Abstract
The molecular basis underlying the pathogenic success of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) is not completely understood, but differential gene expression has been suggested to account at least in part for the high virulence of CA-MRSA strains. Here, we show that the agr gene regulatory system has a crucial role in the development of skin infections in the most prevalent CA-MRSA strain USA300. Importantly, our data indicate that this is due to discrepancies between the agr regulon of CA-MRSA and those of hospital-associated MRSA and laboratory strains. In particular, agr regulation in strain USA300 led to exceptionally strong expression of toxins and exoenzymes, upregulation of fibrinogen-binding proteins, increased capacity to bind fibrinogen, and increased expression of methicillin resistance genes. Our findings demonstrate that agr functionality is critical for CA-MRSA disease and indicate that an adaptation of the agr regulon contributed to the evolution of highly pathogenic CA-MRSA.
INTRODUCTION
Pandemic community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) strains represent a serious problem for the public health system (25). Previously, MRSA infections were reported exclusively in the hospital setting and in immune-compromised or otherwise predisposed patients. In contrast, the recently emerging CA-MRSA strains combine antibiotic resistance with high virulence and transmissibility, enabling them to spread and cause severe infections in otherwise healthy people (13, 33). The most serious CA-MRSA pandemic is seen in the United States, where almost all cases are caused by pulsed-field type USA300 (30), a group of closely related CA-MRSA strains (24) that are now also spreading in other parts of the world (37, 39, 48).
The virulence potential of S. aureus is determined mainly by its ability to evade human innate host defenses, among which the interaction with phagocytes, such as neutrophils, plays a preeminent role (12). S. aureus has developed many mechanisms to subvert destruction by human neutrophils, such as a protective capsule or enzymes that eliminate substances produced by neutrophils to kill the bacteria (18). In addition, S. aureus may secrete toxins that lyse human neutrophils and other immune cells, such as alpha-toxin, phenol-soluble modulins (PSMs), and members of the leukocidin family, which include Panton-Valentine leukocidin (PVL) (12, 18). Furthermore, surface proteins that, among other tasks, facilitate adhesion to host tissue are believed to contribute significantly to the establishment of S. aureus infections (11).
Virtually all S. aureus toxins are under the control of the pivotal virulence regulator agr (31). This system triggers pronounced changes in gene expression at a certain level of cell density by a process called quorum sensing. In addition to toxins, agr is known to upregulate a wide variety of virulence determinants, such as exoenzymes (proteases, lipases, nucleases), and downregulate expression of surface binding proteins. This adaptation is believed to ascertain production of specific sets of virulence determinants of an infection, when they are most needed: binding proteins at the beginning, when cell density is low and adhesion to host tissue is crucial, and toxins and degradative exoenzymes when the infection is established, nutrients need to be acquired from host tissues, and the concomitant activation of the host's immune system requires production of immune evasion factors. The timely activation of agr in vivo and its importance for virulence of S. aureus have been demonstrated (8, 22, 49), even though these studies often used only laboratory strains. The role of agr in clinical strains of S. aureus is less well understood. Particularly, no reports about the impact of agr on the virulence of CA-MRSA have been published.
Previous work indicated a crucial role of differential gene expression in the evolution of CA-MRSA virulence and suggested that agr control has a major function in establishing the exceptional virulence potential of the most predominant CA-MRSA strain USA300 (27, 29, 33, 45). Thus, we investigated the role of agr in the CA-MRSA strain USA300 in detail, using an animal skin infection model and genome-wide analysis of agr-dependent gene expression in comparison with hospital-associated MRSA (HA-MRSA) and laboratory strains. We demonstrate a strong impact of agr on CA-MRSA (USA300) virulence in experimental skin infection and provide evidence indicating that an adaptation of agr-dependent gene regulation contributed to the evolution of virulence in the CA-MRSA strain USA300.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. aureus strains used in this study were LAC (CA-MRSA, pulsed-field type USA300) (6), MW2 (CA-MRSA, pulsed-field type USA400) (1), Sanger 252 (HA-MRSA, pulsed-field type USA200) (21), and RN6390 (laboratory strain, derived from strain 8325) (32). RN6390 and USA300 both belong to sequence type 8. Deletion mutants in the agr regulatory system were produced by transduction of the transposon insertion from strain RN6911 in this study (strain 252) or previously (4). The agr system is entirely deleted in these strains, except for a 3′ part of RNAIII, which is not transcribed owing to the absence of the corresponding promoter. All mutants were verified by analytical PCR, DNA sequencing of the flanking regions, and real-time PCR (RT-PCR) for absence of the RNAIII transcript. Main cultures for microarray analysis and quantitative RT-PCR (qRT-PCR) were inoculated from precultures grown overnight to an optical density at 600 nm (OD600) of 0.1.
RNA isolation, transcriptional profiling, and qRT-PCR.
RNA isolation from cultures grown to the indicated time points, removal of remaining DNA, cleanup, cDNA synthesis, and labeling were performed as previously described (28). Biotinylated S. aureus cDNA was hybridized to custom Affymetrix GeneChips (RMLChip 7) with 100% coverage of chromosomal genes from USA300 and scanned according to standard GeneChip protocols (Affymetrix). Each experiment was replicated three times. Affymetrix GeneChip Operating Software (GCOS; version 1.4) was used to perform the preliminary analysis of the custom GeneChips at the probe set level. Subsequent data analysis was performed as described previously (28). The complete set of microarray data was deposited in NCBI's Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and is accessible through GEO Series accession number GSE18793. Quantitative RT-PCR was performed as described previously (26), with at least three independent samples. All oligonucleotides were synthesized by Sigma (Table 1).
Table 1.
