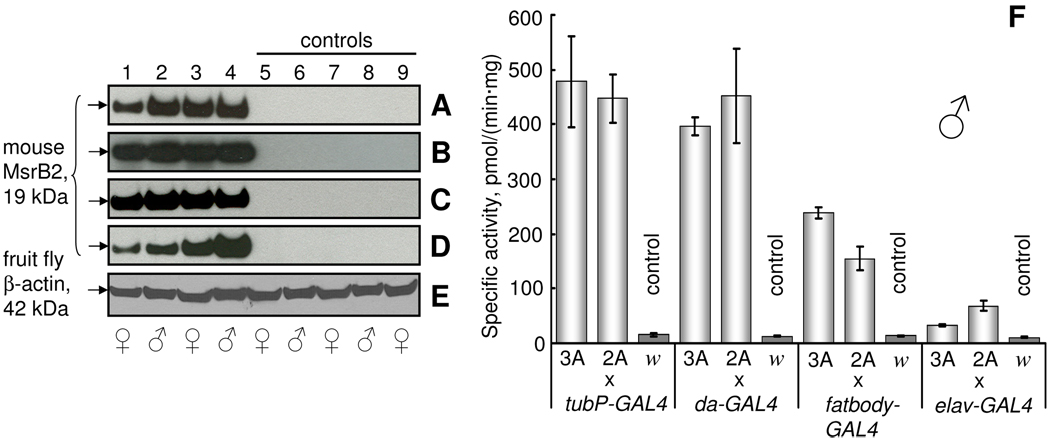
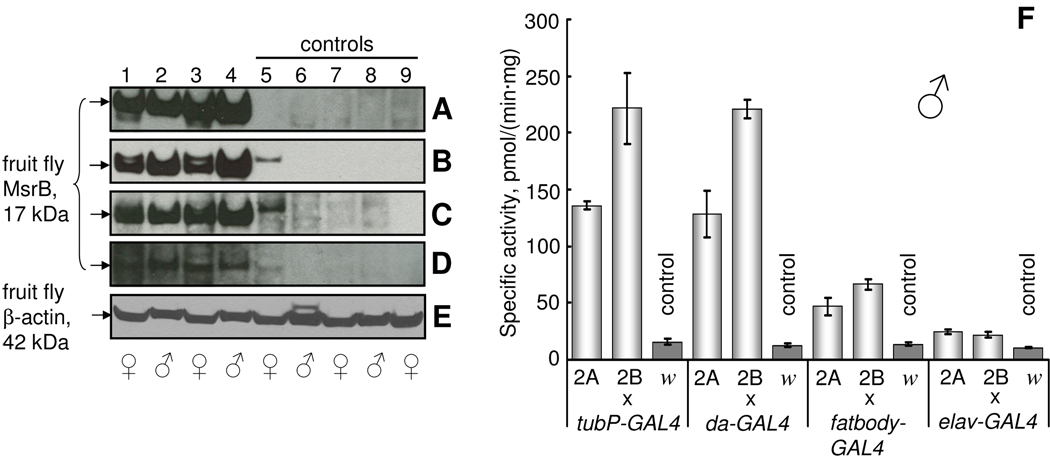
After crossing GAL4-activator lines with UAS-MsrB-responder lines, the overexpression of MsrBs in total body homogenate of resulting progeny was verified by Western blot analyses and MsrB activity assays (Fig. 1 and 2). The bands corresponding to mouse MsrB2 (19 kDa) (Fig. 1A–D, lanes 1–4) and fruit fly MsrB (17 kDa) (Fig. 2A–D, lanes 1–4) were detected in all analyzed flies carrying both GAL4 and UAS transgenes. No bands with the same size were detected in control flies (Figs. 1, 2A–D, lanes 5–9). As control, we used F1 progeny of crosses between corresponding GAL4-activator line and white flies (GAL4-activator/w1118) as well as crosses between flies carrying MsrB transgene and white flies (w1118/UAS-MsrB). Endogenous Drosophila MsrB was present at undetectable level in comparison with MsrB expressed from the transgene (Fig. 2A–D, control lanes 5–9). The lowest level of MsrB expression was observed in the samples prepared from flies expressing Drosophila MsrB in the nervous system (Fig. 2D), and this finding corresponded to only a small increase in MsrB activity driven by the elav-GAL4 driver in these samples (compare cross elav-GAL4/UAS-MsrBs with other crosses on Fig. 2F and Fig. 1F). The low MsrB activity in flies with the transgene expressed in the nervous system reflects the fact that neuronal tissues contribute a smaller part by weight, and the homogenates of flies expressing MsrB in the nervous system show lower levels of this enzyme in comparison, for example, with the homogenates of fruit flies expressing MsrB in the whole body. The highest MsrB activity was found in both ubiquitous GAL4-activator lines. Total body homogenates prepared from flies ubiquitously expressing mouse MsrB2 had almost 40-fold higher MsrB activity than the background activity due to endogenous fruit fly MsrB (Fig. 1F). Homogenates prepared from fruit flies overexpressing Drosophila MsrB in whole body had almost 20-fold increase in activity (Fig. 2F). No difference between two ubiquitous GAL4-activator lines (tubP-GAL4 and da-GAL4) was found with regard to MsrB activity or expression (Fig. 1A, B and F), although the da-GAL4 line is considered as a weaker driver. Fruit flies expressing MsrBs in the nervous system had statistically significant elevation in total MsrB activity (up to 6 times for mouse MsrB2 and up to 2 times for fruit fly MsrB overexpression) in comparison with controls. Clearly, efficient expression of active enzymes was achieved with both mouse MsrB2 and Drosophila MsrB transgenes. Background activity due to endogenous Drosophila MsrB was 13.2±1.9 pmol/(min mg protein).
Fig. 1.
Expression of mouse MsrB2 in fruit flies. (A–D) Western blot analyses of Drosophila total homogenates with anti-mouse MsrB2 antibodies. mMsrB23A (lanes 1, 2) and mMsrB22A (lanes 3, 4) lines were crossed to GAL4-activator lines to drive mouse MsrB2 expression. Control flies were obtained by crossing white flies (w1118) with mMsrB23A (lanes 5, 6), mMsrB22A (lanes 7, 8) and GAL4-activator (lane 9) lines. Expression of mouse MsrB2 (lanes 1–4) in whole body was achieved by crossing mMsrB2 lines and tubP-GAL4 (A) and da-GAL4 (B) drivers, in fatbody by crossing mMsrB2 with fatbody-GAL4 driver (C), and in the nervous system by crossing mMsrB2 with elav-GAL4 driver (D). Migration of the 19 kDa band corresponding to mouse MsrB2 is indicated on the left in panels A–D. (E) Protein loading control (42 kDa Drosophila β-actin) for the Western blotting experiment shown in A. Gender of 6-day-old females and males analyzed is shown by the corresponding symbol below panel E. (F) MsrB activity of total homogenates of male flies used for western blot analyses. Designations 3A, 2A and letter w refer to mMsrB23A, mMsrB22A and w1118 lines that were crossed with the GAL4-driver. Measurements of each sample were performed in triplicates. All data are reported as the means ±S.D.
Fig. 2.
Expression of Drosophila MsrB in fruit flies. (A–D) Western blot analyses of fruit fly total homogenates with anti-Drosophila MsrB antibodies. dMsrB2A (lanes 1, 2) and dMsrB2B (lanes 3, 4) flies were crossed to GAL4-activator lines to overexpress Drosophila MsrB. Controls were obtained by crossing white flies with dMsrB2A (lanes 5, 6), dMsrB2B (lanes 7, 8) and GAL4-activator (lane 9) lines. F1 progeny of crosses between tubP-GAL4 (A), da-GAL4 (B), fatbody-GAL4 and elav-GAL4 (D) drivers and dMsrB (lanes 1–4) and white flies w1118 (lane 9) were analyzed. Adult animals used were 6 days old. Migration of the 17 kDa band corresponding to fruit fly MsrB is shown by arrows on the left in panels A–D. (E) Protein loading control (42 kDa Drosophila β-actin) for the Western blotting experiment presented in A. Gender of 6-day-old females and males analyzed is shown by the corresponding symbols below panel E. (F) MsrB activity of total homogenate of fruit flies used for Western blot analyses. Designations 2A, 2B and letter w refer to mMsrB23A, mMsrB22A and w1118 lines that were crossed with the indicated GAL4-driver. Measurements were performed in triplicate. All data are reported as the means ±S.D.
