ABSTRACT
Uropathogenic Escherichia coli (UPEC) strains, which cause the majority of uncomplicated urinary tract infections (UTIs), carry a unique assortment of virulence or fitness genes. However, no single defining set of virulence or fitness genes has been found in all strains of UPEC, making the differentiation between UPEC and fecal commensal strains of E. coli difficult without the use of animal models of infection or phylogenetic grouping. In the present study, we consider three broad categories of virulence factors simultaneously to better define a combination of virulence factors that predicts success in the urinary tract. A total of 314 strains of E. coli, representing isolates from fecal samples, asymptomatic bacteriuria, complicated UTIs, and uncomplicated bladder and kidney infections, were assessed by multiplex PCR for the presence of 15 virulence or fitness genes encoding adhesins, toxins, and iron acquisition systems. The results confirm previous reports of gene prevalence among isolates from different clinical settings and identify several new patterns of gene associations. One gene, tosA, a putative repeat-in-toxin (RTX) homolog, is present in 11% of fecal strains but 25% of urinary isolates. Whereas tosA-positive strains carry an unusually high number (11.2) of the 15 virulence or fitness genes, tosA-negative strains have an average of only 5.4 virulence or fitness genes. The presence of tosA was predictive of successful colonization of a murine model of infection, even among fecal isolates, and can be used as a marker of pathogenic strains of UPEC within a distinct subset of the B2 lineage.
IMPORTANCE
Escherichia coli is the primary cause of urinary tract infections, the most common bacterial infection of humans. Virulence of a uropathogenic strain is typically defined by the clinical source of the isolate, the ability to colonize the bladder and kidneys in a murine model, the phylogenetic group of the bacterium, and virulence gene content. Here we describe a novel single gene, the repeat-in-toxin gene tosA, the presence of which predicts virulence of E. coli isolates regardless of source. Rapid identification of uropathogenic strains of E. coli may aid in the development of therapeutic and preventive therapies.
Introduction
The majority of urinary tract infections (UTIs) in otherwise healthy individuals are caused by uropathogenic Escherichia coli (UPEC) (1). This unique group of E. coli strains can reside in the lower gastrointestinal tract of healthy adults (2, 3), but upon entry into the urinary tract can ascend to and colonize the bladder, causing cystitis. The infection may be confined to the bladder, or bacteria may ascend the ureters to infect the kidneys causing pyelonephritis. In severe cases, bacteria can further disseminate across the proximal tubular epithelium and capillary endothelium to the bloodstream, causing bacteremia (4). A significant proportion of UTIs occur in patients with no known abnormalities of the urinary tract, so-called “uncomplicated UTIs.” However, certain host characteristics, such as the presence of an indwelling Foley catheter or congenital defect in urinary tract anatomy, are considered complicating factors for UTI, and increase susceptibility to this infection, as well as affecting diagnosis and management (5). Finally, colonization of the bladder in high numbers (>105 CFU/ml of voided urine) may occur without eliciting symptoms from the host, a condition known as asymptomatic bacteriuria (ABU) (6).
As a species, E. coli demonstrates significant genome plasticity, possessing a well-conserved core genome, as well as highly variable and heterogeneous accessory genetic material apparently acquired through multiple horizontal gene transfer events (7). Among UPEC strains, individual mechanisms of pathogenesis have been well characterized (8); however, no single fixed set of virulence factors has been observed in the majority of clinical isolates. Unlike the intestinal pathogenic E. coli strains, distinct pathotypes of UPEC have yet to be defined (9). Without a distinct set of markers to differentiate UPEC from other types of E. coli, most of our knowledge about this important human pathogen comes from studying a few prototype strains. Determination of a more accurate and reliable pathogenic signature for UPEC among the other commensal and pathogenic E. coli strains in the gastrointestinal tract, the natural reservoir of UPEC (2, 3, 10), will be invaluable for investigations of the epidemiology of UPEC or public health concerns, such as the impact of antimicrobial pressure on the natural diversity of UPEC.
To develop means to reliably differentiate UPEC from non-UPEC, we tested E. coli isolates from fecal samples and specific UTI syndromes for the presence of specific genes previously characterized as UPEC virulence factors or suspected to play a role in fitness in the urinary tract, querying whether unique combinations of these genes were enriched in UPEC isolates from distinct clinical syndromes. A single gene, tosA, which encodes an in vivo-induced repeat-in-toxin (RTX) family member, was identified as a candidate UPEC marker. We tested the hypothesis that the presence of tosA correctly predicts the ability of a strain to successfully colonize the urinary tract. Finally, the possibility that tosA-positive strains represent a phylogenetically distinct subset within the overall E. coli population was examined by sequence analysis of representative isolates identified in the study.
RESULTS
Prevalence of virulence and fitness genes increases in strains from clinical scenarios of increasing severity.
A collection of 314 strains of E. coli, representing isolates from fecal samples, ABU, uncomplicated cystitis and pyelonephritis, and complicated UTIs (Table 1), was assessed by multiplex PCR for the presence of 15 virulence or fitness genes (Fig. 1A). The 15 genes, chosen to broadly represent documented virulence and fitness factors and general mechanisms of pathogenesis, have been well characterized, with the exception of tosA (Table 2). These genes are categorized as encoding proteins that (i) mediate adherence to host cells (fimA and papA), (ii) mediate acquisition of the essential micronutrient iron (chuA, hma, iutA, iroN, fyuA, iha, and ireA), or (iii) secrete proteins that elicit toxic effects on host cells (hlyA, cnf1, tosA, sat, picU, and tsh). Eleven of the chosen genes reside on large stretches of DNA within the genome of E. coli CFT073 known as pathogenicity-associated islands (PAI) (11), predicted to be acquired through horizontal gene transfer, while three of the genes are widely dispersed around the genome of the prototypical pyelonephritis strain CFT073 (Fig. 1B). cnf1 is not present in CFT073, but it is prevalent in other strains.
TABLE 1 .
E. coli isolates used in this study
| Clinical group | No. of isolatesa | Source of collectiona |
|---|---|---|
| Fecal | 91 | 13, 35 |
| Compromised host | 41 | 38, 39 |
| Asymptomatic bacteriuria | 68 | 6, 35 |
| Cystitis | 37 | 25, 35, 36 |
| Pyelonephritis | 78 | 35, 37 |
The number of total isolates of each category is given along with the reference to the publication that describes the source of the isolates.
FIG 1 .