Primers and probes used for RT-PCR
| Gene | Forward primer | Reverse primer | Probe |
|---|---|---|---|
| clfB | TTCCAATGCGCAAGGAACTAG | CAGCATTTACTACAGGTTCAGCAACT | AGACTACGTACAGCTCTCGTTCTAACACTT |
| gyrB | CAAATGATCACAGCATTTGGTACAG | CGGCATCAGTCATAATGACGAT | AATCGGTGGCGACTTTGATCTAGCGAAAG |
| hla | AAAAAACTGCTAGTTATTAGAACGAAAGG | GGCCAGGCTAAACCACTTTTG | CCTTCTTCGCTATAAACTCTATATTGACCAGCAAT |
| rnaIII | GTGATGGAAAATAGTTGATGAGTTGTTT | GAATTTGTTCACTGTGTCGATAATCC | TGCACAAGATATCATTTCAACAATCAGTGACTTAGTAAAA |
| sdrD | TCAGATGAGCAAGCTTCACCAA | TTGGTTGAGCATTTACCACTGATT | ATTCTCTTGCAAATCAGGTTGTAACGCTTCTTG |
| rot | ATTTTGCAATTAGAAACACTTTTGG | TCTTCTCTAGACATTTTGTATTCGCTTT | TGACATTAACTCAATTTTCAGCGAGATTG |
| sarH1 | CCACCATAAATACCCTCAAACTGTT | TCATCTTCAGTTGAGCGTTCTTTT | AGCTCTCAATAATTTAAAAAAGCAAGGCTA |
| sdrC | CAACTGCAGATCAGCCTAAAGTGA | TGGTGATTGCATGTTACTACTAGTTTCTT | TGAGTGATAGTGCAACAGTT |
| clfA | AGGTTCTGGTGACGGTATCGA | TCAATTTCACCAGGCTCATCAG | AAACCAGTTGTTCCTGAAC |
| efb | TTTAACGATGGTACATTCGAATATGG | ATCAGTTTTCGCTGCTGGTTTAT | CACGTCCACAATTT |
| mecA | TTCCACATTGTTTCGGTCTAAAATT | AATGCAGAAAGACCAAAGCATACA | CCACGTTCTGATTTTAAA |
| arcA1 | TGCGATCGTATGTCACCACAA | CAATGGAATGATGGCTCAAACA | CCTGGTCGAATACATAAT |
| arcA2 | TCAGCTGCTAACTTCTCAAGGTAAAG | CATTTTGCGCAGGTGCTAAG | ACTTCAACACCCTCTTC |
| spa | CAGCAAACCATGCAGATGCTA | GCTAATGATAATCCACCAAATACAGTTG | CATTACCAGAAACTGGTGAAGAAAATCCATTCATTG |
Fibrinogen adhesion assay.
To measure adhesion to fibrinogen, microtiter plates were coated with 100 μl/well of a 10-μg/ml fibrinogen suspension at 4°C overnight. Afterward, an equal volume of a 5-mg/ml bovine serum albumin (BSA) solution was added and the plates were incubated at 37°C for 2 h. Plates were washed four times with phosphate-buffered saline (PBS), after which 100 μl of bacterial solutions normalized to an OD600 of 1 were added. Plates were incubated at 37°C for 1 h. Adhered bacteria were counted using microscopy.
Mouse subcutaneous infection model.
The subcutaneous abscess model was performed as described previously (45). Briefly, Crl:SKH1-hrBR hairless mice were between 4 and 6 weeks of age at the time of use. Mice were inoculated with S. aureus from mid-exponential growth phase (3 h) with the indicated number of CFU in 50 μl as described previously (44). Animals were examined for skin lesions at 24-h intervals for a total of 14 days. Skin lesion dimensions were measured daily with a caliper. Length (L) and width (W) values were applied to calculate the area of lesions using the formula of L × W. All animals were euthanized after completion of the entire procedure. Animal studies were approved by the Animal Care and Use Committee (IUCAC number ASP LHBP 1E), National Institute of Allergy and Infectious Diseases.
RESULTS
Impact of agr on the outcome of skin infection caused by CA-MRSA.
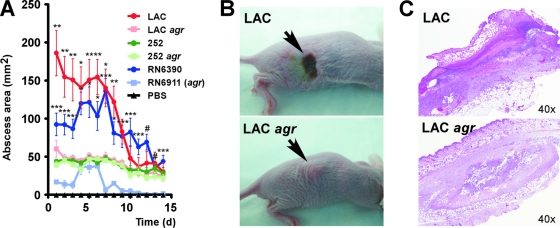
To investigate the role of agr during infection, we performed a mouse subcutaneous infection model, which mimics skin and soft tissue infections as the most common manifestation of disease caused by CA-MRSA (30) (Fig. 1). Subcutaneous injection of the same numbers of bacteria (∼3 × 107) resulted in strong abscess formation using strain LAC (USA300), while abscesses were significantly smaller with the LAC agr, 252, and 252 agr strains (Fig. 1A). Furthermore, animals infected with the LAC strain developed necrosis of the epidermis and dermis with epidermal ulceration (Fig. 1B and C). In contrast, the LAC agr, 252, and 252 agr strains caused chronic, subcutaneous abscesses lacking the dermal and epidermal necrosis noted with the LAC strain. Mice infected with the laboratory strain RN6390 (at 106 CFU) developed abscesses that were dermonecrotic and larger than those in mice infected with RN6911 (agr), which never showed necrosis. Finally, animals injected with PBS control or heat-killed cells of any of the strains used did not develop abscesses, supporting the idea that bacterial production of toxins rather than the host's response to proinflammatory components of the staphylococcal surface plays a major role in the development of CA-MRSA-induced skin disease. These results demonstrate that the agr system has a crucial impact on the development of skin infections by the CA-MRSA strain USA300.
Fig. 1.