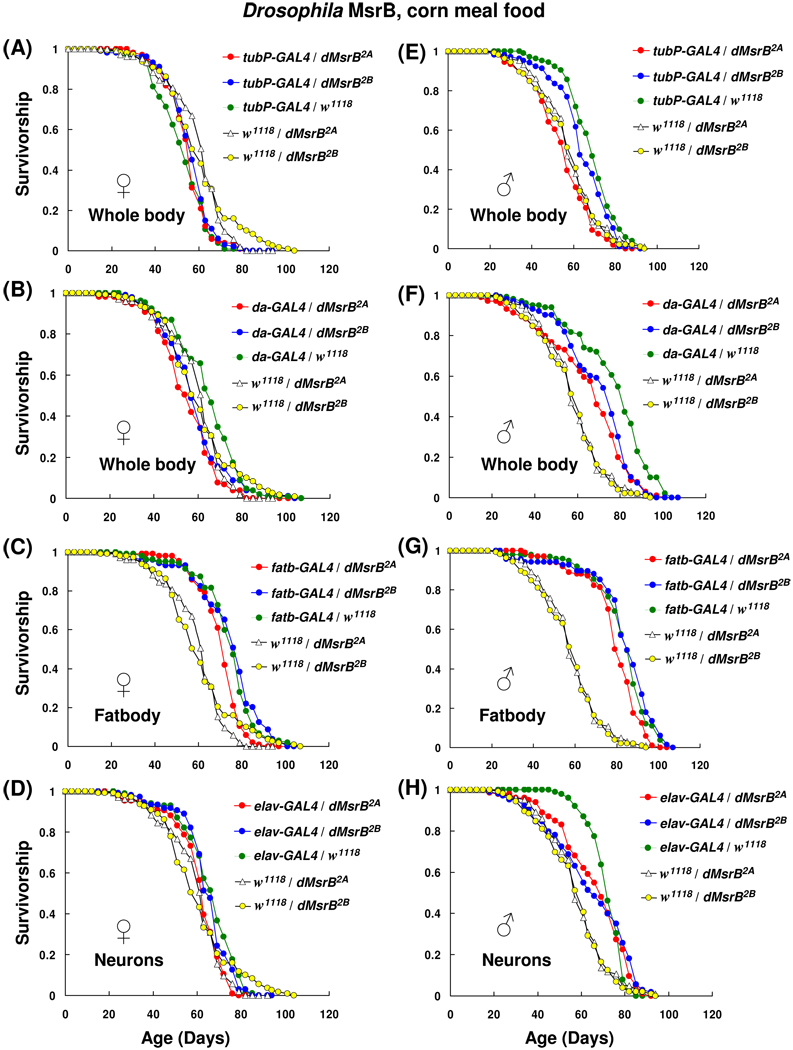
3.2. Overexpression of mouse MsrB2 has no effect on lifespan on corn meal food
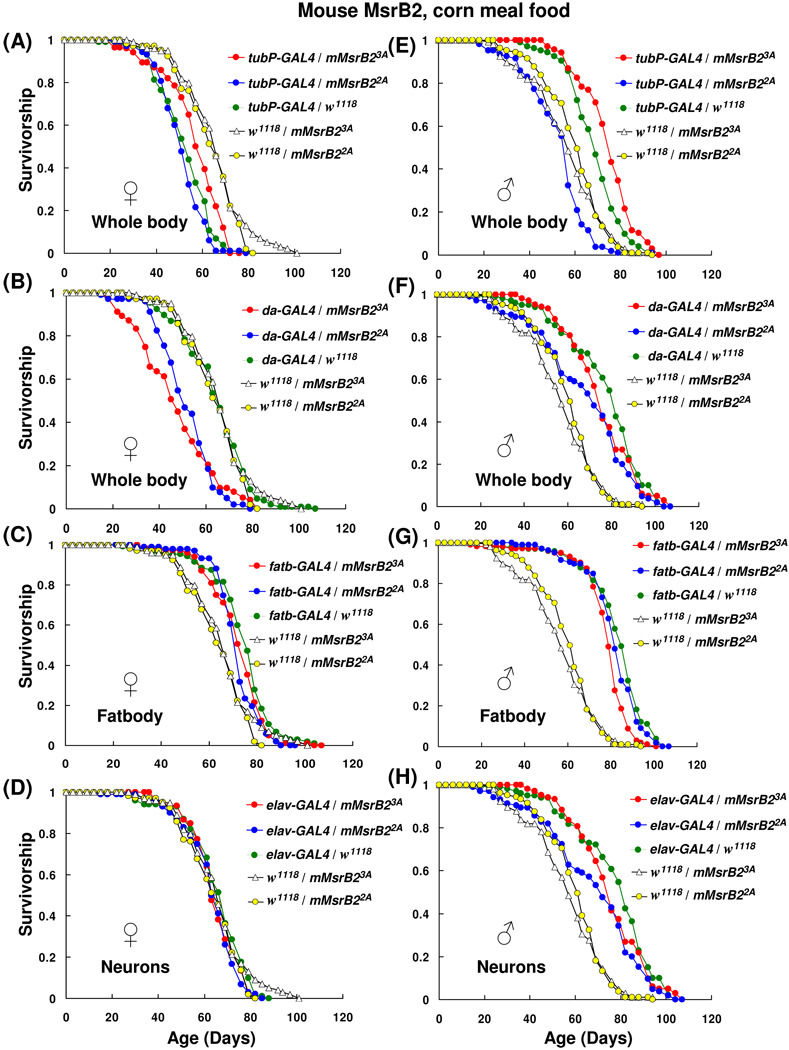
Expression of MsrB2 in whole body had no life-prolonging effect; in contrast, a small reduction in the mean lifespans (Table 1; Fig. 3B, E and F) or no effect (Table 1; Fig. 3A) in MsrB2-expressing flies was observed. The longevity of females in which the expression of mouse MsrB2 driven by tubP-GAL4 activator line (tubP-GAL4/mMsrB23A and tubP-GAL4/mMsrB22A) was not significantly different compared to driver line control tubP-GAL4/w1118 (Fig. 3A; Table 1; column 5). However, male progeny of a cross tubP-GAL4/mMsrB22A showed a reduction (13%) in the mean lifespan in a comparison with tubP-GAL4/w1118 control (χ2 = 68.73, p < 0.0001, Table 1, column 5, and Fig. 3E). These three lines also demonstrated 10–20% reduction (p < 0.0001, Table 1, column 8) in the mean lifespan in comparison with the corresponding responder line controls (females tubP-GAL4/mMsrB23A vs. w1118/mMsrB23A, females tubP-GAL4/mMsrB22A vs. w1118/mMsrB22A and males tubP-GAL4/mMsrB23A vs. w1118/mMsrB23A). One male line tubP-GAL4/mMsrB23A displayed an 8% increase (χ2 = 10.76, p = 0.001) and 30% increase (χ2 = 68.73, p < 0.0001) in mean lifespan in comparison with driver line control and responder line control, respectively (Fig. 3A, Table 1, columns 5 and 8). This was the only line in this study that showed lifespan extension with both controls.
Table 1.
Statistical analysis of survivor curves presented in Fig. 3 for mouse MsrB2-expressing flies and their controls.
| 1. Panel in Fig. 3 |
2. Genotype |
3. Sex |
4. Mean, days |
5. % vs driver/ w1118 |
6. χ2 |
7. p > χ2 |
8. % vs responder/w1118 |
9. χ2 |
10. p > χ2 |
|---|---|---|---|---|---|---|---|---|---|
| w1118/mMsrB23A | F | 64.4 | |||||||
| w1118/mMsrB22A | F | 65.0 | +0.01* | 0.23* | 0.62* | ||||
| A | tubP-GAL4/mMsrB23A | F | 58.5 | +1 | 1.69 | 0.19 | −10 | 25.91 | <0.0001 |
| A | tubP-GAL4/mMsrB22A | F | 52.5 | −1 | 3.89 | 0.05 | −19 | 68.51 | <0.0001 |
| A | tubP-GAL4/w1118 | F | 54.1 | ||||||
| B | da-GAL4/mMsrB23A | F | 49.2 | −24 | 39.42 | <0.0001 | −24 | 28.25 | <0.0001 |
| B | da-GAL4/mMsrB22A | F | 53.2 | −19 | 51.78 | <0.0001 | −18 | 48.17 | <0.0001 |
| B | da-GAL4/w1118 | F | 65.5 | ||||||
| C | fatb-GAL4/mMsrB23A | F | 73.7 | −1 | 2.47 | 0.12 | +14 | 6.66 | 0.0099 |
| C | fatb-GAL4/mMsrB22A | F | 72.8 | −4 | 3.21 | 0.06 | +12 | 7.28 | 0.0012 |
| C | w1118/mMsrB22A | F | |||||||
| D | elav-GAL4/mMsrB23A | F | 66.1 | −1 | 2.86 | 0.09 | +3 | 0.19 | 0.66 |
| D | elav-GAL4/mMsrB22A | F | 64.9 | −1 | 4.26 | 0.04 | −0.01 | 0.21 | 0.95 |
| D | elav-GAL4/w1118 | F | 67.1 | ||||||
| w1118/mMsrB23A | M | 57.7 | |||||||
| w1118/mMsrB22A | M | 61.4 | +6* | 6.66* | 0.0099* | ||||
| E | tubP-GAL4/mMsrB23A | M | 75.8 | +8 | 10.76 | 0.001 | +30 | 66.71 | <0.0001 |
| E | tubP-GAL4/mMsrB22A | M | 54.4 | −13 | 68.73 | <0.0001 | −11 | 16.69 | <0.0001 |
| E | tubP-GAL4/w1118 | M | 70.5 | ||||||
| F | da-GAL4/mMsrB23A | M | 76.5 | −3 | 1.09 | 0.29 | +32 | 67.56 | <0.0001 |
| F | da-GAL4/mMsrB22A | M | 69.3 | −12 | 7.52 | 0.0061 | +13 | 28.89 | <0.0001 |
| F | da-GAL4/w1118 | M | 78.4 | ||||||
| G | fatb-GAL4/mMsrB23A | M | 79.6 | −6 | 2.31 | 0.11 | +38 | 114.72 | <0.0001 |
| G | fatb-GAL4/mMsrB22A | M | 83.4 | −2 | 0.25 | 0.62 | +36 | 107.56 | <0.0001 |
| G | fatb-GAL4/w1118 | M | 84.7 | ||||||
| H | elav-GAL4/mMsrB23A | M | 73.7 | 0 | 4.44 | 0.04 | +28 | 58.10 | <0.0001 |
| H | elav-GAL4/mMsrB22A | M | 69.3 | −6 | 0.42 | 0.52 | +13 | 30.38 | <0.0001 |
| H | elav-GAL4/w1118 | M | 73.9 |
Column 1 indicates the letter of the corresponding panel in Fig. 3. The genotypes and gender (female, F; males, M) are shown in columns 2 and 3, respectively. The mean lifespan is shown in column 4. The percent change in the mean lifespan compared with corresponding driver controls (GAL4-driver/w1118) is displayed in column 5 and compared with corresponding responder control in column 8. Comparison of two types (transgenic and control) of survivor curves (three curves in each group, 105 flies total) was performed with SAS software (see Section 2.12). Statistics of non-parametrical log rank test (χ2) for comparison with driver control lines is shown in column 6; p-value in column 7 and for comparison with responder control line in columns 9, 10. Stars marks a comparison of two responder lines.