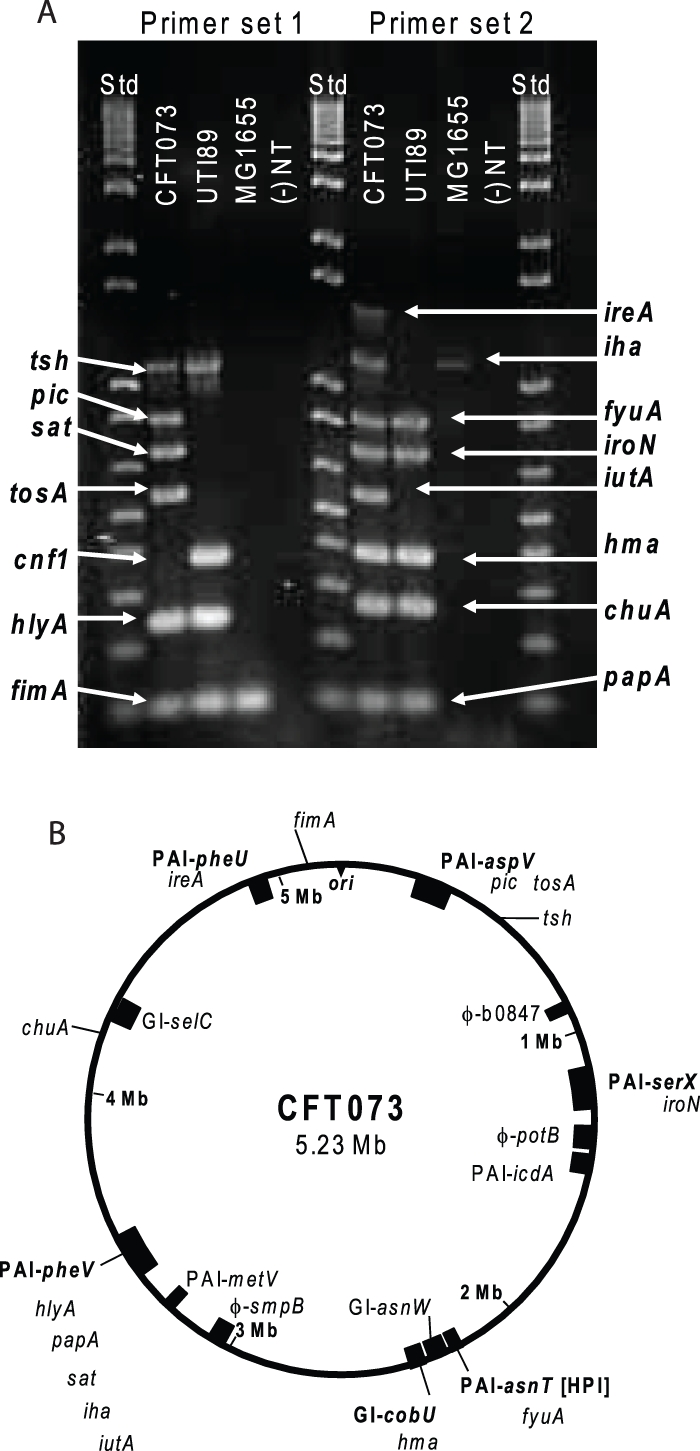
Virulence and fitness genes assessed in this study. (A) Multiplex PCR results using genomic DNA purified from CFT073, UTI89, and MG1655. Primers specific for each virulence or fitness gene were used for PCR amplification of gene fragments from a boiled lysate of the indicated E. coli strain. If there was no band observed at the expected size, the result was counted as negative for the presence of a gene (−). NT, no-template control samples; Std, DNA ladder standards. (B) Schematic of CFT073 genome with the location of each gene under study indicated. The origin of replication is indicated by ori. Previously identified pathogenicity-associated islands (PAI) or genomic islands (GI) containing one or more genes under study are indicated according to the nomenclature of Lloyd et al. (11). cnf1, which is not present in strain CFT073, is not shown. Phage insertion sites (φ). Three genes, tsh, chu, and fimA, are not found in a PAI or GI.
TABLE 2 .
Prevalence of virulence-associated genes among E. coli isolates from different clinical settings
| Isolate type | % of isolates with genea |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adhesins |
Toxins |
Outer membrane iron receptors |
|||||||||||||
| fimA | papA | hlyA | cnf1 | tosA | sat | pic | tsh | chuA | hma | iutA | iroN | fyuA | iha | ireA | |
| Fecal | 98.9 | 26.4 | 13.2 | 7.7 | 11.0 | 19.8 | 12.1 | 20.9 | 39.6 | 26.4 | 31.9 | 20.9 | 47.3 | 31.9 | 14.3 |
| CHb | 97.6 | 51.2 | 19.5 | 4.9 | 19.5 | 41.5 | 12.2 | 48.8 | 75.6 | 34.1 | 58.5 | 26.8 | 78.0 | 46.3 | 24.4 |
| ABUc | 98.5 | 54.4 | 35.3 | 33.8 | 26.5 | 33.8 | 23.5 | 63.2 | 73.5 | 51.5 | 45.6 | 41.2 | 83.8 | 35.3 | 16.2 |
| Cystitis | 97.3 | 67.6 | 48.6 | 32.4 | 16.2 | 32.4 | 18.9 | 54.1 | 83.8 | 81.1 | 43.2 | 48.6 | 89.2 | 37.8 | 10.8 |
| Pyelonephritis | 98.7 | 63.6 | 48.1 | 13.0 | 28.6 | 51.9 | 29.9 | 67.5 | 89.6 | 61.0 | 66.2 | 41.6 | 93.5 | 55.8 | 27.3 |
| tosA -positive strains | 98.4 | 59.4 | 70.3 | 37.5 | 100.0 | 67.2 | 89.1 | 96.9 | 98.4 | 92.2 | 67.2 | 59.4 | 98.4 | 67.2 | 20.3 |
| tosA-negative strains | 98.4 | 47.2 | 21.6 | 12.0 | 0.0 | 26.8 | 2.0 | 36.8 | 61.6 | 36.4 | 43.2 | 28.0 | 69.6 | 34.4 | 18.4 |
Gene prevalence is expressed as the percentage of isolates that scored on multiplex PCR.
CH, compromised host anatomy.
ABU, asymptomatic bacteriuria isolates.
Screening of the 314 clinical and fecal E. coli isolates for UPEC-associated virulence or fitness genes identified a wide range of prevalence values for the 15 targeted virulence genes, in certain instances with considerable variation according to source group. The average percentage of strains that scored positive for a given gene per isolate group is shown in Table 2.
tosA is a marker for the presence of other virulence or fitness genes.
In a previous study, our group identified tosA as the main contributing factor of PAICFT073-aspV to enhanced fitness of UPEC strain CFT073 in a murine model of ascending UTIs (12). Here, we explored associations among tosA and other virulence genes by comparing the prevalence of other virulence genes in isolates positive versus negative for a given virulence gene (Table 3). In this analysis, tosA-positive strains overall showed a combination of carrying the highest number of other virulence genes, the greatest differential in overall virulence gene content, the greatest differential in percent virulence gene prevalence, and the ability of strains from a fecal source marked by that gene to colonize both the murine bladder and kidney (Table 3), and thus, the following analysis focused on this gene.
TABLE 3 .
Characteristics of isolates marked by the presence of a single virulence or fitness gene
| Gene | Avg no. of virulence genes present in isolatea |
ΔVFb | Avg % change in gene prevalencec |
t test result ford: |
||
|---|---|---|---|---|---|---|
| Positive | Negative | Bladder | Kidney | |||
| tosA | 11.2 | 5.4 | 5.9 | 34.7 | <0.0001 | 0.0071 |
| picU | 11.2 | 5.4 | 5.8 | 34.6 | <0.0001 | NS |
| hlyA | 10.1 | 4.9 | 5.2 | 30.2 | NS | NS |
| cnf1 | 10.1 | 5.8 | 4.3 | 23.7 | NS | NS |
| sat | 9.5 | 5.0 | 4.5 | 25.2 | NS | NS |
| hma | 9.4 | 4.0 | 5.4 | 31.6 | <0.0001 | 0.0074 |
| tsh | 9.4 | 3.9 | 5.5 | 32.1 | <0.0001 | NS |
| iroN | 9.0 | 5.3 | 3.7 | 19.5 | NS | NS |
| iha | 9.0 | 4.9 | 4.1 | 22.1 | NS | NS |
| papA | 9.0 | 4.2 | 4.8 | 26.9 | 0.014 | NS |
| ireA | 8.7 | 6.1 | 2.7 | 11.8 | 0.0002e | 0.0155e |
| iutA | 8.7 | 4.6 | 4.1 | 22.1 | 0.0405e | NS |
| chuA | 8.4 | 2.4 | 6.0 | 35.4 | 0.0003 | NS |
| fyuA | 8.1 | 1.7 | 6.4 | 38.6 | 0.0017 | NS |
| fimA | 6.6 | 6.6 | 0.0 | −7.5 | ||
Average number of the 15 virulence factors present in isolates that contain (positive) or lack (negative) the indicated gene.