Effect of agr on virulence in a mouse subcutaneous infection model. (A) Abscess sizes. Bacteria of the indicated strains were injected subcutaneously in hairless mice at ∼3 × 107 CFU (except RN6390 and RN6911, 1 × 106 CFU), and abscess dimensions were measured every day. Statistical significance of differences between abscess sizes on each day for each of the three wild-type/agr mutant comparisons was determined using unpaired t tests. *, P < 0.05; **, P < 0.01; ***, P < 0.001; #, all values achieved with the agr mutant strain were 0. (B) Development of dermonecrotic lesions. Mice infected with the LAC and RN6390 strains, but not the other strains, commonly developed open dermonecrotic lesions as shown in the upper panel. The lower panel shows a characteristic closed abscess formed by the LAC agr strain. (C) Histopathological evaluation of abscesses formed by the LAC and LAC agr strains. Histopathology of abscesses formed by RN6390 was similar to that of abscesses formed by strain LAC, and histopathologies of abscesses formed by the 252, 252 agr, and RN6911 strains were similar to those of abscesses formed by the LAC agr strain.
agr activity.
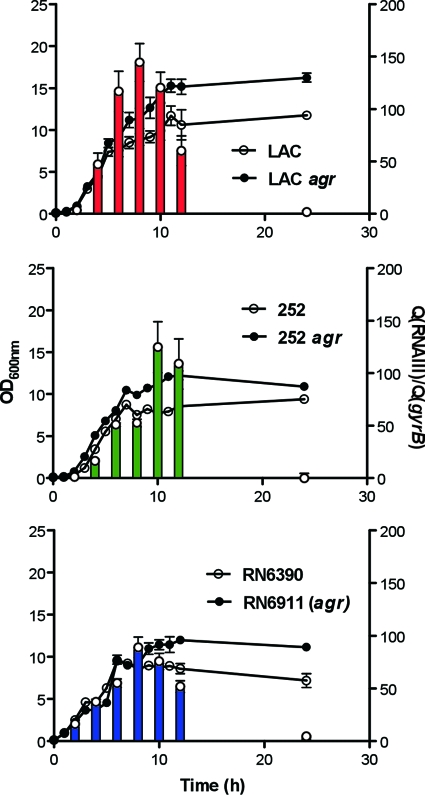
To investigate the mechanistic basis of the impact that the agr system has on the virulence of strain USA300, we first determined agr activities during in vitro growth (32, 35). While maximal agr activity in strain 252 was reached 2 h later than that in strains USA300 and RN6390, most likely owing to slower growth and delayed onset of the postexponential growth phase in that strain, we found that maximal activities of agr in the LAC and 252 strains were not significantly different (P = 0.55, Student's t test) (Fig. 2). This indicates that the significantly lower virulence potential of strain 252 and the observed differential impact of agr on virulence in these two strains are not due primarily to differences in agr activities.
Fig. 2.
Growth-dependent expression of agr. Expression of agr was determined using qRT-PCR of RNAIII during the growth of S. aureus strains and their isogenic agr mutants. Lines represent OD600, plotted on the left y axis. Bars represent the relative expression of RNAIII compared to that of the housekeeping gene gyrB (control), plotted on the right y axis. Expression of RNAIII in agr-negative strains was not detectable in any strain at any time point.
agr-dependent gene regulation in CA-MRSA: overview, toxins, and exoenzymes.
It has been reported that the agr regulon in clinical strains may differ from the scheme of agr regulation that had been established using laboratory strains (2, 35). This prompted us to evaluate the hypothesis that the agr regulons rather than agr activities may account for the differential impact agr has on the virulence potential in CA- versus HA-MRSA. To that end, we analyzed the agr regulons of the LAC, 252, and RN6390 strains by genome-wide transcriptional profiling.
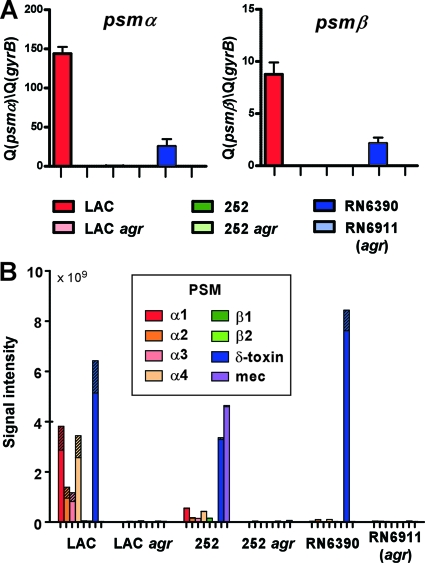
Analysis of the total up- and downregulated genes did not reveal pronounced differences: strain 252 showed 360 differentially regulated genes, strain LAC showed 262, and strain RN6390 showed 228. Forty-three genes were differentially regulated in all three strains. Genes coding for proteases, lipases, phenol-soluble modulins (PSMs), and other toxins were strongly upregulated by agr in all three strains (Table 2; see also Table S1 in the supplemental material). The most strongly regulated toxin genes in strain LAC were those coding for the Panton-Valentine leukocidin (PVL) and PSMs. While the microarray contained only samples for the psmβ genes, owing to the small size of the other psm genes, qRT-PCR demonstrated exceptionally strong agr control and exceptionally high expression in strain LAC also for the psmα genes (Fig. 3 A). Furthermore, the strong impact of agr on PSM expression was verified on the protein level for all PSMs (Fig. 3B). In accordance with previous results (45), expression of PSMs was overall lower in the 252 strain compared to strain LAC. Production of the PSMα peptides, the major contributors among PSMs to CA-MRSA skin infection and bacteremia (45), was considerably higher in strain LAC than in strains 252 and RN6390, in both absolute and relative terms. Moreover, upregulation by agr of secreted degradative enzymes, such as lipases and proteases, was much stronger in strain LAC than in strain 252, while further virulence genes, such as lrgA and lrgB, involved in bacterial programmed cell death (36) and staphylokinase (sak) (3) were downregulated by agr in strain 252 but not in strain LAC. While the hla (alpha-toxin) gene is not functional in strain 252 owing to a nonsense mutation (5), it is still interesting to note that hla was upregulated by agr in strain LAC while downregulated in strain 252. Data achieved using qRT-PCR and Western blot analysis confirmed a strong impact of agr on hla expression in strain LAC (Fig. 4). Remarkably, the relative abundances of hla transcript and alpha-toxin in the six investigated strains were similar to the differences in virulence that we observed in the skin infection model, in keeping with the notion that alpha-toxin is a key virulence determinant in CA-MRSA skin infections (23) and serving as an explanation for the high virulence observed for strain RN6390.