Fig. 3.
Survivorship curves of mouse MsrB2 expressing flies on corn meal food. 35 female and 35 male animals were housed together in plastic cages; three replica cages were used for each lifespan curve. Survivorship distribution presented shows the average of those independent replicas (105 females and 105 males for each curve). Fresh corn meal food was supplied and surviving animals were counted every 3–4 days. Expression of mouse MsrB2 in whole body was activated by using tubP-GAL4 (A and E) or da-GAL4 (B and F) activator lines; in fatbody by using fatbody-GAL4 driver (C and G); in the nervous system by using elav-GAL4 line (D and H). Genotypes and sexes are indicated on the plots. All trials were performed concurrently.
When tubP-GAL4 activator line was replaced with da-GAL4 line, females expressing mouse MsrB2 lived significantly shorter (p < 0.0001) than driver control da-GAL4/w1118. Experimental flies had 24% decrease in mean lifespan for cross da-GAL4/mMsrB23A vs. da-GAL4/w1118 (χ2 = 39.42, p < 0.0001) and 19% decrease for cross da-GAL4/mMsrB22A vs. da-GAL4/w1118 (χ2 = 51.78, p < 0.0001) (Fig. 3B and Table 1, column 5). In males, respective 3% and 12% reductions in mean lifespan were observed (Fig. 3F and Table 1, column 5). However, when a larger group was used (i.e., 210 males instead of 105) in the experiment with using sugar yeast (DR SY) food, no significant decrease in the mean lifespan for males with the same genotypes was observed (Fig. 5B, Table 3).
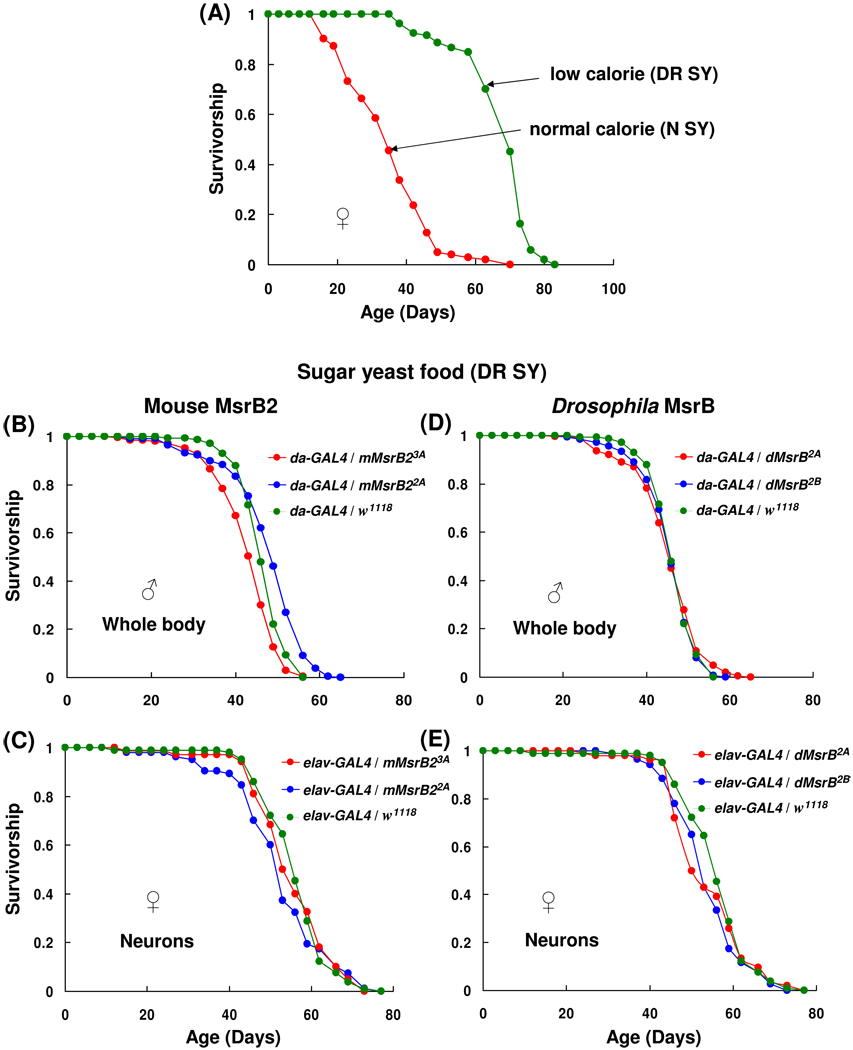
Fig. 5.
Survivorship curves of mouse MsrB2- and Drosophila MsrB-overexpressing flies on sugar yeast food. (A) Survivorship curves of control flies (da-GAL4/w1118) on normal and dietary restriction sugar yeast food (N SY and DR SY, respectively). (B–E) Survivorship of flies overexpressing mouse MsrB2 (B and C) and Drosophila MsrB (D and E) on DR SY food. Overexpression of MsrBs in whole body (B and D) and nervous system (C and E) was driven by da-GAL4 and elav-GAL4 activator lines, respectively. Males and females were kept together (A, C and E) or separate (B and D) during the lifespan study. 210 males were tested for each survivorship curve in panels B and D, and 105 animals for all other curves. Genotypes and sexes are marked on the plots. All trials were performed concurrently.
Table 3.
Statistical analysis of survivor curves presented in Fig. 5 for flies overexpressing mouse MsrB2 and Drosophila MsrB and their controls.
| 1. Panel on Fig. 5 |
2. Genotype |
3. Sex |
4. Mean, days |
5. % vs driver/ w1118 |
6. Max, days |
7. % vs driver/ w1118 |
8. χ2 |
9. p > χ 2 |
|---|---|---|---|---|---|---|---|---|
| A | da-GAL4/w1118 N SY | F | 36.3 | −52 | 64.4 | −15 | 187.7 | <0.0001 |
| A | da-GAL4/w1118 DR SY | F | 69.2 | 76.1 | ||||
| B | da-GAL4/mMsrB23A | M | 44.7 | −5 | 51.8 | 0 | 12.7 | 0.0004 |
| B | da-GAL4/mMsrB22A | M | 49.7 | +6 | 59.9 | +16 | 35.99 | <0.0001 |
| B | da-GAL4/w1118 | M | 46.9 | 51.8 | ||||
| D | da-GAL4/dMsrB2A | M | 47.5 | −1 | 59.9 | +16 | 3.12 | 0.08 |
| D | da-GAL4/dMsrB2B | M | 46.2 | −1 | 54.5 | +5 | 0.75 | 0.39 |
| D | da-GAL4/w1118 | M | 46.9 | 51.8 | ||||
| C | elav-GAL4/mMsrB23A | F | 52.6 | −9 | 70.7 | 0 | 3.64 | 0.06 |
| C | elav-GAL4/mMsrB22A | F | 51.4 | −11 | 67.9 | −4 | 41.25 | <0.0001 |
| C | elav-GAL4/w1118 | F | 57.8 | 70.7 | ||||
| E | elav-GAL4/dMsrB2A | F | 54.2 | −6 | 67.1 | −5 | 49.7 | 0.02 |
| E | elav-GAL4/dMsrB2B | F | 55.0 | −5 | 70.7 | 0 | 4.7 | 0.26 |
| E | elav-GAL4/w1118 | F | 57.8 | 70.7 |
Column 1 indicates panel in Fig. 5. The genotypes and gender (female, F; males, M) of flies are shown in columns 2 and 3, respectively. The mean lifespan is indicated in column 4 and percent change in the mean lifespan compared with their genetically matched controls (GAL4-driver/w1118) is displayed in column 5. Maximum lifespan and percent change of the maximum lifespan are shown in columns 6 and 7, respectively. Comparison of two types (transgenic and control) of survivor curves (three curves in each group, 210 flies for panels B, D and 105 flies for other panels) was performed with SAS software (see Section 2.12). Statistics of non-parametrical log rank test (χ2) is shown in column 8; p-value in column 9.
Although two ubiquitous GAL4-activator lines were used, conflicting data obtained does not allow making definitive conclusions with regard to negative influence of mouse MsrB2 overexpression in whole body on longevity of fruit flies. One of the possible reasons of such variations in lifespan could be the hybrid effect of two genetic backgrounds. Although both UAS-mMsr2 flies and GAL4-drivers had white genetic background, they were not isogenic with experimental flies. The substantial effect of variations in genetic background was observed on responder line controls (Table 1, column 8). Although responder line controls had similar mean lifespan (≈65 days for females, χ2 = 0.23, p = 0.62; 58 and 61 days for males, χ2 = 6.66, p = 0.0099; Table 1), they showed statistically significant difference (χ2 >20, p < 0.0001) in mean lifespan in comparison with the GAL4-activator line control and experimental flies. The responder line controls w1118/UAS-mMsrB2 had white genetic background. However, both the GAL4-activator line control and experimental flies had the same mixture (50:50) of GAL4-driver and white line chromatids. Therefore, an identical genetic background was achieved only in the driver line control, which should be viewed as the best control for the analysis of the effect of mMsrB2 expression on longevity. It was shown previously that genetic background itself could influence lifespan (Toivonen et al., 2007); our data obtained for ubiquitous drivers are consistent with this idea. It should be noted that life-prolonging effect of MsrA overexpression in whole body (40% increase in median lifespan in comparison with homozygous GAL4-activator line Ubi-GAL4 and 80% increase in comparison with homozygous UAS-MsrA responder line) was obtained on non-backcrossed animals. Also, no responder line and driver line controls were analyzed in comparison with the flies overexpressing MsrA in the whole body (Ruan et al., 2002).