ΔVF, difference in the number of virulence factors between positive and negative.
The percentage change in gene prevalence of all 15 genes is averaged for each group.
Data in Fig. 2B and C were grouped according to the presence or absence of each gene and reanalyzed; the P value of each group that was found to be significant is reported. NS, not significant.
These genes were found to be negatively correlated with CFU/g of tissue. The presence of ireA was found previously (44) to negatively correlate with lethality in a murine model of peritonitis.
tosA-positive isolates collectively exhibited a higher prevalence of almost every virulence gene than was observed in any of the clinical isolate groups (Tables 2 and 3). For example, hlyA was present in 48% of pyelonephritis isolates (the highest-prevalence clinical group) but in fully 70% of tosA-positive isolates. Likewise, the heme receptor chuA was present in 89.6% of pyelonephritis isolates (again, the highest-prevalence clinical group) but in 98.4% of tosA-positive isolates. tosA-positive isolates contained, on average, 11.2 of the 15 studied virulence or fitness genes, whereas tosA-negative isolates averaged only 5.4 virulence or fitness genes (Table 3). This trend was observed regardless of from which strain collection the tosA-positive isolate originated. When data were analyzed independently in this way for every virulence gene, strains grouped first by tosA or second by picU were associated with the highest number of other virulence genes assayed for in this study (Table 3). Both tosA and picU genes reside within the same pathogenicity-associated island, PAICFT073-aspV (Fig. 1B), and thus may be genetically linked in strains within the collection.
Other toxin genes.
The category of secreted toxins showed a similar pattern, with the prevalence of a given gene generally increasing in relation to increasing clinical severity of the source group. The ABU isolates were a notable exception. Although by definition, ABU strains elicit no symptoms from the host, the prevalence of the gene sequences in this category were comparable among ABU strains to those observed among cystitis and pyelonephritis isolates. In the case of cnf1 (cytotoxic necrotizing factor 1), for example, ABU isolates were the highest-prevalence group. However, the presence of a specific gene sequence does not necessarily indicate the presence of an intact gene and functional gene product. Accordingly, additional experiments were conducted to determine if multiplex PCR results for another toxin gene, hlyA, which encoded the secreted toxin α-hemolysin, were predictive of its distinctive phenotype.
The presence or absence of the secreted protein toxin gene hlyA among strains in this study is highly correlated with the in vitro hemolysis phenotype. When the 314 strains under study were cultured on 5% sheep blood agar plates, a positive PCR result predicted a hemolytic pattern on blood agar plates with a sensitivity of 97.9%. A negative PCR result for this gene was determined to be 98.1% specific for a lack of hemolysis seen on blood agar plates. Previous research has established that the presence of hemolysin is predictive of the level of pathology observed in tissue culture systems (13) and correlated to the severity of pathology observed in the murine model of an ascending UTI (14). The high sensitivity and specificity of the multiplex PCR assay for predicting the hemolysin phenotype indicated that the data on a gene’s presence or absence provided insight into the pathogenic potential of each strain. This appealing concept was further investigated with several statistical tools in an analysis of the entire data set to gain a more detailed understanding of how the molecular mechanisms of pathogenesis are distributed among natural isolates of E. coli (see the section on testing of three models).
Adhesin and iron acquisition genes.
fimA encoding the type 1 fimbria major structural subunit was almost universally present in strains from each clinical group regardless of source, with >98% of isolates testing positive. In contrast, papA (coding for the structural subunit of pyelonephritis-associated pili), although less prevalent overall (56.5%), was significantly more prevalent among all urine isolates combined (59.2%) than among fecal isolates (26.4%) (P < 0.0001). Both of these findings are in general agreement with results from previous studies (4).
The genes encoding outer membrane iron receptors (chuA, hma, iutA, iroN, fyuA, iha, and ireA) were less common among fecal isolates (average prevalence, 30.3%; range, 14.3 to 47.3%) and more common among strains isolated from a symptomatic UTI (average prevalence of 59.3% and range of 10.8 to 93.5%), with the prevalence rates of certain genes, such as fyuA in pyelonephritis isolates, exceeding 90%. This result is also in agreement with previously published findings (15).
tosA-positive strains colonize the murine urinary tract regardless of clinical source.
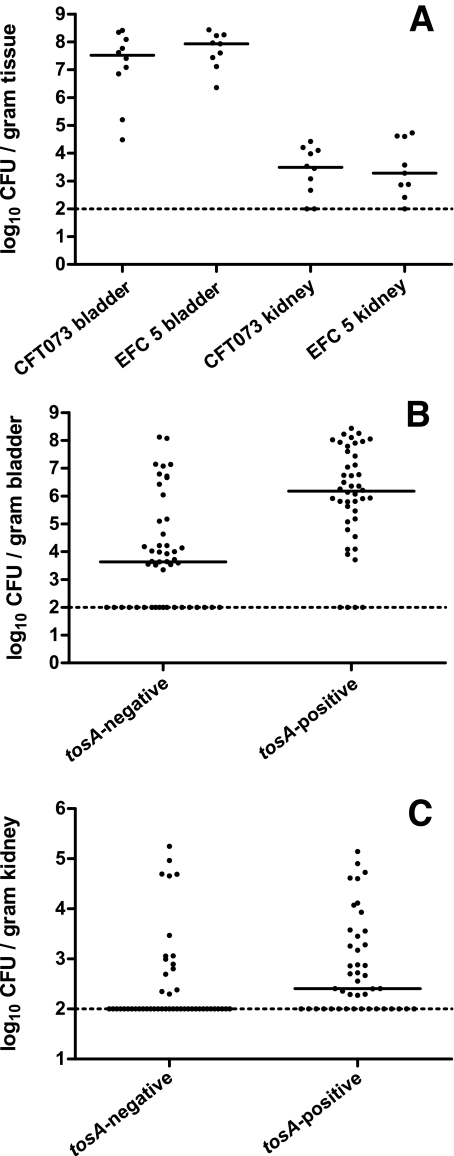
If one hypothesized that the presence of tosA predicts virulence regardless of clinical source, then the presence of tosA should predict successful colonization of a tosA-positive fecal E. coli isolate in the murine model of an ascending urinary tract infection. For example, EFC5, a fecal isolate from a healthy human volunteer (13) determined in this study to be tosA positive, was transurethrally inoculated into the bladder of female C57BL/6 mice. Forty-eight hours postinoculation, mice were euthanized, bladder and kidney tissue was removed, and the CFU/g of tissue was determined for each mouse to assess whether or not this fecal isolate could colonize and survive in the host’s urinary tract. This was repeated with strain CFT073, a human pyelonephritis isolate that is also tosA positive (Fig. 2A). There were no differences observed among colonization levels for both bladder and kidney tissue between these two strains, indicating that the fecal isolate EFC5 was a successful colonizer in this model system.