Table 2.
Selected agr-regulated genes in strains LAC, 252, and RN6390
| Gene type and no. | Gene | Function | Fold change in gene expression (wild-type/agr strain)a |
||
|---|---|---|---|---|---|
| LAC | 252 | RN6390 | |||
| Virulence | |||||
| SAUSA300_0113 | spa | Immunoglobulin G binding protein A | 0.12 | 0.04 | 0.03 |
| SAUSA300_0256 | lrgA | Murein hydrolase exporter | NC | 0.35 | |
| SAUSA300_0257 | lrgB | Murein hydrolase export regulator | 0.09 | ||
| SAUSA300_0320 | geh | Lipase (EC 3.1.1.3) | 6.31 | 2.49 | 3.13 |
| SAUSA300_0424 | Low-affinity zinc transport protein | 7.33 | |||
| SAUSA300_0546 | sdrC | Neurexin binding protein | 0.46 | ||
| SAUSA300_0547 | sdrD | Surface binding protein | 0.17 | NP | 0.06 |
| SAUSA300_0548 | sdrE | Surface binding protein | |||
| SAUSA300_0630 | Multidrug resistance ABC transporter | 7.51 | 10.26 | 5.70 | |
| SAUSA300_0772 | clfA | Clumping factor A (fibrinogen binding protein) | 2.08 | 2.57 | |
| SAUSA300_0774 | empbp | Extracellular matrix binding protein/fibrinogen binding protein | 0.36 | ||
| SAUSA300_0776 | nuc | Thermonuclease (EC 3.1.31.1) | 0.18 | 0.27 | |
| SAUSA300_0835 | dltA | d-Alanine-activating enzyme (EC 6.3.2.-) | 0.33 | 0.46 | |
| SAUSA300_0836 | dltB | Protein DltB | 0.33 | 0.33 | |
| SAUSA300_0837 | dltC | d-Alanyl carrier protein | 0.43 | 0.41 | |
| SAUSA300_0838 | dltD | Protein DltD | 0.27 | 0.39 | 0.49 |
| SAUSA300_0949 | sspC | Hypothetical protein | 6.79 | 27.33 | |
| SAUSA300_0950 | sspB | Staphopain (EC 3.4.22.-) | 9.74 | 11.33 | |
| SAUSA300_0951 | sspA | Glutamyl endopeptidase (EC 3.4.21.19) | 11.30 | 9.59 | |
| SAUSA300_1055 | efb | Fibrinogen binding protein | 3.00 | ||
| SAUSA300_1058 | hla | Alpha-hemolysin | 2.17 | 0.25 | 23.68 |
| SAUSA300_1067 | psmβ1 | Phenol-soluble modulin beta 1 | 173.35 | 61.61 | 41.55 |
| SAUSA300_1068 | psmβ2 | Phenol-soluble modulin beta 2 | 63.24 | NP | 26.07 |
| SAUSA300_1381 | lukF-PV | Leukocidin F subunit | 7.14 | NP | NP |
| SAUSA300_1382 | lukS-PV | Leukocidin S subunit | 6.52 | NP | NP |
| SAUSA300_1753 | splF | Serine protease (EC 3.4.21.-) | 14.96 | 3.32 | 89.05 |
| SAUSA300_1754 | splE | Serine protease (EC 3.4.21.-) | 11.11 | 33.61 | |
| SAUSA300_1755 | splD | Serine protease (EC 3.4.21.-) | 10.88 | 44.83 | |
| SAUSA300_1756 | splC | Serine protease (EC 3.4.21.-) | 9.91 | 46.21 | |
| SAUSA300_1757 | splB | Serine protease (EC 3.4.21.-) | 10.71 | 64.80 | |
| SAUSA300_1758 | splA | Serine protease (EC 3.4.21.-) | 7.99 | 27.45 | |
| SAUSA300_1759 | Hypothetical protein | 69.53 | NC | 4.69 | |
| SAUSA300_1918 | hlb | Sphingomyelin phosphodiesterase (EC 3.1.4.12), truncated beta-toxin | 6.39 | 8.85 | |
| SAUSA300_1922 | sak | Staphylokinase | 0.49 | ||
| SAUSA300_2440 | fnbB | Fibronectin binding protein | 0.24 | NP | |
| SAUSA300_2441 | fnbA | Fibronectin binding protein | 2.87 | ||
| SAUSA300_2603 | lip | Lipase (EC 3.1.1.3) | 12.54 | 7.53 | 13.29 |
| Resistance | |||||
| SAUSA300_0032 | mecA | MecA protein | 4.70 | NC | |
| SAUSA300_0033 | mecR1 | Methicillin resistance protein | 3.02 | NC | |
| SAUSA300_0928 | comK | Competence transcription factor | 10.50 | 2.44 | 7.25 |
| Metabolism | |||||
| SAUSA300_0061 | arcC1 | Carbamate kinase (EC 2.7.2.2) (ACME) | NP | NC | |
| SAUSA300_0062 | arcB1 | Ornithine carbamoyltransferase (EC 2.1.3.3) (ACME) | 0.46 | NP | NC |
| SAUSA300_0063 | Transcription regulator, crp family (ACME) | 0.45 | NP | NC | |
| SAUSA300_0064 | arcD1 | Arginine/ornithine antiporter (ACME) | 0.40 | NP | NC |