Mouse MsrB2 ectopic expression in fatbody mediated by fatbody-GAL4 activator line had no effect on lifespan in comparison with GAL4-acivator line control (Fig. 3C and G; Table 1, column 5). Male and female progeny of MsrB2-expressing flies had the similar mean lifespan as GAL4-fatbody/w1118 controls and their survivor curves were undistinguishable by statistical analysis (Table 1, column 5). The fatbody-GAL4 flies (Grönke et al., 2003) were generated on the basis of long-lived flies with yellow and white genetic background, y w. Long-lived flies expressing transgenes usually show more reproducible and reliable data than their short-lived counterparts (Orr et al, 2003). Two heterozygous controls (w1118/mMsrB23A and w1118/mMsrB22A) were tested concurrently with the experimental flies; no lifespan extension for MsrB2-expressing flies compared with their genetically matched w1118/mMsrB23A and w1118/mMsrB22A controls was observed (Table 1, column 8). The positive effect of combination of two different genetic backgrounds (w1118 and y w) on lifespan was observed. The heterozygotes combined w1118 and y w chromosomes lived for approximately 13% (females) and 37% (males) longer than the responder line control with only white genetic background.
Ectopic expression of mouse MsrB2 in neurons by elav-GAL4 driver had not revealed any effect of MsrB2 expression on lifespan in comparison with the GAL4-activator line control, elav-GAL4/w1118 (Fig. 3D and H; Table 1, column 5). No statistically significant difference in survivor curves was observed (Table 1, column 5). Survivor curves of two other control lines, which do not express MsrB2 (w1118/mMsrB23A and w1118/mMsrB22A), were also obtained in a parallel experiment. Female responder line controls had the mean lifespans that were similar to those of the tested lines (Table 1, column 8). Consequently, we conclude that neither ubiquitous nor ectopic expression of mitochondrial mouse MsrB2 in flies had the dramatic and consistent effect on lifespan on corn meal food.
3.3. Overexpression of Drosophila MsrB has no effect on lifespan on corn meal food
Similar results were obtained with flies overexpressing Drosophila MsrB (Fig. 4 and Table 2, columns 5 and 8). Variations in mean lifespan (reduction up to 20%) were observed between MsrB-overexpressing flies and their controls for both ubiquitous drivers (Fig. 4A, B, E, F and Table 2). The most significant decrease in the mean lifespan in comparison with GAL4-activator line control was found for crosses da-GAL4/dMsrB2A and da-GAL4/dMsrB2B. Females with respective genotype had 14% and 8% reduction in the mean lifespan, and males had 20% and 6% reduction in the mean lifespan (Fig. 4B, F and Table 2, column 5). However, this effect was not seen with female progeny of a cross between UAS-dMsrBs lines and another ubiquitous driver tubP-GAL4 (compare Fig. 4A and B; Table 2, column 5). In addition, when more da-GAL4/dMsrBs males were studied on DR SY food (210 males instead of 105), no changes in mean lifespan of the experimental and the driver line control flies were found (Fig. 5D; Table 3). The survivor curves of animals overexpressing Drosophila MsrB in fatbody as well as the mean lifespans were not changed significantly when compared with fatbody-GAL4/w1118 control (Fig. 4C, G; Table 2, column 5). As in the case with expression of mouse MsrB2, responder line controls for fatbody-GAL4/dMsrB lines showed statistically significant (p < 0.0001) difference with experimental flies (Table 2, column 8). No dramatic effect of MsrB expression in the nervous system was in comparison with driver line and responder line controls were detected (Fig. 4D and H; Table 2 (columns 5 and 8). In conclusion, strong host MsrB overexpression in the nervous system, fatbody or whole body of flies had no dramatic and consistent lifespan-prolonging effect on corn meal food.
Fig. 4.
Survivorship curves of Drosophila MsrB-overexpressing flies on corn meal food. Males and females were kept together during lifespan study. Each survivorship curve represents 105 flies. Expression of Drosophila MsrB in whole body was driven by using tubP-GAL4 (A, E) or da-GAL4 (B, F) activator lines; in fatbody by using fatbody-GAL4 driver (C, G); in the nervous system by using elav-GAL4 line (D, H). Genotypes and sexes are shown on the plots. All trials were performed concurrently.
Table 2.
Statistical analysis of survivor curves presented in Fig. 4 for flies overexpressing Drosophila MsrB and their controls.
| 1. Panel in Fig. 4 |
2. Genotype |
3. Sex |
4. Mean, days |
5. % vs driver / w1118 |
6. χ2 |
7. p > χ2 |
8. % vs responder/ w1118 |
9. χ2 |
10. p > χ2 |
|---|---|---|---|---|---|---|---|---|---|
| w1118/dMsrB2A | F | 60.9 | |||||||
| w1118/dMsrB2B | F | 58.2 | −5* | 3.17* | 0.12* | ||||
| A | tubP-GAL4/dMsrB2A | F | 56.7 | +5 | 0.75 | 0.39 | −7 | 11.75 | 0.0006 |
| A | tubP-GAL4/dMsrB2B | F | 57.6 | +6 | 2.84 | 0.09 | −2 | 45.37 | 0.0003 |
| A | tubP-GAL4/w1118 | F | 54.1 | ||||||
| B | da-GAL4/dMsrB2A | F | 56.2 | −14 | 25.9 | <0.0001 | −8 | 9.88 | 0.0017 |
| B | da-GAL4/dMsrB2B | F | 60.1 | −8 | 7.08 | 0.01 | +3 | 11.74 | 0.0006 |
| B | da-GAL4/w1118 | F | 65.5 | ||||||
| C | fatb-GAL4/dMsrB2A | F | 71.2 | −6 | 14.2 | 0.0002 | +17 | 21.41 | <0.0001 |
| C | fatb-GAL4/dMsrB2B | F | 76.2 | +1 | 1.74 | 0.19 | +30 | 79.68 | <0.0001 |
| C | fatb-GAL4/w1118 | F | 75.5 | ||||||
| D | elav-GAL4/dMsrB2A | F | 62.6 | −7 | 2.86 | 0.08 | +3 | 1.49 | 0.22 |
| D | elav-GAL4/dMsrB2B | F | 66.2 | −1 | 1.51 | 0.22 | +0.1 | 1.51 | 0.22 |
| D | elav-GAL4/w1118 | F | 67.1 | ||||||
| W1118/dMsrB2A | M | 60.2 | |||||||
| w1118//dMsrB3A | M | 58.6 | −3* | 0.03* | 0.86* | ||||
| E | tubP-GAL4/dMsrB2A | M | 56.4 | −20 | 49.7 | <0.0001 | −7 | 1.56 | 0.2114 |
| E | tubP-GAL4/dMsrB2B | M | 66.1 | −6 | 4.7 | 0.03 | +9 | 17.47 | <0.0001 |
| E | tubP-GAL4/w1118 | M | 70.5 | ||||||
| F | da-GAL4/dMsrB2A | M | 66.8 | −15 | 22.89 | <0.0001 | +11 | 16.8 | <0.0001 |
| F | da-GAL4/dMsrB2B | M | 71.5 | −9 | 16.85 | <0.0001 | +22 | 39.74 | <0.0001 |
| F | da-GAL4/w1118 | M | 78.4 | ||||||
| G | fatb-GAL4/dMsrB2A | M | 80.8 | −5 | 2.31 | 0.11 | +34 | 82.4 | <0.0001 |
| G | fatb-GAL4/dMsrB2B | M | 77.2 | −9 | 0.25 | 0.62 | +31 | 73.4 | <0.0001 |
| G | fatb-GAL4/w1118 | M | 84.7 | ||||||
| H | elav-GAL4/dMsrB2A | M | 56.6 | −23 | 4.44 | 0.04 | −6 | 1.18 | 0.28 |
| H | elav-GAL4/dMsrB2B | M | 65.5 | −11 | 0.42 | 0.52 | +11 | 20.67 | <0.0001 |
| H | elav-GAL4/w1118 | M | 73.9 |
Column 1 indicates the letter of the corresponding panel in Fig. 3. The genotypes and gender (female, F; males, M) are shown in columns 2 and 3, respectively. The mean lifespan is shown in column 4. The percent change in the mean lifespan compared with corresponding driver controls (GAL4-driver/w1118) is displayed in column 5 and compared with corresponding responder control in column 8. Comparison of two types (transgenic and control) of survivor curves (three curves in each group, 105 flies total) was performed with SAS software (see Section 2.12). Statistics of non-parametrical log rank test (χ2) for comparison with driver control lines is shown in column 6; p-value in column 7 and for comparison with responder control line in columns 9, 10. Stars marks a comparison of two responder lines.
3.4. Overexpression of MsrBs has no effect on lifespan on sugar yeast food
A link between overexpression of host MsrB in yeast and extended lifespan under caloric restriction was previously reported (Koc et al., 2004). Under calorie restriction conditions, MsrB-overexpressing cells divided twice as many times (lifespan was assessed by replicative assays of yeast aging) (Koc et al., 2004). Our initial purpose was to examine whether or not dietary restriction (DR) influences the lifespan of MsrB transgenic flies. It was demonstrated (Chapman and Partridge, 1996; Mair et al., 2005; Kabil et al., 2007) that sugar yeast diet (SY) regulated lifespan of flies in response to dietary dilution of sugar, but yeast had a stronger effect on lifespan (Mair et al., 2005). Dietary dilution provided up to 50% increase in the median lifespan for Dahomey females (the effect was less pronounced for males) (Mair et al., 2005; Chapman and Partridge, 1996). To confirm that dietary restriction works for our flies, the diet containing twice less sugar and yeast (DR SY food, section 2.3 “Fly culture media”) in comparison with normal sugar yeast food (N SY) was tested for da-GAL4/w1118 female flies. As expected, these females had 52% increase in the mean lifespan (χ2 = 187.7, p < 0.0001, Table 1, column 5, and Fig. 5A). However, the mean lifespan of these flies on N SY food was significantly lower than that on corn meal food tested previously (36.3 days on N SY diet against 65.5 days on corn meal diet). The mean lifespan of tested females on DR SY (69.2 days) diet was comparable to that on corn meal food. We have not compared the nutritional composition and caloric content of corn meal diet and DR SY diet by analytical methods, but flies lifespan showed that both foods had approximately the same calorie content. This finding led us to an additional test of MsrB-expressing flies on the other diet, however, even if we kept designation of this diet (DR SY) as accepted in the literature, this diet could not be considered as dietary restriction diet in comparison with corn meal food.