FIG 2 .
tosA presence predicts successful colonization of the murine urinary tract. (A) Mice were transurethrally inoculated with 108 CFU of either CFT073 or EFC5. Forty-eight hours postinoculation, bladders and kidneys were removed, and the numbers of CFU/g of tissue were determined. Each dot represents the CFU load for one mouse. Panels B and C show combined results for 10 tosA-negative and 7 tosA-positive strains for bladder and kidney tissue, respectively. Each dot represents one mouse (n = 44 and 48 for tosA-positive and tosA-negative, respectively).
The murine model is recognized as capable of differentiating fecal and uropathogenic strains of E. coli (13, 16). However, fecal E. coli isolates that carry tosA colonize the murine model in higher numbers in both bladder and kidney tissue than tosA-negative fecal isolates. Seven tosA-positive fecal strains, including EFC5, were tested in the murine model and compared to 10 tosA-negative fecal isolates for CFU/g of bladder (Fig. 2B) or kidney tissue (Fig. 2C) at 48 h postinfection. Statistically significant differences were observed using a two-tailed Student’s t test with higher average CFU levels in the tosA-positive group for both bladder (P < 0.001) and kidney (P < 0.01) tissue.
The presence of tosA also predicted a greater CFU load in bladder and kidney tissue of infected mice regardless of the host species from which a strain was isolated. One tosA-positive strain (ECOR 57) and one tosA-negative strain (ECOR 70) isolated from gorillas were compared for the ability to colonize in the murine model described above. A second pair of tosA-positive and tosA-negative fecal isolates from orangutans (ECOR 52 and ECOR 7) was also compared. Both tosA-positive isolates colonized both bladder and kidney, while only ECOR 7, a tosA-negative strain isolated from an orangutan, colonized the bladder and kidneys (data not shown). An additional tosA-positive fecal isolate collected from a lion (ECOR 58) was tested and found to colonize both bladder and kidneys in the murine model. ECOR 58 is unusual as this strain only contained three additional genes tested for in this study (fimA, picU, and iroN). These results demonstrate that the tosA gene is a marker for strains that can successfully colonize the murine model, including fecal E. coli isolates from other animals as well as for isolates that contained relatively few of the genes that have been well studied in the context of pathogenesis in the urinary tract.
tosA-positive strains comprise a distinct subset of the B2 lineage of E. coli.
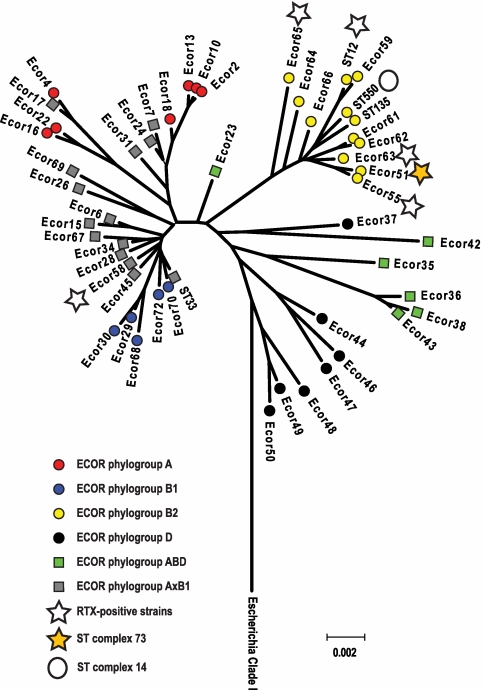
A multiplex assay (17) to place E. coli into one of the four main ECOR phylogenetic group assignments, A, B1, B2, or D, was applied to each strain in this study (Table 3). E. coli fecal isolates were frequently placed within group A (42.9% of isolates), but this group less commonly contained isolates cultured from the urinary tract. For example, only 7.7% of strains isolated from cases of pyelonephritis localized to group A. On the other hand, 70% of isolates from cases of cystitis and pyelonephritis were found to be members of group B2, which was the most common group among isolates from the urinary tract. In contrast to this, 63 of the 64 (98.4%) tosA-positive strains in this study were determined to reside in the B2 phylogenetic group of E. coli. A single tosA-positive isolate (ECOR 58) localized to the B1 phylogenetic group. The hypothesis that these isolates comprised a closely related subset of UPEC was then examined further by sequence analysis
The majority of tosA-positive isolates (8 of 12 [66.6%]) examined by multilocus sequence typing (MLST) belonged to a single sequence type (ST) complex, ST complex 73. In addition to the ECOR strain collection, the strains examined in the murine model were assigned to a ST by standard protocols (18), and this collection of sequences was used to build a dendrogram illustrating the relationship of these groups to each other (Fig. 3). The ECOR strain collection provided a framework of sequence types associated with each of the four main E. coli groups as well as a source of several groups of E. coli previously determined to have undergone extensive genetic recombination. The only isolate that contained tosA outside of the B2 group (ECOR 58) was previously determined to belong to a group of E. coli strains that appear to have undergone extensive recombination with other groups of E. coli (18). Further examination of the MLST database showed that the majority of strains in ST complex 73 (29 of 48 [60.4%]) are UPEC isolates and were collected in the setting of a human UTI (18). Based on the MLST and ECOR typing, tosA appears to be a marker for a group of strains that are enriched for UPEC and present in a few outlier groups that may have acquired this gene by extensive genetic recombination.
FIG 3 .
Phylogenetic relationship of tosA-positive strains. Representative tosA-positive strains included in this study are marked with stars. The four main groups of E. coli (A, B1, B2, and D) and two groups of recombinants (ABD and AxB1) were assigned according to Wirth et al. (18). MLST complexes 73 and 14 are indicated next to representative isolates from each group. Sequences from Escherichia clade 1 isolates that do not group with phylogenetic groups A, B1, and B2 (D) or the hybrid groups AxB and ABD were used to root the dendrogram (42).
Testing of three models for assessing pathogenic potential.
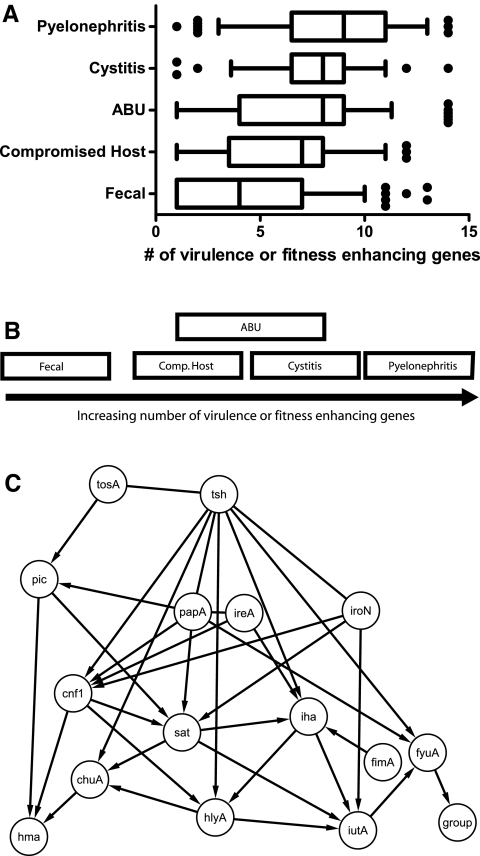
While we have discovered that the presence of tosA predicts virulence regardless of clinical source, can we also use the full virulence and fitness gene prevalence data set to differentiate strains by clinical source alone? Fecal isolates of E. coli on average had significantly fewer virulence genes (mean of 4.2) than isolates from the urinary tract (mean of 7.5) (Table 4). One simple model that could explain the difference in the distribution of the genes under study is that isolates that caused the most severe clinical disease contain more genes and mechanisms linked to pathogenesis than those that cause milder clinical cases of disease or those that were obtained from healthy individuals. This model was tested by averaging the number of genes detected as present in a group of strains and compared across all groups by one-way analysis of variance (ANOVA) (Fig. 4A). The global test statistic was significant (P < 0.001); however, pairwise testing was only significant for fecal isolates compared to the other groups and for comparison of pyelonephritis isolates with isolates from patients with compromised urinary tract anatomy. This indicates that fecal isolates can be differentiated from strains of E. coli isolated from the urinary tract based on the average number of virulence-associated genes present, but that this is insufficient to fully differentiate between urinary tract isolates from specific clinical scenarios of different severities. Isolates that were presumed to have limited pathogenic potential, such as ABU isolates, were indistinguishable from isolates known to be highly pathogenic and elicit strong symptoms from the host, such as pyelonephritis isolates, based on this test. While it is possible that ABU isolates are true pathogens and the lack of symptoms is the result of host innate immune defects in the infected individual (19), it is also possible that mutations exist within virulence gene operons, rendering their products inactive (20).