| SAUSA300_0065 | arcA1 | Arginine deiminase (EC 3.5.3.6) (ACME) | 0.41 | NP | NC |
| SAUSA300_0220 | pflB | Formate acetyltransferase (EC 2.3.1.54) | 0.10 | ||
| SAUSA300_0221 | pflA | Pyruvate formate-lyase-activating enzyme (EC 1.97.1.4) | 0.19 | ||
| SAUSA300_0311 | Ribokinase (EC 2.7.1.15) | 0.05 | 0.06 | 0.16 | |
| SAUSA300_0312 | Sugar kinase | 0.05 | 0.04 | 0.26 | |
| SAUSA300_0313 | Nucleoside permease nupC | 0.05 | 0.05 | 0.19 | |
| SAUSA300_0863 | argH | Argininosuccinate lyase (EC 4.3.2.1) | 58.19 | ||
| SAUSA300_0864 | argG | Argininosuccinate synthase (EC 6.3.4.5) | 54.46 | ||
| SAUSA300_1062 | argF | Ornithine carbamoyltransferase (EC 2.1.3.3) | 0.07 | 0.12 | 0.31 |
| SAUSA300_1063 | arcC3 | Carbamate kinase (EC 2.7.2.2) | 0.08 | 0.19 | 0.30 |
| SAUSA300_1712 | ribH | 6,7-Dimethyl-8-ribityllumazine synthase (EC 2.5.1.9) | 31.50 | ||
| SAUSA300_1713 | ribBA | GTP cyclohydrolase II (EC 3.5.4.25)/3,4-dihydroxy-2-butanone-4-phosphate synthase (EC 4.1.2.-) | 15.58 | ||
| SAUSA300_1714 | ribE | Riboflavin synthase alpha chain (EC 2.5.1.9) | 42.18 | ||
| SAUSA300_1715 | ribD | Diaminohydroxyphosphoribosylaminopyrimidine deaminase (EC 3.5.4.26)/5-amino-6-(5-phosphoribosylamino)uracil reductase (EC 1.1.1.193) | 24.48 | ||
| SAUSA300_2567 | arcC2 | Carbamate kinase (EC 2.7.2.2) | 0.15 | 0.08 | |
| SAUSA300_2568 | arcD2 | Arginine/ornithine antiporter | 0.15 | 0.21 | |
| SAUSA300_2569 | arcB2 | Ornithine carbamoyltransferase (EC 2.1.3.3) | 0.18 | 0.11 | |
| SAUSA300_2570 | arcA2 | Arginine deiminase (EC 3.5.3.6) | 0.22 | 0.05 | |
| Regulation | |||||
| SAUSA300_0605 | sarA | Staphylococcal accessory regulator | 2.77 | ||
| SAUSA300_0690 | saeS | Sensory transduction protein kinase (EC 2.7.3.-) | 2.61 | ||
| SAUSA300_0691 | saeR | Two-component response regulator | 2.17 | ||
| SAUSA300_1708 | rot | Staphylococcal accessory regulator | 0.64 | 3.18 | |
| Transport | |||||
| SAUSA300_2453 | ABC transporter ATP binding protein | 0.05 | 0.09 | 0.20 | |
| SAUSA300_2454 | ABC transporter ATP binding protein | 0.06 | 0.06 | 0.15 | |
NC, empty fields, no significant change; NP, gene not present in that strain.
Fig. 3.
Effect of agr on PSM expression in various strain backgrounds. (A) Expression of the psmα and psmβ operons was determined using qRT-PCR as previously described (35). (B) Concentrations of all S. aureus PSMs (see legend) in culture filtrates were measured as described previously (35). Samples for measurements were taken from 8-h (strains LAC and RN6390) or 10-h (strain 252) cultures. Striped parts of bars represent the N-deformylated fraction of a particular PSM. Note that strain 252 contains the psm-mec gene in contrast to strains LAC and RN6390.
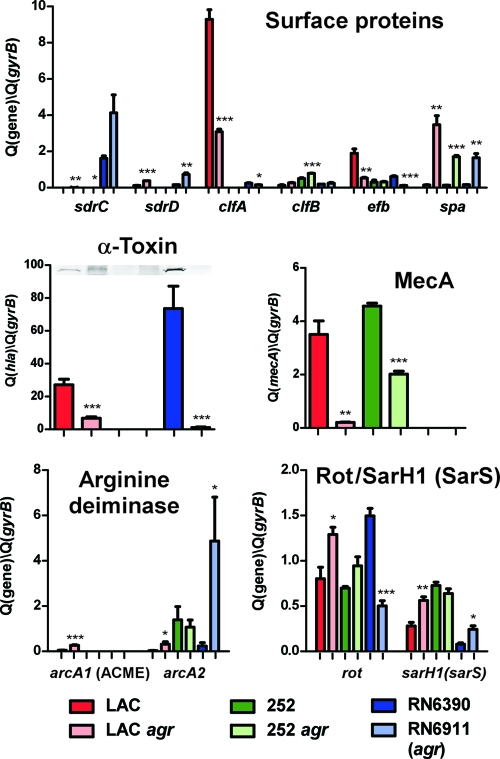
Fig. 4.
Effect of agr on the expression of selected surface protein and regulatory genes in various strain backgrounds. The graphs show expression of selected genes as determined by qRT-PCR at maximal expression of agr in the LAC, LAC agr, RN6390, and RN6911 strains for 8 h and the 252 and 252 agr strains for 10 h. Data shown on the y axis represent expression relative to that of the housekeeping gene gyrB (control). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (t tests comparing agr-negative to wild-type strain samples). Expression levels of alpha-toxin were confirmed on the protein level by Western blots with specific alpha-toxin antiserum and are shown above the qRT-PCR data.