Overexpression of mouse MsrB2 or host MsrB in the nervous system (Fig. 5C and E; Table 3) or the whole body (Fig. 5B and D; Table 3) had no significant effect on lifespan on DR SY food. Slight reduction in the mean lifespan of elav-GAL4/mMsrB22A females (11 %, χ2 = 41.25, p < 0.0001, Table 3, column 5) was not observed on corn meal food (compare Fig. 3D and Fig. 5C) and therefore appeared not to be the result of mouse MsrB2 expression. Again, the responder control lines showed a statistically significant differences (p < 0.0001) in mean lifespan (data not shown). Differences in metabolic pathways, MsrB expression levels, and localization of the MsrB protein function between flies and yeast could possibly explain different outcomes of MsrB expression in these organisms.
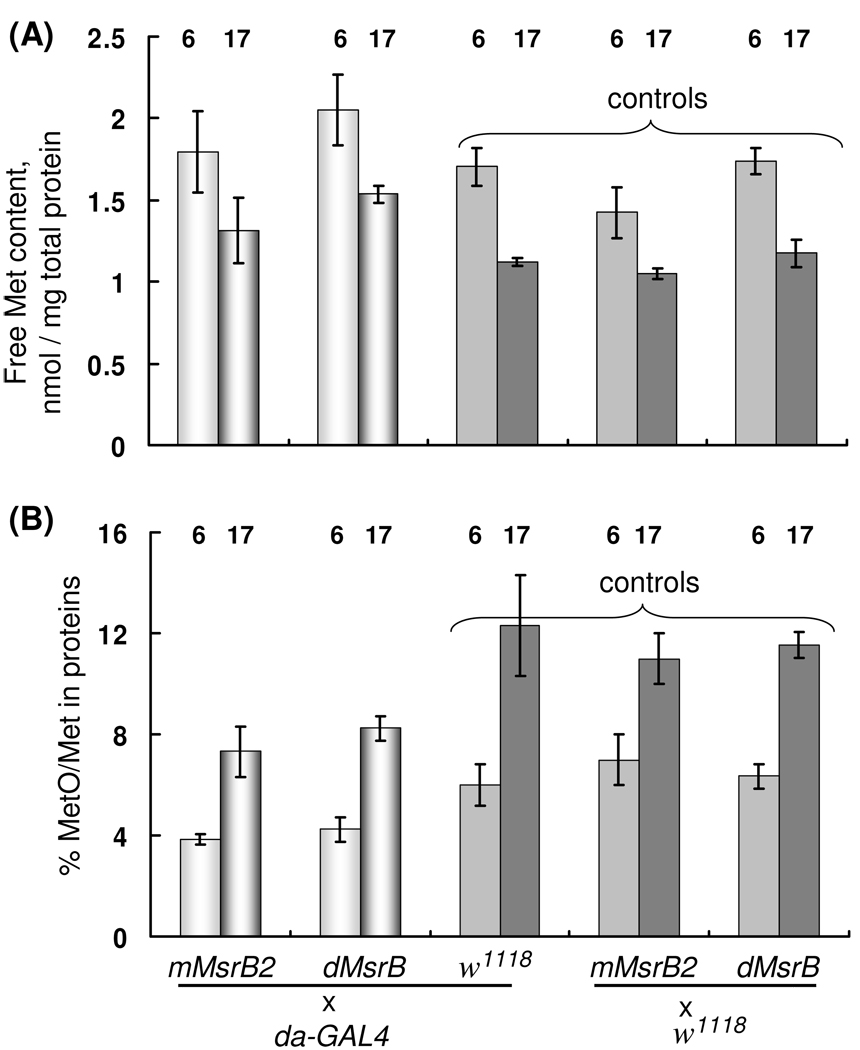
3.5. Methionine/methionine sulfoxide content of MsrBs-overexpressing flies
Methionine sulfoxide content in proteins is considered as a measure of oxidative damage in cell (Stadtman et al., 2005), but for Drosophila there are no direct experimental data confirming this observation. To compare experimental and control flies, we prepared 6- and 17-day-old da-GAL4/dMsrB and da-GAL4/mMsrB2 flies, their corresponding driver line control da-GAL4/w1118, and responder line controls w1118/dMsrB and w1118/mMsrB2. Proteins in homogenate were precipitated by trichloroacetic acid and methionine content in a protein-free form was measured (Fig. 6A). The pellet was subjected to hydrolysis with p-toluenensulfonic acid followed by analysis of Met and MetO content (Fig. 6B). Significant reduction of free methionine during aging (up to 35% decline, Fig. 6A) was observed for both experimental and control flies. Our data are in agreement with the previously reported decline of Met content during aging (Rebrin et al., 2004). Remarkably, 17-day-old flies expressing Drosophila MsrB showed approximately 30% increase in methionine content in comparison with both control flies. Mouse MsrB2 expressing flies had the same content of free methionine (within error) as control flies.
Fig. 6.
Analysis of Met and MetO content in 6- and 17-day-old flies. (A) Met content of the protein-free fraction of fruit fly homogenates. Met content is given in nmoles/mg of protein in homogenates. (B) Ratio of MetO to Met in a protein fraction. Flies (6- and 17- day-old) overexpressing mouse MsrB2 or Drosophila MsrB in whole body (da-GAL4/dMsrB and da-GAL4/mMsrB2), their corresponding driver line control da-GAL4/w1118, and responder line controls w1118/dMsrB and w1118/mMsrB were homogenized and proteins were precipitated. The supernatant was used to determine protein-free Met (A), and the pellet was used for protein hydrolysis and analysis of MetO and Met content. 70 females were used for each sample. Two independent preparations of each transgenic fly line as well as control flies were analyzed. Measurements for each sample were done in triplicate. All data are reported as means ±S.D. The age of flies (in days) is shown for each column. Genotypes are indicated.
Analysis of methionine sulfoxide content in hydrolyzed proteins revealed the presence of both diastereoisomers in equimolar ratio ([Met-RO]/[Met-SO] = 1.11 ± 0.04). We explain this finding by racemization of methionine sulfoxide during hydrolysis that was performed at 110 °C for 22 h. However, our experimental flies had a decreased methionine sulfoxide/methionine ratio in comparison with both types of controls (Fig. 6B) suggesting the expression of functional MsrB. We observed an almost 40% increase in the MetO/Met ratio during aging for the flies with the same genotype. However, it should be noted that MetO in proteins still stays at low levels even in 17-day old flies. The MetO/Met ratio in hydrolyzed protein samples was almost 50% higher in 17-day-old flies than it in 6-day-old flies with the same genotype (Fig. 6B). This finding is consistent with the idea that methionine residues in proteins become more oxidized during aging.
3.6. MsrB-expressing flies have the same stress resistance as controls
Drosophila longevity, when influenced by genetic interventions, DR, and selection for lines with postponed senescence, often positively correlates with enhanced resistance of animals to different stresses (e.g., oxidative stress, starvation, desiccation, heat) (Parkes et al., 1998; Ruan et al., 2002; Lin et al, 1998; Orr et al., 2005; Mockett et al, 2001; Arking et al., 1991; Vermeulen and Loeschcke, 2007). However, molecular mechanisms for such correlation remain elusive and there were several examples when oxidative stress response was not involved directly in aging (Bayne et al., 2005; Vermeulen and Loeschcke, 2007). For example, the lifespan of mitochondrial Mn-SOD/catalase transgenic flies was decreased up to 43%, but their resistance to oxidative stress caused by H2O2 treatment or exposure to 100% oxygen was elevated (Bayne et al., 2005). Similarly, although MsrB expression had no influence on lifespan, this is an important antioxidant enzyme and its protein repair function could presumably be uncoupled from its influence on lifespan. Thus, we examined resistance of transgenic MsrB flies to oxidative stress induced by dietary paraquat, which upon intake generates superoxide radicals (Arking et al., 1991) that oxidize methionine to MetO (Ruan et al., 2002).
17-day-old males overexpressing MsrBs in the nervous system were fed with 5% sucrose and 20 mM paraquat (Fig. 7A) or were starved for 6 h to minimize the variations in paraquat intake and then fed with 5% sucrose and 30 mM paraquat (Fig. 7B). No statistically significant difference in survivorship between experimental (elav-GAL4/mMsrB22A and elav-GAL4/dMsrB2A) and control (elav-GAL4/w1118) flies were observed on 20 mM paraquat (Fig. 7A). The 17 h exposure to 30 mM paraquat killed 80% of experimental (mMsrB2/elav-GAL4 and dMsrB/elav-GAL4) and control males (w1118/UAS-MsrBs); only 50% of other control flies (elav-GAL4/w1118) were dead at these conditions (Fig. 7B). These data demonstrate no protective effect of MsrB overexpression against oxidative stress induced by paraquat. All control animals (elav-GAL4/w1118) fed with the diet containing 5% sucrose in the absence of paraquat were alive after 6 days.
Fig. 7.