TABLE 4 .
Average number of virulence-associated genes and ECOR group membership
| Clinical group | Avg no. of virulence genes |
% of isolates that belong to ECOR phylogenetic groupa: |
|||
|---|---|---|---|---|---|
| A | B1 | B2 | D | ||
| Fecal | 4.2 | 42.9 | 18.7 | 20.9 | 17.6 |
| Compromised host | 6.4 | 17.1 | 7.3 | 51.2 | 24.4 |
| Asymptomatic bacteriuria | 7.2 | 15.9 | 11.1 | 58.7 | 14.3 |
| Cystitis | 7.6 | 16.7 | 0.0 | 71.4 | 11.9 |
| Pyelonephritis | 8.4 | 7.7 | 2.6 | 70.5 | 19.2 |
| tosA-positive strains | 11.2 | 0.0 | 1.6 | 98.4 | 0.0 |
| tosA-negative strains | 5.4 | 27.6 | 11.2 | 39.2 | 22.0 |
ECOR group membership expressed as % of isolates that belong to the four main ECOR phylogenetic groups.
FIG 4 .
Models demonstrating differences and similarities between E. coli isolates from different clinical settings. (A) Under the first model, groups are placed according to the average number of virulence factors as described in a box-and-whisker plot in which the mean is indicated by a vertical line, the 25th and 75th percentiles are indicated by the left and right borders of the box, and the 10th and 90th percentiles are indicated by the whiskers. Dots represent outliers in the analysis. (B) The third model used multivariate ANOVA of the full presence or absence data for each gene to test for differences in gene prevalence rates between all five groups of isolates. Groups shown to overlap cannot be readily differentiated based on the data collected in this study, while those that do not overlap were found to contain different combinations of the genes under study. (C) Bayesian model of unique connections between virulence and fitness factor genes. The presence or absence of each gene in each of 314 strains was modeled to identify a consensus network of the genes. Knowledge of the presence or absence of a given gene predicts the presence or absence of another gene. Arrows indicate directed edges linking two variables together in the network.
A second model that could explain the distribution of the genetic mechanisms of virulence, based on classes of virulence determinants present in the urinary tract, was explored. UPEC strains are genetically heterogeneous and often exhibit multiple molecular mechanisms for causing disease, such as secreting a series of protein toxins (8) or synthesizing multiple acquisition systems for the uptake of iron and iron-containing compounds (15). The second model assumed that success in the urinary tract and the production of symptoms in the host were the sum of three activities—the production of large secreted protein toxins or RTX family member (cnf1, hlyA, and tosA), the production of secreted proteolytic autotransporter proteins (sat, pic, and tsh), and the acquisition of iron (chuA, hma, iutA, iroN, fyuA, iha, and ireA). Under this model, the isolates from different clinical groups may be differentiated by comparing the potential of the strains to achieve each of these three activities. After averaging the number of genes in each category per strain and adjusting for the mean and standard deviation of each category, a multivariate version of an ANOVA design (MANOVA) was used to seek differences between the clinical groupings. Similar to the first model, although the global test was significant (P < 0.001), pairwise testing only clearly differentiated fecal isolates from the other clinical isolates. The urine-source groups were statistically indistinguishable, regardless of disease severity or host compromise status.
The third model, which was able to differentiate between groups of urinary tract isolates, assumed that each genetic mechanism of survival and pathogenesis (defined by the presence of distinct virulence or fitness genes) confers a unique advantage in the host. Under this model, the level of success of a strain of E. coli in different patient populations and sites within the urinary tract would be partly determined by the unique contribution of each genetic mechanism of pathogenesis. While this study could not assess the entire collection of known virulence factors described in the literature, the 15 genes assayed by PCR provided sufficient information to allow a multivariate ANOVA model using the full presence or absence data for each gene to test for differences in gene prevalence rates between all five groups of isolates. Similar to the first two models, the global test was significant (P < 0.001), and fecal isolates were significantly different from other strain collections. However, significant differences were also observed among E. coli isolates from patients with different clinical scenarios. Under this model, all groups of urinary tract isolates were distinguished from each other, except for an overlap between ABU isolates and isolates from patients with compromised urinary tract anatomy and between ABU isolates and cystitis isolates. Figure 4B presents a visual representation of the results of the ANOVA and MANOVA testing and demonstrates that, even after allowing each of the 15 genes studied to count as separate variables, urinary tract isolates of E. coli from the four different classes of clinical severity exhibited more similarities to other urinary tract isolates than to fecal isolates.
tosA-positive isolates are significantly different from tosA-negative strains in all three statistical models tested.
Due to the high similarity noted among the gene prevalence data, the three models described above were retested with the strains grouped by the presence or absence of tosA. Because only two groups (tosA positive and tosA negative) were being tested, the original ANOVA was replaced by a Student’s t test for evaluation of the average number of genes present. Both multivariate tests were conducted as before, and in each case, tosA-positive isolates were significantly different from tosA-negative isolates (P < 0.01).
Bayesian network modeling reveals extensive connections between virulence and fitness genes.
Finally, to seek relationships between the combinations of virulence genes present in specific strains, Bayesian network (BN) modeling was employed. This technique allows the influence of the presence of a gene on the presence of the other genes to be assessed visually without the need for hundreds of individual pairwise statistical comparisons. A consensus network compiled from the best scoring models discovered numerous connections between genes (Fig. 4C). For example, three genes, tosA, tsh, and iroN, are at the top of the hierarchical structure within the consensus network. These genes appear to influence or closely associate with the presence or absence of the other genes. This model illustrates that only the yersiniabactin receptor (fyuA) was directly linked to a particular clinical setting (“group”). Indeed, fyuA is less common in fecal isolates (47.3%) but steadily increases in presence as the severity of the disease increases, reaching a maximum prevalence of 93.5% in pyelonephritis isolates (Table 2). Our model also indicates that fyuA is closely associated with and possibly influenced by three other genes: tsh, papA, and iutA. Table 2 indicates that these three genes have a similar pattern to fyuA. However, based on the available binary data, our modeling predicted that fyuA has strong direct association with the clinical setting.