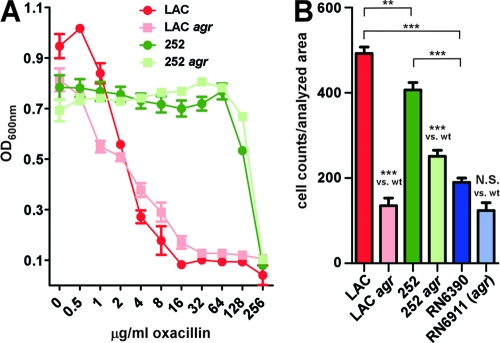
Methicillin resistance.
So far, no report describing an impact of agr on methicillin resistance exists, except for our previous observation of agr-dependent expression of mecA in the CA-MRSA strain MW2 (USA400) (35). Here, we found that genes involved in the resistance to methicillin (mecA, mecR1) were strongly upregulated by agr in the CA-MRSA strain but much less in the HA-MRSA strain, which was confirmed by qRT-PCR of the mecA gene (Fig. 4). In accordance with the idea that agr impacts methicillin resistance in strain LAC, oxacillin had a stronger growth impact on the LAC agr than the LAC wild-type strain at low concentrations, while no impact of agr on methicillin resistance was detectable for strain 252 (Fig. 5 A). These results show that agr has a gene regulatory effect on the expression of methicillin resistance genes and the methicillin resistance phenotype in CA-MRSA.
Fig. 5.
Effect of agr on methicillin resistance and fibrinogen binding phenotypes. (A) MIC assays with oxacillin. Strains were inoculated from precultures and grown with the addition of oxacillin at different concentrations in microtiter plates for 24 h. (B) Fibrinogen binding capacity. Bacteria were grown for 16 h and assayed for fibrinogen binding capacity on fibrinogen-coated microtiter plates after an incubation period of 2 h. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Surface binding proteins and fibrinogen binding capacity.
According to microarray and qRT-PCR analyses, expression of surface binding protein genes in strain LAC differed significantly from that of the other strains and only partially followed the general dogma of agr regulation, which predicts a negative effect of agr on the expression of those genes. First, transcription of sdrC, sdrE, fnbA, fnbB, and empbp was either only moderately downregulated by agr in strain LAC or not affected (Table 2). Only with the protein A gene spa did we observe the predicted strong negative agr effect (Fig. 4). Notably, strong expression and positive rather than negative regulation by agr of the fibrinogen binding protein genes clfA and efb in strain LAC were in clear contrast to the low expression of surface binding protein genes in the other wild-type strains and the notion about commonly negative regulation of this class of genes by agr. Fibrinogen binding assays confirmed that the observed differences in clfA and efb expression translated to a high capacity of strain LAC to bind fibrinogen and a strong effect of agr on that capacity, which was much lower or absent in the other strains (Fig. 5B). Thus, expression and agr-dependent regulation of surface protein genes in strain LAC are profoundly different from those in HA-MRSA and laboratory strains, allowing for a simultaneous expression of surface binding proteins, in particular those involved in binding of fibrinogen (ClfA, Efb), and universally agr-upregulated virulence determinants, such as toxins and exoenzymes.
Metabolism and arginine deiminase.
The arc operon responsible for arginine deiminase activity has been discussed as a potential contributor to pH homeostasis and immune evasion, impacting colonization and virulence properties (17, 41). In addition to the core genome-encoded arc operon (arc2), the arc operon present on the arginine catabolic mobile element (ACME), arc1, which among CA-MRSA is uniquely found in USA300 clones, has drawn particular interest, as it may contribute to the exceptional virulence and transmissibility features of these predominant CA-MRSA strains (14, 15, 17). The core genome and ACME-encoded arc operons were downregulated by agr in USA300, similar to a single, third arc gene (arcC3), but expression of the arc operons was low overall (Table 2, Fig. 4). Interestingly, the sum of arcA1 and arcA2 transcript levels in strain LAC was not as high as the level of arcA2 transcript in strain 252, which is inconsistent with the idea that arginine deiminase activity is responsible for the higher pathogenic potential of LAC than 252. Nevertheless, ACME-encoded arc may be important in overcoming to some extent the very limited core genome-encoded arc activity in strain LAC.
Additionally, we observed several pronounced differences between the strains regarding agr-dependent expression of further genes involved in metabolism (Table 2). While the present evaluation is not meant to focus on metabolism, we noted, for example, a strong upregulation of argininosuccinate lyase and synthase only in strain RN6390 but not in the other strains, while enzymes involved in riboflavin synthesis were upregulated only in strain 252. Formate acetyltransferase pflB and pyruvate formate-lyase-activating pflA genes were agr regulated only in strain 252.
Virulence regulators.
Expression of several regulatory systems that are reportedly involved in virulence gene regulation was different among the tested strains (Table 2). The sarA regulator (9) was affected in the laboratory but not the clinical strains, while upregulation of the saeRS regulator (20) occurred only in strain LAC. Interestingly, expression of the sarA homolog rot, an important mediator of the agr regulatory effect (19, 38), differed substantially among the strains. It was not significantly affected in strain LAC, moderately downregulated in strain 252, and upregulated in strain RN6390. While agr-dependent control of Rot function occurs predominantly on the posttranscriptional level (19), upregulation of rot transcript levels in an agr-dependent fashion has been observed previously (19), in accordance with our results. Opposite effects of agr on rot transcript levels between laboratory and clinical strains were confirmed by qRT-PCR (Fig. 4), an observation with potentially great importance for the difference in virulence gene regulation between those strains.