Stress resistance of mouse MsrB2- and Drosophila MsrB-overexpressing flies. 20-day-old male flies overexpressing MsrBs in the nervous system were examined for resistance to oxidative stress and starvation. Newly eclosed flies were mated, sex-segregated and maintained on corn meal food before the day of the tests. (A, B) Oxidative stress was induced by 20 mM (A) or 30 mM (B) dietary paraquat. (A) 80 animals were tested for each genotype (shown in the legend). Flies were transferred to test vials (5% sucrose, 20 mM paraquat) without preliminary starvation and dead animals were counted at fixed time points. 80 control flies (elav-GAL4/w1118) were kept on 5 % sucrose without paraquat (marked as “no paraquat” in the legend). (B) 360 animals were tested for flies overexpressing mouse MsrB2 (elav-GAL4/mMsrB2), Drosophila MsrB (elav-GAL4/dMsrB), and control flies (MsrBs/w1118), except for control elav-GAL4/w1118 where 180 males were studied. Genotypes are shown in the Y-axis legend; 180 flies with genotype elav-GAL4/w1118 were kept on food without paraquat and noted in Fig. 7B as “no paraquat”. All flies were starved for 6 h before the test and then fed with or without 30 mM paraquat and 5% sucrose for 17 h; animals alive were counted and normalized to the total number of animals used for each genotype. (C, D) Resistance to starvation was studied with flies expressing mouse MsrB2 (C) and Drosophila MsrB (D) in the nervous system. Flies were maintained in vials with 1% agar; dead animals were counted every 8 h. Total number of males tested for each genotype was 140. Genotypes are shown in the legends.
Although we have shown that both types of MsrB-expressing flies have low concentration of methionine sulfoxide in protein-bound form and Drosophila MsrB-expressing flies have a lower decline of free Met, the protective affect of methionine was not detected in assay with paraquat. The exact mechanism of paraquat in vivo toxicity is not known, even it is evident that paraquat generates superoxide radicals. Probably, maintaining of higher methionine level in MsrB-overexpressing flies is not enough to overcome consequences of superoxide radicals formation upon paraquat-induced oxidative stress.
As in the case of MsrA-overexpressing flies, MsrB overexpression did not protect against starvation (Fig. 7C, D). The median lifespan was approximately 60 h for MsrBs-expressing and control flies. The observation of no influence of MsrA and MsrB overexpression on starvation seems logical, as both types of enzymes are unlikely to have direct influence on metabolic pathways linked to resistance against starvation (e.g., increase in lipid content).
3.7. MsrB-expressing flies show no changes in reproduction and physical activity compared to controls
Aging in many species is associated with decline in physical activity and fertility. The long-lived MsrA-overexpressing flies with transgene expression predominantly in the nervous system had a markedly delayed age-related decline in general activity and reproduction (Ruan et al., 2002). In contrast, we found that MsrB-expressing flies had similar physical characteristics and showed no changes in the number of developed pupa.
Timing of development of transgenic flies expressing MsrB from eggs to hatching was the same as in the parental and heterozygous control flies that did not express MsrB transgenes (9 to 10 days at 25 °C). Qualitative observation of flight, courtship, and feeding behavior revealed no significant differences between transgenic MsrB flies and their controls in early life. The average body weight of 10-day-old males expressing MsrBs in the nervous system (elav-GAL4/mMsrB23A and elav-GAL4/dMsrB2A) was indistinguishable from the body weight of controls (elav-GAL4/w1118 and w1118/UAS-MsrBs); animals weighted 0.89 ± 0.02 mg (Supplementary Table S1). The average locomotor activity (mean ± S.E.) of 8-day-old males expressing MsrBs in whole body was similar with controls (Supplementary Fig. S1).
In a further experiment characterizing overall reproductive vigor, we counted the number of pupa that developed from eggs laid by one female over 24 h (Fig. 8). We used female animals expressing MsrBs in the nervous system (elav-GAL/mMsrB23A and elav-GAL/dMsrB2A) and the offspring of a cross between elav-GAL4 and white flies (elav-GAL4/w1118) as a control. Pupal progeny developed into adult flies in more than 95% cases. No changes in the number of developed pupa were observed (Fig. 8).
Fig. 8.
Age-associated changes in pupa production. Five females were kept with five males in a vial with corn meal food. Changes in pupa production were determined from counts of pupa developed from eggs laid by five female in 24 h; the developed pupa was counted after eclosion. The Y axis represents the number of pupa produced by one female in one vial. 10 replica of each vial were tested. Offsprings of crosses used (elav-GAL4/mMsrB23A, elav-GAL4/dMsrB2A or elav-GAL4/w1118) are shown in the legend.
Discussion
Expression of GFP-fused bovine MsrA in the adult nervous system of fruit flies resulted in a greatly increased median lifespan (~70%) (Ruan et al., 2002). To explain this finding, it was proposed that Drosophila has high levels of ROS and/or low levels of endogenous MsrA in the adult neurons, and that MsrA function might be a factor that limits the lifespan of flies (Ruan et al., 2002). Other studies reported the low level of Cu,Zn-SOD in adult nervous system of fruit flies (Klichko et al, 1999); and overexpression of human Cu,Zn-SOD in adult motorneurons using GAL4-UAS system extended the mean lifespan of fruit flies up to 40% (Parkes et al., 1998). An attempt to reproduce this result using elav-GAL4 driver failed (Ruan et al., 2002). Based on this observation and on a greater life-prolonging effect of MsrA overexpression (70% against published 40% for Cu,Zn-SOD), it was suggested that the antioxidant mechanism involving MsrA-dependent reduction of oxidized methionine residues in cellular proteins may be more robust than the superoxide dismutase/catalase system with regard to lifespan (Ruan et al., 2002). Since MsrA is specific for Met-SO, whereas Met-RO is reduced by MsrB, the analysis of fruit flies overexpressing MsrB provided an opportunity to better understand the unique role of MsrA and/or MetO reduction in influencing animal lifespan. In addition, since MsrA and MsrB catalyze the same reaction (but with different diastereomers of MetO), one would expect that the lifespan extension due to MsrA overexpression would be reproduced in flies overexpressing MsrB.
However, we found that overexpression of either mouse mitochondrial MsrB2 or Drosophila cytosolic MsrB in neurons of fruit flies had no dramatic and consistent lifespan-prolonging influence in these animals. Our transgenic flies expressed high levels of catalytically active enzymes, and these animals were examined with four different GAL4-activator lines as well as in regular and calorie restriction diets. None of the conditions showed clear lifespan extension. Since both MsrA and MsrB catalyze the reduction of MetO residues in proteins, it is puzzling how one enzyme could provide such a dramatic effect on lifespan by reducing half of MetO residues, and the other enzyme that reduces the other half of MetO residues could be ineffective under all conditions tested.
Apparently, MsrA has broader substrate specificity than MsrB. Besides Met-SO residues in proteins, this enzyme has activity for free Met-SO (Lee et al., 2008). In yeast, MsrA transcriptional regulation clusters it with genes involved in methionine metabolism and sulfur acquisition, whereas MsrB does not show this expression pattern (Koc et al., 2004). Moreover, MsrA deletion has a more significant effect on the growth of cells on free MetO than the deletion of MsrB (Kryukov et al., 2002). We hypothesize that the role of MsrA in sulfur or methylation pathways is responsible for the observed role of this enzyme on lifespan extension and methionine metabolism.
Methionine serves as a source of sulfur for cysteine synthesis via the transsulfuration pathway (Stipanuk, 2004). Cysteine may influence cellular redox homeostasis and serves as an intermediate in GSH biosynthesis. The GSH/GSSG ratio determines an intracellular redox potential as GSH is a major antioxidant in the cell. In D. melanogaster, the GSH/GSSG ratio is decreased during aging (Rebrin et al., 2004; Mockett et al., 1999). Addition of GSH to food significantly increased resistance of fruit flies to oxidative stress induced by paraquat (Bonilla et al., 2006). The importance of glutathione balance for Drosophila aging was previously shown in flies overexpressing glutamate-cysteine ligase, which catalyzes a rate-limiting reaction in GSH biosynthesis. These flies, expressing transgene in the nervous system, had extended mean and maximum lifespans up to 50% (Orr et al., 2005).
Overexpression of MsrA in flies may influence GSH synthesis or methylation pathways by providing cells with an additional source of free methionine. Additionally, Drosophila lacks glutathione reductase and GSSG reduction is carried out by the thioredoxin reductase/thioredoxin system (Kanzok et al., 2001) that may be affected during aging. Thus, it is possible that an altered metabolism of sulfur-containing compounds in MsrA-overexpressing flies may be one of the reasons for their delayed aging. Recent data on the role of MsrA in the aging process appear to support this idea. It was found that MsrA and a dietary supplement of S-methyl- L-cysteine prevent Parkinson’s like symptoms in a fruit fly model (Wassef et al., 2007). In this model, S-methyl-L-cysteine could enhance MsrA function, presumably through its cyclic oxidation by ROS and reduction by MsrA. MsrB is not an efficient reductant for this oxidized substrate. Importance of free Met-RO reduction was demonstrated in human liver carcinoma SK-Hep1 cells expressing yeast free methionine-R-sulfoxide reductase (Lee et al., 2008). These cells had an increased resistance against oxidative stress induced by hydrogen peroxide as they had an additional source of methionine derived from Met-RO.