DISCUSSION
Virulence-associated gene content among E. coli isolates from the urinary tract demonstrated more similarities to other UPEC strains than to fecal isolates. While a general trend of increasing numbers of virulence factors was observed in strains obtained from the more severe clinical settings (Table 4 and Fig. 4A), each group of isolates could only be differentiated by multivariate analysis of the entire virulence gene prevalence data set (Fig. 4B). In contrast, strains that carried tosA were easily differentiated from tosA-negative strains by several measures. tosA, a putative RTX family member gene (12), co-occurred on average with more than 10 of the other 14 genes assayed for in this study, and thus its presence was predictive of highly virulent strains (Tables 2 and 3). This gene represents a marker for E. coli belonging to the B2 phylogenetic group and is enriched in sequence type complex 73, a group of E. coli strains enriched for UPEC isolates, including the prototype human pyelonephritis strain CFT073 (18) (Fig. 3).
UPEC strains reside in the gastrointestinal tract and thus can occasionally be isolated among commensal strains cultured from fecal samples (2, 3, 21). Indeed, the presence of tosA predicted successful colonization of all but one of seven tosA-positive fecal E. coli isolates tested in the murine model of an ascending UTI (Fig. 2B and C). ECOR 51, the only tosA-positive fecal isolate that did not colonize the animal model, lacked the gene fimA, the major structural subunit of type 1 fimbriae. While type 1 fimbriae are critical for the success of UPEC in the murine model (22–24), recent studies have questioned the importance of this factor in human infections (25, 26) and raise the possibility that this isolate could potentially colonize the human urinary tract. While it was beyond the scope of this study to identify a specific role for this gene in bacterial pathogenesis, the results presented here indicate that success of tosA-positive strains may rely in part on the co-occurrence of known virulence factors of UPEC. With the exception of papA and ireA, the highest prevalence of the 12 other genes included in this study among all groups of isolates was observed among tosA-positive strains (Table 2). However, the observation that ECOR 58, a fecal E. coli isolate that lacked many of the best-characterized UPEC virulence factors, demonstrates that tosA can nevertheless serve as a marker both for UPEC strains that are predicted to cause disease through well-known mechanisms as well as for strains that may contain unique and as yet uncharacterized approaches to colonizing the host urinary tract.
One goal of the study was to determine whether we could differentiate strains based on clinical source and virulence and fitness gene content. First, it was straightforward to statistically separate fecal strains from strains isolated from the urinary tract by using the three models (Fig. 4). However, separation of isolates by specific clinical populations proved more difficult. We found seemingly minimal differences in the numbers of virulence or fitness genes among UPEC isolates from different patient populations (Fig. 4A). Using model 3, however, these differences were sufficient to differentiate between strains from patients with complicated UTI (compromised host urinary tract anatomy) and those from uncomplicated symptomatic infections (cystitis and pyelonephritis) (Fig. 4B).
ABU strains in this study, on the other hand, failed to coalesce into a discrete group and instead revealed a spectrum of isolates that ranged from strains that contained only 1 of the 15 genes assayed to strains that contained 14 of the same genes (Fig. 4A). These strains, by our analysis, closely resemble fecal commensal strains that failed to colonize the murine model (that is, with low virulence potential) and those strains that more closely resemble human cystitis isolates (with higher virulence potential), respectively. That these strains colonize but elicit no symptoms could be explained in at least three ways. (i) Low-virulence strains, unrelated to typical UPEC strains, could colonize a healthy individual if there were additional as yet undetermined colonization genes present (27, 28). Lack of virulence factors would not damage the host and thus cause no symptoms. (ii) Strains with a high number of virulence genes could contain mutations within these genes that are nevertheless present and detectable by PCR (20, 29). E. coli strain 83972, for example, may represent such a strain that contains virulence genes but also carries mutations within some of these genes (20, 29). (iii) Truly high-virulence strains with intact virulence genes could colonize a host with defects in innate immunity who cannot respond properly to infection (19). The broad overlap of ABU isolates with strains obtained from hosts with compromised urinary tract anatomy and the overlap with cystitis isolates observed in this study could be explained by a combination of these three scenarios.
Bayesian network analysis is a powerful statistical method that infers potential relationships among a set of variables and provides an alternative to analyzing hundreds of individual pairwise comparisons (30, 31). In the present study, such a network indicates that the knowledge of the presence or absence of a given gene can predict the presence or absence of another gene. Before conducting this analysis, we predicted that genes contained in the same pathogenicity-associated island (PAI), large stretches of DNA thought to be acquired en masse through horizontal gene transfer, would be linked together in the consensus network (Fig. 4C). For example, PAICFT073-pheV contains papA, sat, iha, iutA, and hlyA; these genes are all serially connected in the consensus network. What was not expected was that nearly every gene included in the study would demonstrate some level of connectivity. As an example, the gene tsh, which encodes an autotransporter expressed during both in vivo and in vitro growth of CFT073 (32), is not associated with any known PAI (Fig. 1B). Despite this, tsh contains direct links to 8 other genes in the consensus network. This result indicates that gene association between established UPEC virulence factors may not be confined to those genes contained in the same PAI. In addition, the gene content of a PAI, the location of additional copies of the same genes outside of the archetypal PAI, and differences in number of PAIs have been previously documented between strains of UPEC (7, 33). The Bayesian network results (Fig. 4C) are consistent with the hypothesis that such genetic reassortments of virulence factors are common among UPEC strains.
Given the previous observations that Bayesian network analysis is sufficiently robust to screen out noise in large gene expression arrays (30, 34), it is surprising that only the yersiniabactin receptor gene fyuA, and not tosA or other genes, was determined to directly influence the probability that a given strain originated in a particular clinical group. Indeed, our Bayesian network modeling found that the presence or absence of fyuA was directly associated with or influenced by three other genes (tsh, papA, and iutA), and the presence of tosA appears to play a leading role in predicting the presence of many of the other genes (Fig. 4C). This was somewhat surprising since tosA was found in only one-fourth of strains. Taken together, these results may suggest that distinct pathotypes of UPEC exist, but identification of these groups will require knowledge about the diverse mechanisms of pathogenesis found within each of the different subpopulations of UPEC.
We define a UPEC strain as one that infects the human urinary tract. Our tools to define such an isolate include virulence and fitness gene content, colonization of the murine urinary tract, and the original clinical source. We observed here that clinical source alone is not necessarily predictive of UPEC status, nor is the presence of a single gene. Not all isolates that have the potential to colonize the urinary tract, for example, carry the tosA gene. Only 24.3% (28/115) of cystitis and pyelonephritis isolates, in the present study, contain this gene. These isolates are most likely to resemble ST complex 73 strains such as CFT073, but other groups of UPEC exist. Strains such as E. coli fecal control 24 (EFC24), one of the tosA-negative strains investigated in this study that successfully colonized the bladders of infected mice, represent one of the groups that together comprise the remaining 75% of UPEC isolates. EFC24 was determined in this study to belong to ST complex 14, a group of B2 isolates highly enriched for UPEC. In fact, 10 of the 13 (76.9%) isolates that make up ST complex 14 were isolated from the human urinary tract (18). While many of the tosA-negative fecal strains that were successful in the murine model appear to belong to larger groups of urinary tract isolates, none of the genes included in this study could differentiate these isolates from those tosA-negative isolates that failed to colonize the host urinary tract. Additional studies will be necessary to identify a reliable marker for groups of UPEC that lack the tosA gene.
Despite these limitations, the presence or absence of tosA provides useful information in assessing the pathogenic potential of a given E. coli isolate. As this gene is found in one-quarter of UPEC strains that cause symptomatic infections in normal human hosts, assaying for this one gene would enable larger studies that have previously not been feasible, such as studies that compare UPEC carriage rates in patient populations with different rates of urinary tract infections. When combined with other virulence and fitness genes such as hma, picU, and fyuA, the urovirulence potential of these isolates could be accurately predicted. Given the heterogeneity in pathogenetic mechanisms observed among UPEC, such an understanding may prove critical to future efforts to develop novel therapeutic strategies to combat this widespread human pathogen.