DISCUSSION
One of the most challenging tasks in current research on staphylococcal diseases is to elucidate the basis of the exceptional virulence potential of CA-MRSA strains, particularly the most epidemiologically successful clone USA300 (15). Acquisition of mobile genetic elements (MGEs), such as ΦSLT, which carries the genes encoding PVL, and ACME, which comprises two operons involved in arginine metabolism and oligopeptide transport, has been deemed a crucial step during the evolution of the virulence of USA300 (15, 16, 42). While ACME is found only in USA300, the lukSF genes encoding PVL are also found in many other CA-MRSA strains, and a correlation between CA-MRSA infection and the presence of the lukSF genes has been noted (42). However, more recent research has questioned the importance of PVL as a major contributor to CA-MRSA virulence (43, 44, 46, 47). Furthermore, while ACME may have other roles that promote the epidemiological success of USA300 (15), such as in facilitating colonization, the contribution of ACME to virulence is minor (17). Alternatively, or additionally, genetic rearrangements leading to increased expression of core genome-encoded gene products may have contributed to the evolution of CA-MRSA virulence (15). Indeed, recent results from our laboratory have emphasized the importance of differential gene expression for the evolution of virulence within the clonal complex 8 subtree that includes USA300 (27). In the present study, we observed a dramatic impact of agr on the development of experimental skin disease caused by strain USA300, confirming our hypothesis on the presumed key role of agr in CA-MRSA skin infections (27). Of note, these results underline the potential of drugs interfering with the function of agr (22, 34). Furthermore, we demonstrated that the agr regulon of strain USA300 shows important discrepancies compared to those of HA-MRSA and laboratory strains, indicating a possible role of these adaptations in the contribution of agr to CA-MRSA pathogenesis.
Transcriptional profiling analysis and qRT-PCR indicated strong agr regulation of important toxins in the CA-MRSA USA300 strain, including the most frequently proposed candidates for proteins with a major contribution to CA-MRSA virulence: alpha-toxin, PVL, and PSMs. Furthermore, we found differences in the agr regulons that may explain strain differences in virulence even at the observed comparable overall activity of agr between standard CA- and HA-MRSA strains. These included much more pronounced upregulation of toxins and lytic enzymes in USA300 compared to that of the HA-MRSA 252 strain (Table 2 and Fig. 3 and 4). Moreover, in contrast to the established scheme of agr-dependent gene regulation, high activity of agr did not cause, or only resulted in moderate, downregulation of surface protein gene expression, with the exception of the protein A gene spa. Of particular interest, the clfA and efb fibrinogen binding protein genes were highly expressed and upregulated by agr in strain USA300, as was fibrinogen binding capacity in that strain. Positive regulation by agr is in accordance with the described roles of fibrinogen-binding proteins, such as ClfA, in the early dissemination phase of S. aureus infection (7) and high activity of agr in the early infection phase (49). Comparatively high expression of those proteins in CA-MRSA may thus contribute to the exceptional capacity to infect and spread systemically, a hypothesis that needs to be further evaluated.
We previously noted that the mecA gene responsible for methicillin resistance in MRSA is under the control of agr in the CA-MRSA strain MW2 (35). In the present study, we observed similar control in the CA-MRSA LAC strain, with the effect of agr on mecA expression and methicillin resistance being more pronounced than in the HA-MRSA 252 strain. The regulatory interdependence of the mec and agr systems may contribute to balancing these two important energy-consuming pathogenesis mechanisms.
Taken together, our findings suggest that evolution of virulence of USA300 proceeded via adaptation of the agr regulatory network, resulting in the potential to maintain production of commonly agr-downregulated factors, such as surface proteins, while simultaneously ensuring secretion of aggressive toxins and promoting methicillin resistance, thereby optimizing gene expression for immune evasion, tissue adhesion, and antibiotic resistance. At present, the molecular nature of these regulatory adaptations is not clear. Comparing strains USA300 and 252, we did not detect differences that could clearly be made responsible for the agr regulon changes at the level of the regulator molecules, notably including both the RNAIII- and AgrA-regulated parts of the agr regulon (35). Namely, in strain USA300 compared to strain 252, there was no difference in the AgrA amino acid sequence: only one conservative amino acid exchange (I20M), which was not unique among S. aureus strains for either strain, in the Rot sequence, and only a 1-base-pair change within RNAIII (deletion of one T in a low-complexity 8-T sequence). We previously speculated that SarH1 (SarS) (40), the only SarA paralog (10) found to be agr regulated in strain MW2, might substitute for the pivotal role of Rot in agr-dependent target gene regulation (35). However, sarH1 (sarS) expression data did not confirm the hypothesis that agr regulon diversity is due to differential expression of SarH1/SarS (Fig. 4). Together with the microarray data, these observations suggest that adaptations may have occurred at the target gene level. In the future, a detailed investigation of regulatory networks and regulated targets in USA300 will be needed to further elucidate the interesting phenomenon of agr regulon diversity and its impact on the evolution of virulence in CA-MRSA.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), The National Institutes of Health (NIH).
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 14 March 2011.