An indirect proof of an important role of MsrA in processes other than reduction of Met-SO residues in proteins came from our experiment on the resistance of MsrB-overexpressing flies to oxidative stress induced by paraquat. It was previously shown (Ruan et al., 2002) that long-lived flies expressing MsrA in the nervous system had an enhanced resistance to oxidative stress caused by dietary paraquat. Based on these data, the authors suggested that the lifespan extension is offered by the antioxidant action of MsrA (Ruan et al., 2002). We used the same system for MsrB overexpression and the GAL4-driver with the same genotype as that in the study that examined MsrA overexpression. Our data demonstrated no protective effect of MsrBs overexpression in the fruit fly nervous system against paraquat-generated oxidative stress. We propose that an altered metabolism of sulfur-containing compounds in MsrA-overexpressing flies could provide an additional protection to oxidative stress in comparison with metabolism of MsrBs-overexpressing flies. In any case, our data suggest further studies are needed to explain how MsrA increases resistance to oxidative stress.
MsrA expression in whole body (Ruan et al., 2002) offered an approximately 40% increase in median lifespan in comparison with homozygous GAL4-activator line Ubi-GAL4 and 80% increase in comparison with homozygous UAS-MsrA responder line. It is possible that the extension effect was caused by a combination of different genetic backgrounds from UAS-MsrA line and Ubi-GAL4. In our study, we used heterozygous lines as controls that combined genetic backgrounds of lines carrying GAL4 and UAS transgenes (GAL4-activator/w1118 and UAS-MsrBs/w1118). In the experiments with whole body MsrBs expression, two different drivers (tubP-and da-GAL4) and two different enzymes (mouse MsrB2 and Drosophila MsrB) were studied. No lifespan extension was observed at any conditions tested.
Overall, our study highlights an important difference in the aging process between two enzymes that catalyze the same reaction, MetO reduction, with different stereospecificity. Apparently, MsrA is unique in providing a remarkable lifespan extension, whereas MsrB is more in line with most other antioxidant enzymes in that its overexpression does not significantly influence lifespan in the fruit fly model. Since MsrA can also reduce free Met-SO, it would be important to examine the contribution of this process to Drosophila aging in future studies.
Supplementary Material
Acknowledgments
The authors are very grateful to Dr. Daryl A. Travnicek (Department of Statistics, University of Nebraska-Lincoln, USA) for help with statistical analyses. This work was supported by the NIH (National Institute of Health) grants AG021518 (to VNG) and DK074136 (to LGH), and the KOSEF (Korea Science and Engineering Foundation) grant M10642040001-07N4204-00111 (to HYK).
Abbreviations
- Msr
methionine sulfoxide reductase
- GSH
glutathione
- GSSG
oxidized glutathione
- MetO
methionine sulfoxide
- Met-SO
methionine-S-sulfoxide
- Met-RO
methionine-R-sulfoxide
- ROS
reactive oxygen species
- SY
sugar yeast food
- DR
dietary restriction
- DR SY
dietary restriction sugar yeast food
- N SY
normal sugar yeast food
- ORF
open reading frame
- PBS
phosphate buffer saline
- HPLC
high performance liquid chromatography
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arking R, Buck S, Berrios A, Dwyer S, Baker GT., 3rd Elevated paraquat resistance can be used as a bioassay for longevity in a genetically based long-lived strain of Drosophila. Dev. Genet. 1991;12:362–370. doi: 10.1002/dvg.1020120505. [DOI] [PubMed] [Google Scholar]
- Bayne A-CV, Mockett RJ, Orr WC, Sohal RS. Enhanced catabolism of mitochondrial superoxide/hydrogen peroxide and aging in transgenic Drosophila. Biochem. J. 2005;391:277–284. doi: 10.1042/BJ20041872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer EA, Zalis MG, Wilchek M. 3-(N-Maleimido-propionyl)biocytin: a versatile thiol-specific biotinylating reagent. Anal. Biochem. 1985;149:529–536. doi: 10.1016/0003-2697(85)90609-8. [DOI] [PubMed] [Google Scholar]
- Beckman KB, Ames BN. The free radical theory of aging matures. Physiol. Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- Bonilla E, Medina-Leendertz S, Villalobos V, Molero L, Bohorquez A. Paraquat-induced oxidative stress in Drosophila melanogaster: effects of melatonin, glutathione, serotonin, minocycline, lipoic acid and ascorbic acid. Neurochem. Res. 2006;31:1425–1432. doi: 10.1007/s11064-006-9194-8. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–405. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brot N, Weissbach H. Peptide methionine sulfoxide reductase: biochemistry and physiological role. Biopolymers. 2000;55:288–296. doi: 10.1002/1097-0282(2000)55:4<288::AID-BIP1002>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Chapman T, Partridge L. Female fitness in Drosophila melanogaster: an interaction between the effect of nutrition and of encounter rate with males. Proc. Biol. Sci. 1996;263:755–759. doi: 10.1098/rspb.1996.0113. [DOI] [PubMed] [Google Scholar]
- Chavous DA, Jackson FR, O’Connor CM. Extension of the Drosophila lifespan by overexpression of a protein repair methyltransferase. Proc. Natl. Acad. Sci. USA. 2001;98:14814–14818. doi: 10.1073/pnas.251446498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönke S, Beller M, Fellert S, Ramakrishnan H, Jäckle H, Kühnlein RP. Control of fat storage by a Drosophila PAT domain protein. Curr. Biol. 2003;13:603–606. doi: 10.1016/s0960-9822(03)00175-1. [DOI] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J. Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Häcker U, Kaufmann E, Hartmann C, Jürgens G, Knöchel W, Jäckle H. The Drosophila fork head domain protein crocodile is required for the establishment of head structures. EMBO J. 1995;14:5306–5317. doi: 10.1002/j.1460-2075.1995.tb00215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfand SL, Inouye SK. Aging, life span, genetics and the fruit fly. Clin. Neurosci. Res. 2003;2:270–278. [Google Scholar]
- Holmgren A. Antioxidant enzymes. The thioredoxin system. In: Banerjee R, editor. Redox biochemistry. Hoboken, New Jersey: John Wiley & Sons, Inc; 2008. pp. 68–74. [Google Scholar]
- Kabil H, Partridge L, Harshman LG. Superoxide dismutase activities in long-lived Drosophila melanogaster females: chico1 genotypes and dietary dilution. Biogerontology. 2007;8:201–208. doi: 10.1007/s10522-006-9065-3. [DOI] [PubMed] [Google Scholar]
- Kanzok SM, Fechner A, Bauer H, Ulschmid JK, Müller H-M, Botella-Munoz J, Schneuwly S, Schirmer RH, Becker K. Substitution of the thioredoxin system for glutathione reductase in Drosophila melanogaster. Science. 2001;291:643–646. doi: 10.1126/science.291.5504.643. [DOI] [PubMed] [Google Scholar]
- Kim HY, Gladyshev VN. Methionine sulfoxide reductases: selenoprotein forms and roles in antioxidant protein repair in mammals. Biochem J. 2007;407:321–329. doi: 10.1042/BJ20070929. [DOI] [PubMed] [Google Scholar]
- Kim HY, Gladyshev VN. Selenium and methionine sulfoxide reduction. In: Hatfield DL, Berry MJ, Gladyshev VN, editors. Selenium, its molecular biology and role in human health. New York: Springer; 2006. pp. 123–133. [Google Scholar]
- Kim HY, Gladyshev VN. Different Catalytic Mechanisms in Mammalian Selenocysteine and Cysteine-Containing Methionine-R-Sulfoxide Reductases. PLoS Biol. 2005;3:2080–2089. doi: 10.1371/journal.pbio.0030375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Gladyshev VN. Methionine sulfoxide reduction in mammals: characterization of methionine-R-sulfoxide reductases. Mol. Biol. Cell. 2004;15:1055–1064. doi: 10.1091/mbc.E03-08-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby K, Hu J, Hilliker AJ, Phillips JP. RNA-interference-mediated silencing of Sod2 in Drosophila leads to early adult-onset mortality and elevated endogenous oxidative stress. Proc. Natl. Acad. Sci. USA. 2002;99:16162–16167. doi: 10.1073/pnas.252342899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klichko VI, Radyuk SN, Orr WC. CuZn-SOD promoter-driven expression in the Drosophila central nervous system. Neurobiol. Aging. 1999;20:537–543. doi: 10.1016/s0197-4580(99)00090-1. [DOI] [PubMed] [Google Scholar]
- Koc A, Gasch AP, Rutherford JC, Kim H-Y, Gladyshev VN. Methionine sulfoxide reductase regulation of yeast lifespan reveals reactive oxygen species-dependent and-independent components of aging. Proc. Natl. Acad. Sci. USA. 2004;101:7999–8004. doi: 10.1073/pnas.0307929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Kumar RA, Koc A, Sun Z, Gladyshev VN. Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proc. Natl. Acad. Sci. USA. 2002;99:4245–4250. doi: 10.1073/pnas.072603099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RA, Koc A, Cerny RL, Gladyshev VN. Reaction mechanism, evolutionary analysis, and role of zinc in Drosophila methionine-R-sulfoxide reductase. J. Biol. Chem. 2002;277:37527–37535. doi: 10.1074/jbc.M203496200. [DOI] [PubMed] [Google Scholar]