MATERIALS AND METHODS
Strain collection and culture conditions.
A total of 314 strains of E. coli was collected from a variety of clinical settings (Table 1) (6, 13, 20, 25, 35–39). The collection covered a range of natural isolates, from nonpathogenic strains that reside in the lower gastrointestinal tract to highly pathogenic strains that were isolated from the blood of human patients with clinical cases of pyelonephritis complicated by bacteremia. This collection included a broad range of human fecal isolates and isolates from nonhuman primates and other mammalian hosts of E. coli (13, 35). In addition to isolates recovered from human patients with lower urinary tract infections (cystitis) (25, 35, 36) and upper urinary tract infections (pyelonephritis) (35, 37), this collection included samples taken from two additional patient populations. The compromised host anatomy group included strains from patients with abnormal urinary tract anatomy, due either to a congenital defect leading to clinical problems like vesicoureteral reflux (39), or to extrinsic factors such as the presence of an indwelling Foley catheter (38). This patient group is at an increased risk for UTIs from a variety of opportunistic pathogens (5). Finally, the collection also included samples taken from patients with ≥105 CFU/ml of urine, but without symptoms of cystitis (6), a condition known as asymptomatic bacteriuria. Frozen glycerol stocks were streaked onto LB agar plates and cultured aerobically at 37°C overnight for PCR testing.
Hemolysin phenotype.
Blood agar plates were prepared as described previously (40). For determination of the presence or absence of hemolytic activity, a colony of each strain was streaked onto a blood agar plate for isolation of single colonies and incubated overnight at 37°C. Plates were examined the next day for a visible clearing surrounding isolated single colonies. If no clearing was observed, part of a colony was pushed aside with a sterile toothpick to look for clearing underneath the colony. Any strain that showed clearing around or underneath isolated colonies was scored as positive, and all others were scored as negative. Strains were tested using the same batch of blood agar plates to minimize variation between samples.
Multiplex PCR.
PCR primers were designed based on DNA sequences for each of the 15 genes under study. Each group of sequences was aligned and conserved regions of homology for each gene were chosen to create primers that would generate PCR products of a unique length with a ≥100-bp difference in size from the other PCR products in the multiplex assay. Two sets of multiplex primers were created to maximize the amount of difference in expected product size (Fig. 1A). Multiplex primer sets were validated by amplifying purified genomic DNA from five sequenced strains of E. coli (MG1655, HS, UTI89, 536, and CFT073) using DNeasy columns (Qiagen). Multiplex PCRs used 1 µl of DNA template, 12.5 µl multiplex PCR master mix (Qiagen), 1.25 µl PCR primer mix (concentration of stock, 2 pmol/µl/primer), and 10.25 µl nuclease-free water, for a final volume of 25 µl. The thermal cycling protocol was (i) 95°C for 15 min, (ii) 94°C for 0.5 min, (iii) 62°C for 1.5 min, (iv) 72°C for 1.5 min, (v) repeat steps ii to iv 29 times, and (vi) 72°C for 10 min. PCR products were separated by electrophoresis on 1.5% UltraPure agarose (Invitrogen) Tris-actetate-EDTA (TAE) gels. Gels were stained with ethidium bromide and photographed under UV transillumination using a Gel Doc XR system (Bio-Rad).
For multiplex screening, a single colony from each strain was scraped from an LB agar plate with a sterile toothpick and resuspended in 50 µl sterile water. Cell suspensions were heated to 100°C for 10 min, cooled, and frozen until use. PCRs were carried out in 25-µl volumes as described above, substituting 1 µl of cell boil preparation for the DNA template.
Phylogenetic typing.
The major E. coli phylogenetic group (A, B1, B2, and D) was determined by an established triplex PCR method (17), using PCR reagents and cycling conditions as described above for virulence genotyping.
MLST.
Multilocus sequence typing (MLST) data were retrieved from the database maintained at the University College of Cork (UCC) website (http://mlst.ucc.ie) for the 72 strains of the ECOR collection. The remaining strains tested in the murine model of ascending UTIs were typed according to standard protocols (18). Briefly, genomic DNA was extracted from overnight cultures of each strain and used in separate PCRs with primers for each of the seven genes used in the database (adk, fumC, gyrB, icd, mdh, purA, and recA). PCR products were purified and submitted for DNA sequencing using the same primers. Data for each gene were aligned with reference sequences in the database and submitted to the online analysis program at the UCC website for determination of the sequence type for each strain.
Representative DNA sequences for the ECOR collection and the strains for which MLST data were generated in this study were aligned for each of the seven gene fragments in Lasergene 8 software (DNASTAR). Alignments were imported to MEGA4 (41), and genes were aligned in frame using in silico translation of codons. Using concatenated sequences of the seven gene fragments, a neighbor-joining dendrogram was constructed in MEGA4 by the maximum composite likelihood method. An E. coli clade I strain was used to root the dendrogram (42).
Murine model of ascending urinary tract infection.
The murine model of ascending UTI was followed as previously described (43). Briefly, female C57BL/6 mice, 4 to 6 weeks of age were transurethrally inoculated with 108 CFU of the strain of E. coli being tested. Following this, mice were allowed to recover and returned to the animal care facility. Forty-eight hours postinoculation, animals were euthanized and bladder and kidneys were aseptically removed, weighed, and placed into separate test tubes filled with 3 ml of sterile PBS. Organs were homogenized, and dilutions of the homogenate were plated onto LB agar plates with a Spiral plater (Spiral Biotech) and incubated overnight at 37°C. The following day, colony counts for each plate were enumerated, and the numbers of CFU/g of tissue for each mouse were determined. If no colonies were recovered, the sample was assigned a value of 100 CFU/g of tissue, the limit of detection for this assay.
Statistical and data analysis.
Images of agarose gels containing multiplex PCR products were scored for the presence or absence of each gene under study for each strain. One-way ANOVA and Student’s t tests were carried out using Prism software (GraphPad). Multivariate ANOVA testing using the ANOSIM protocol was conducted using PRIMER version 6 (Primer-E).
Bayesian network analysis.
The Bayesian network (BN) analysis was performed using a web-based MARIMBA system (http://marimba.hegroup.org/) (30). Investigators (Y.H. and A.P.H.) were blinded to the putative roles of the 15 genes in virulence. Binary data (0 or 1) were generated for each gene, representing the presence or absence of this gene for individual strains. A new variable called “group” was created to represent five different clinical settings for each strain: (i) fecal, (ii) compromised host, (iii) ABU, (iv) cystitis, and (v) pyelonephritis. Simulated annealing was used to search for candidate network structures where each network has a unique set of connections between variables. In total, 750 million BNs were searched. A consensus network was calculated by considering the best 11 BN models sharing the top log posterior probability.
ACKNOWLEDGMENTS
We thank Seth Walk and Vince Young for contributions to the MLST data analysis, including providing protocols and assistance with dendrogram construction.
This work was supported in part by Public Health Service grant AI043363 from the National Institutes of Health.
Footnotes
Citation Vigil PD, et al. 2011. Presence of putative repeat-in-toxin gene tosA in Escherichia coli predicts successful colonization of the urinary tract. mBio 2(3):e00066-11. doi:10.1128/mBio.00066-11.
REFERENCES
- 1. Warren J. 1996. Urinary tract infections: molecular pathogenesis and clinical management. ASM Press, Washington, DC [Google Scholar]
- 2. Foxman B, et al. 2002. Uropathogenic Escherichia coli are more likely than commensal E. coli to be shared between heterosexual sex partners. Am. J. Epidemiol. 156:1133–1140 [DOI] [PubMed] [Google Scholar]
- 3. Yamamoto S, et al. 1997. Genetic evidence supporting the fecal-perineal-urethral hypothesis in cystitis caused by Escherichia coli. J. Urol. 157:1127–1129 [PubMed] [Google Scholar]
- 4. Mobley HLT, Donnenberg MS, Hagan EC. 2009. Uropathogenic Escherichia coli. In Böck A, EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC [Google Scholar]
- 5. Foxman B. 2002. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am. J. Med. 113(Suppl. 1A):5S–13S [DOI] [PubMed] [Google Scholar]
- 6. Hooton TM, et al. 2000. A prospective study of asymptomatic bacteriuria in sexually active young women. N. Engl. J. Med. 343:992–997 [DOI] [PubMed] [Google Scholar]
- 7. Welch RA, et al. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:17020–17024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nielubowicz GR, Mobley HL. 2010. Host-pathogen interactions in urinary tract infection. Nat. Rev. Urol. 7:430–441 [DOI] [PubMed] [Google Scholar]
- 9. Marrs CF, Zhang L, Foxman B. 2005. Escherichia coli mediated urinary tract infections: are there distinct uropathogenic E. coli (UPEC) pathotypes? FEMS Microbiol. Lett. 252:183–190 [DOI] [PubMed] [Google Scholar]
- 10. Johnson JR, Clabots C, Kuskowski MA. 2008. Multiple-host sharing, long-term persistence, and virulence of Escherichia coli clones from human and animal household members. J. Clin. Microbiol. 46:4078–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lloyd AL, Rasko DA, Mobley HL. 2007. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J. Bacteriol. 189:3532–3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lloyd AL, Henderson TA, Vigil PD, Mobley HL. 2009. Genomic islands of uropathogenic Escherichia coli contribute to virulence. J. Bacteriol. 191:3469–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mobley H, et al. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58:1281–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smith YC, Rasmussen SB, Grande KK, Conran RM, O’Brien AD. 2008. Hemolysin of uropathogenic Escherichia coli evokes extensive shedding of the uroepithelium and hemorrhage in bladder tissue within the first 24 hours after intraurethral inoculation of mice. Infect. Immun. 76:2978–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hagan EC, Mobley HL. 2007. Uropathogenic Escherichia coli outer membrane antigens expressed during urinary tract infection. Infect. Immun. 75:3941–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hagberg L, et al. 1983. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect. Immun. 40:273–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wirth T, et al. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hawn TR, et al. 2009. Genetic variation of the human urinary tract innate immune response and asymptomatic bacteriuria in women. PLoS One 4:e8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Klemm P, Roos V, Ulett GC, Svanborg C, Schembri MA. 2006. Molecular characterization of the Escherichia coli asymptomatic bacteriuria strain 83972: the taming of a pathogen. Infect. Immun. 74:781–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moreno E, et al. 2008. Relationship between Escherichia coli strains causing acute cystitis in women and the fecal E. coli population of the host. J. Clin. Microbiol. 46:2529–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bahrani-Mougeot FK, et al. 2002. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol. Microbiol. 45:1079–1093 [DOI] [PubMed] [Google Scholar]
- 23. Connell I, et al. 1996. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. U. S. A. 93:9827–9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gunther NW, et al. 2002. Assessment of virulence of uropathogenic Escherichia coli type 1 fimbrial mutants in which the invertible element is phase-locked on or off. Infect. Immun. 70:3344–3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hagan EC, Lloyd AL, Rasko DA, Faerber GJ, Mobley HL. 2010. Escherichia coli global gene expression in urine from women with urinary tract infection. PLoS Pathog. 6:e1001187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lim JK, et al. 1998. In vivo phase variation of Escherichia coli type 1 fimbrial genes in women with urinary tract infection. Infect. Immun. 66:3303–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klemm P, Hancock V, Schembri M. 2007. Mellowing out: adaptation to commensalism by Escherichia coli asymptomatic bacteriuria strain 83972. Infect. Immun. 75:3688–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zdziarski J, Svanborg C, Wullt B, Hacker J, Dobrindt U. 2008. Molecular basis of commensalism in the urinary tract: low virulence or virulence attenuation? Infect. Immun. 76:695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roos V, Schembri MA, Ulett GC, Klemm P. 2006. Asymptomatic bacteriuria Escherichia coli strain 83972 carries mutations in the foc locus and is unable to express F1C fimbriae. Microbiology 152:1799–1806 [DOI] [PubMed] [Google Scholar]
- 30. Hodges AP, et al. 2010. Bayesian network expansion identifies new ROS and biofilm regulators. PLoS One 5:e9513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hodges AP, Woolf P, He Y. 2010. BN+1 Bayesian network expansion for identifying molecular pathway elements. Commun. Integr. Biol. 3:549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heimer SR, Rasko DA, Lockatell CV, Johnson DE, Mobley HL. 2004. Autotransporter genes pic and tsh are associated with Escherichia coli strains that cause acute pyelonephritis and are expressed during urinary tract infection. Infect. Immun. 72:593–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brzuszkiewicz E, et al. 2006. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc. Natl. Acad. Sci. U. S. A. 103:12879–12884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Friedman N, Linial M, Nachman I, Pe’er D. 2000. Using Bayesian networks to analyze expression data. J. Comput. Biol. 7:601–620 [DOI] [PubMed] [Google Scholar]
- 35. Ochman H, Selander RK. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stapleton A, Moseley S, Stamm WE. 1991. Urovirulence determinants in Escherichia coli isolates causing first-episode and recurrent cystitis in women. J. Infect. Dis. 163:773–779 [DOI] [PubMed] [Google Scholar]
- 37. Warren JW, et al. 1983. A randomized, controlled trial of cefoperazone vs. cefamandole-tobramycin in the treatment of putative, severe infections with gram-negative bacilli. Rev. Infect. Dis. 5(Suppl. 1):S173–S180 [DOI] [PubMed] [Google Scholar]
- 38. Warren JW, Tenney JH, Hoopes JM, Muncie HL, Anthony WC. 1982. A prospective microbiologic study of bacteriuria in patients with chronic indwelling urethral catheters. J. Infect. Dis. 146:719–723 [DOI] [PubMed] [Google Scholar]
- 39. Lomberg H, Hellstrom M, Jodal U, Svanborg Eden C. 1989. Secretor state and renal scarring in girls with recurrent pyelonephritis. FEMS Microbiol. Immunol. 1:371–375 [DOI] [PubMed] [Google Scholar]
- 40. Welch RA, Hull R, Falkow S. 1983. Molecular cloning and physical characterization of a chromosomal hemolysin from Escherichia coli. Infect. Immun. 42:178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 42. Walk ST, et al. 2009. Cryptic lineages of the genus Escherichia. Appl. Environ. Microbiol. 75:6534–6544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sivick KE, Schaller MA, Smith SN, Mobley HL. 2010. The innate immune response to uropathogenic Escherichia coli involves IL-17A in a murine model of urinary tract infection. J. Immunol. 184:2065–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Johnson JR, et al. 2006. Experimental mouse lethality of Escherichia coli isolates, in relation to accessory traits, phylogenetic group, and ecological source. J. Infect. Dis. 194:1141–1150 [DOI] [PubMed] [Google Scholar]