- Lee BC, Le DT, Gladyshev VN. Mammals reduce methionine-S-sulfoxide with MsrA are unable to reduce methionine-R-sulfoxide, and this function can be restored with a yeast reductase. J. Biol. Chem. 2008;283:28361–28369. doi: 10.1074/jbc.M805059200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legan SK, Rebrin I, Mockett RJ, Radyuk SN, Klichko VI, Sohal RS, Orr WC. Overexpression of glucose-6-phosphate dehydrogenase extends the life span of Drosophila melanogaster. J. Biol. Chem. 2008;283:32492–32499. doi: 10.1074/jbc.M805832200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RL, Stadtman ER. Protein modification with aging. In: Scheneider EL, Rower JW, editors. Handbook of the biology of aging. Orlando, FL: Academic Press; 1996. pp. 184–197. [Google Scholar]
- Levine RL, Moskovitz J, Stadtman ER. Oxidation of methionine in proteins: roles in antioxidant defense and cellular regulation. IUBMB Life. 2000;50:301–307. doi: 10.1080/713803735. [DOI] [PubMed] [Google Scholar]
- Lin YJ, Seroude L, Benzer S. Extended life-span and stress resistance in the Drosophila mutant methuselah. Science. 1998;282:943–946. doi: 10.1126/science.282.5390.943. [DOI] [PubMed] [Google Scholar]
- Lou MF. Antioxidant enzymes. The glutathione system. In: Banerjee R, editor. Redox biochemistry. Hoboken, New Jersey: John Wiley & Sons, Inc; 2008. pp. 74–84. [Google Scholar]
- Mair W, Piper MD, Partridge L. Calories do not explain extension of life span by dietary restriction in Drosophila. PLoS Biol. 2005;3:1305–1311. doi: 10.1371/journal.pbio.0030223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockett RJ, Sohal RS, Orr WC. Overexpression of glutathione reductase extends survival in transgenic Drosophila melanogaster under hyperoxia but not normoxia. FASEB J. 1999;13:1733–1742. doi: 10.1096/fasebj.13.13.1733. [DOI] [PubMed] [Google Scholar]
- Mockett RJ, Orr WC, Rahmandar JJ, Sohal BH, Sohal RS. Antioxidant status and stress resistance in long- and short-lived lines of Drosophila melanogaster. Exp. Gerontol. 2001;36:441–463. doi: 10.1016/s0531-5565(00)00258-8. [DOI] [PubMed] [Google Scholar]
- Moskovitz J. Methionine sulfoxide reductases: ubiquitous enzymes involved in antioxidant defense, protein regulation, and prevention of aging-associated diseases. Biochim. Biophys. Acta. 2005;1703:213–219. doi: 10.1016/j.bbapap.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Moskovitz J, Berlett BS, Poston JM, Stadtman ER. The yeast peptide-methionine sulfoxide reductase functions as an antioxidant in vivo. Proc. Natl. Acad. Sci. USA. 1997;94:9585–9589. doi: 10.1073/pnas.94.18.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz J, Bar-Noy S, Williams WM, Requena J, Berlett BS, Stadtman ER. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc. Natl. Acad. Sci. USA. 2001;98:12920–12925. doi: 10.1073/pnas.231472998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz J, Flescher E, Berlett BS, Azare J, Poston JM, Stadtman ER. Overexpression of peptide-methionine sulfoxide reductase in Saccharomyces cerevisiae and human T cells provides them with high resistance to oxidative stress. Proc. Natl. Acad. Sci. USA. 1998;95:14071–14075. doi: 10.1073/pnas.95.24.14071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr WC, Sohal RS. The effects of catalase gene overexpression on life span and resistance to oxidative stress in transgenic Drosophila melanogaster. Arch. Biochem. Biophys. 1992;297:35–41. doi: 10.1016/0003-9861(92)90637-c. [DOI] [PubMed] [Google Scholar]
- Orr WC, Sohal RS. Extension of life-span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science. 1994;263:1128–1130. doi: 10.1126/science.8108730. [DOI] [PubMed] [Google Scholar]
- Orr WC, Sohal RS. Does overexpression of Cu,Zn-SOD extend life span in Drosophila melanogaster? Exp. Gerontol. 2003;38:227–230. doi: 10.1016/s0531-5565(02)00263-2. [DOI] [PubMed] [Google Scholar]
- Orr WC, Mockett RJ, Benes JJ, Sohal RS. Effects of overexpression of copper-zinc and manganese superoxide dismutases, catalase, and thioredoxin reductase genes on longevity in Drosophila melanogaster. J. Biol. Chem. 2003;278:26418–26422. doi: 10.1074/jbc.M303095200. [DOI] [PubMed] [Google Scholar]
- Orr WC, Arnold LA, Sohal RS. Relationship between catalase activity, lifespan and some parameters associated with antioxidant defenses in Drosophila melanogaster. Mech. Ageing Dev. 1992;63:287–296. doi: 10.1016/0047-6374(92)90006-y. [DOI] [PubMed] [Google Scholar]
- Orr WC, Radyuk SN, Prabhudesai L, Toroser D, Benes JJ, Luchak JM, Mockett RJ, Rebrin I, Hubbard JG, Sohal RS. Overexpression of glutamate-cysteine ligase extends life span in Drosophila melanogaster. J. Biol. Chem. 2005;280:37331–37338. doi: 10.1074/jbc.M508272200. [DOI] [PubMed] [Google Scholar]
- Parkes TL, Elia AJ, Dickinson D, Hilliker AJ, Phillips JP, Boulianne GL. Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nat. Genet. 1998;19:171–174. doi: 10.1038/534. [DOI] [PubMed] [Google Scholar]
- Petropoulos I, Mary J, Perichon M, Friguet B. Rat peptide methionine sulphoxide reductase: cloning of the cDNA, and down-regulation of gene expression and enzyme activity during aging. Biochem. J. 2001;355:819–825. doi: 10.1042/bj3550819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JP, Campbell SD, Michaud D, Charbonneau M, Hilliker AJ. Null mutation of copper/zinc superoxide dismutase in Drosophila confers hypersensitivity to paraquat and reduced longevity. Proc. Natl. Acad. Sci. USA. 1989;86:2761–2765. doi: 10.1073/pnas.86.8.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picot CR, Perichon M, Cintrat JC, Friguet B, Petropoulos I. The peptide methionine sulfoxide reductases, MsrA and MsrB (hCBS-1), are downregulated during replicative senescence of human WI-38 fibroblasts. FEBS Lett. 2004;558:74–78. doi: 10.1016/S0014-5793(03)01530-8. [DOI] [PubMed] [Google Scholar]
- Rebrin I, Bayne AC, Mockett RJ, Orr WC, Sohal RS. Free aminothiols, glutathione redox state and protein mixed disulphides in aging Drosophila melanogaster. Biochem. J. 2004;382:131–136. doi: 10.1042/BJ20040506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, Engels WR. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan H, Tang XD, Chen ML, Joiner ML, Sun G, Brot N, Weissbach H, Heinemann SH, Iverson L, Wu CF, Hoshi T. High-quality life extension by the enzyme peptide methionine sulfoxide reductase. Proc. Natl. Acad. Sci. USA. 2002;99:2748–2753. doi: 10.1073/pnas.032671199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Seto NO, Hayashi S, Tener GM. Overexpression of Cu-Zn superoxide dismutase in Drosophila does not affect life-span. Proc. Natl. Acad. Sci. USA. 1990;87:4270–4274. doi: 10.1073/pnas.87.11.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal RS, Mockett RJ, Orr WC. Mechanisms of aging: an appraisal of the oxidative stress hypothesis. Free Radic. Biol. Med. 2002;33:575–586. doi: 10.1016/s0891-5849(02)00886-9. [DOI] [PubMed] [Google Scholar]
- Sohal RS. Role of oxidative stress and protein oxidation in the aging process. Free Radic. Biol. Med. 2002;33:37–44. doi: 10.1016/s0891-5849(02)00856-0. [DOI] [PubMed] [Google Scholar]
- Sohal RS, Agarwal S, Dubey A, Orr WC. Protein oxidative damage is associated with life expectancy of houseflies. Proc. Natl. Acad. Sci. USA. 1993;90:7255–7259. doi: 10.1073/pnas.90.15.7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söndergaard L. Homology between the mammalian liver and the Drosophila fat body. Trends Genet. 1993;9:193. doi: 10.1016/0168-9525(93)90113-v. [DOI] [PubMed] [Google Scholar]
- Stadtman ER, Van Remmen H, Richardson A, Wehr NB, Levine RL. Methionine oxidation and aging. Biochim. Biophys. Acta. 2005;1703:135–140. doi: 10.1016/j.bbapap.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Stadtman ER. Pathological processes related to redox. Protein modification. In: Banerjee R, editor. Redox biochemistry. Hoboken, New Jersey: John Wiley & Sons, Inc; 2008. pp. 184–194. [Google Scholar]
- Stipanuk MH. Sulfur amino acid metabolism: pathways for production and removal of homocysteine and cysteine. Annu. Rev. Nutr. 2004;24:539–577. doi: 10.1146/annurev.nutr.24.012003.132418. [DOI] [PubMed] [Google Scholar]
- Sun J, Tower J. FLP recombinase-mediated induction of Cu/Zn-superoxide dismutase transgene expression can extend the life-span of adult Drosophila melanogaster flies. Mol. Cell. Biol. 1999;19:216–228. doi: 10.1128/mcb.19.1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Folk D, Bradley TJ, Tower J. Induced overexpression of mitochondrial Mn-superoxide dismutase extends the life span of adult Drosophila melanogaster. Genetics. 2002;161:661–672. doi: 10.1093/genetics/161.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Molitor J, Tower J. Effects of simultaneous over-expression of Cu/ZnSOD and MnSOD on Drosophila melanogaster life span. Mech. Ageing Dev. 2004;125:341–349. doi: 10.1016/j.mad.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Toivonen JM, Walker GA, Martinez-Diaz P, Bjedov I, Driege Y, Jacobs HT, Gems D, Partridge L. No influence of Indy on lifespan in Drosophila after correction for genetic and cytoplasmic background effects. PLoS Genet. 2007;3:e95. doi: 10.1371/journal.pgen.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen CJ, Loeschcke V. Longevity and the stress response in Drosophila. Exp. Gerontol. 2007;42:153–159. doi: 10.1016/j.exger.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Wassef R, Haenold R, Hansel A, Brot N, Heinemann SH, Hoshi T. Methionine sulfoxide reductase A and a dietary supplement S-methyl-L-cysteine prevent Parkinson's-like symptoms. J. Neurosci. 2007;27:12808–12816. doi: 10.1523/JNEUROSCI.0322-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.