REFERENCES
- 1. Baba T., et al. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827 [DOI] [PubMed] [Google Scholar]
- 2. Blevins J. S., Beenken K. E., Elasri M. O., Hurlburt B. K., Smeltzer M. S. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70:470–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bokarewa M. I., Jin T., Tarkowski A. 2006. Staphylococcus aureus: staphylokinase. Int. J. Biochem. Cell Biol. 38:504–509 [DOI] [PubMed] [Google Scholar]
- 4. Booth M. C., et al. 1997. Staphylococcal accessory regulator (sar) in conjunction with agr contributes to Staphylococcus aureus virulence in endophthalmitis. Infect. Immun. 65:1550–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cassat J. E., et al. 2005. Comparative genomics of Staphylococcus aureus musculoskeletal isolates. J. Bacteriol. 187:576–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. CDC 2003. Outbreaks of community-associated methicillin-resistant Staphylococcus aureus skin infections—Los Angeles County, California, 2002-2003. MMWR Morb. Mortal. Wkly. Rep. 52:88. [PubMed] [Google Scholar]
- 7. Cheng A. G., et al. 2009. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 23:3393–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheung A. L., et al. 1994. Diminished virulence of a sar−/agr− mutant of Staphylococcus aureus in the rabbit model of endocarditis. J. Clin. Invest. 94:1815–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheung A. L., Projan S. J. 1994. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J. Bacteriol. 176:4168–4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheung A. L., Zhang G. 2002. Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family. Front. Biosci. 7:d1825–d1842 [DOI] [PubMed] [Google Scholar]
- 11. Clarke S. R., Foster S. J. 2006. Surface adhesins of Staphylococcus aureus. Adv. Microb. Physiol. 51:187–224 [DOI] [PubMed] [Google Scholar]
- 12. DeLeo F. R., Diep B. A., Otto M. 2009. Host defense and pathogenesis in Staphylococcus aureus infections. Infect. Dis. Clin. North Am. 23:17–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. DeLeo F. R., Otto M., Kreiswirth B. N., Chambers H. F. 2010. Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375:1557–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Diep B. A., et al. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739 [DOI] [PubMed] [Google Scholar]
- 15. Diep B. A., Otto M. 2008. The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol. 16:361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Diep B. A., et al. 2008. Contribution of Panton-Valentine leukocidin in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. PLoS One 3:e3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Diep B. A., et al. 2008. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 197:1523–1530 [DOI] [PubMed] [Google Scholar]
- 18. Foster T. J. 2005. Immune evasion by staphylococci. Nat. Rev. Microbiol. 3:948–958 [DOI] [PubMed] [Google Scholar]
- 19. Geisinger E., Adhikari R. P., Jin R., Ross H. F., Novick R. P. 2006. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol. Microbiol. 61:1038–1048 [DOI] [PubMed] [Google Scholar]
- 20. Giraudo A. T., Cheung A. L., Nagel R. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 168:53–58 [DOI] [PubMed] [Google Scholar]
- 21. Holden M. T., et al. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. U. S. A. 101:9786–9791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ji G., Beavis R., Novick R. P. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027–2030 [DOI] [PubMed] [Google Scholar]
- 23. Kennedy A. D., et al. 2010. Targeting of alpha-hemolysin by active or passive immunization decreases severity of USA300 skin infection in a mouse model. J. Infect. Dis. 202:1050–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kennedy A. D., et al. 2008. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc. Natl. Acad. Sci. U. S. A. 105:1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Klevens R. M., et al. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771 [DOI] [PubMed] [Google Scholar]
- 26. Li M., et al. 2007. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol. Microbiol. 66:1136–1147 [DOI] [PubMed] [Google Scholar]
- 27. Li M., et al. 2009. Evolution of virulence in epidemic community-associated MRSA. Proc. Natl. Acad. Sci. U. S. A. 106:5883–5888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li M., et al. 2007. Gram-positive three-component antimicrobial peptide-sensing system. Proc. Natl. Acad. Sci. U. S. A. 104:9469–9474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Montgomery C. P., et al. 2008. Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia. J. Infect. Dis. 198:561–570 [DOI] [PubMed] [Google Scholar]
- 30. Moran G. J., et al. 2006. Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 355:666–674 [DOI] [PubMed] [Google Scholar]
- 31. Novick R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429–1449 [DOI] [PubMed] [Google Scholar]
- 32. Novick R. P., et al. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967–3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Otto M. 2010. Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu. Rev. Microbiol. 64:143–162 [DOI] [PubMed] [Google Scholar]
- 34. Park J., et al. 2007. Infection control by antibody disruption of bacterial quorum sensing signaling. Chem. Biol. 14:1119–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Queck S. Y., et al. 2008. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32:150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rice K. C., Bayles K. W. 2003. Death's toolbox: examining the molecular components of bacterial programmed cell death. Mol. Microbiol. 50:729–738 [DOI] [PubMed] [Google Scholar]
- 37. Ruppitsch W., et al. 2007. Occurrence of the USA300 community-acquired Staphylococcus aureus clone in Austria. Euro Surveill. 12:E071025.1. [DOI] [PubMed] [Google Scholar]
- 38. Said-Salim B., et al. 2003. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185:610–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shibuya Y., et al. 2008. Emergence of the community-acquired methicillin-resistant Staphylococcus aureus USA300 clone in Japan. J. Infect. Chemother. 14:439–441 [DOI] [PubMed] [Google Scholar]
- 40. Tegmark K., Karlsson A., Arvidson S. 2000. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 37:398–409 [DOI] [PubMed] [Google Scholar]
- 41. Thomas J. B., Holtsberg F. W., Ensor C. M., Bomalaski J. S., Clark M. A. 2002. Enzymic degradation of plasma arginine using arginine deiminase inhibits nitric oxide production and protects mice from the lethal effects of tumour necrosis factor alpha and endotoxin. Biochem. J. 363:581–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vandenesch F., et al. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Villaruz A., et al. 2009. A point mutation in the agr locus rather than expression of the Panton-Valentine leukocidin caused previously reported phenotypes in Staphylococcus aureus pneumonia and gene regulation. J. Infect. Dis. 200:724–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Voyich J. M., et al. 2006. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J. Infect. Dis. 194:1761–1770 [DOI] [PubMed] [Google Scholar]
- 45. Wang R., et al. 2007. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13:1510–1514 [DOI] [PubMed] [Google Scholar]
- 46. Wardenburg J. B., Bae T., Otto M., DeLeo F. R., Schneewind O. 2007. Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat. Med. 13:1405–1406 [DOI] [PubMed] [Google Scholar]
- 47. Wardenburg J. B., Palazzolo-Ballance A. M., Otto M., Schneewind O., Deleo F. R. 2008. Panton-Valentine leukocidin is not a virulence determinant in murine models of community-associated methicillin-resistant Staphylococcus aureus disease.. J. Infect. Dis. 198:1166–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Witte W., Strommenger B., Cuny C., Heuck D., Nuebel U. 2007. Methicillin-resistant Staphylococcus aureus containing the Panton-Valentine leucocidin gene in Germany in 2005 and 2006. J. Antimicrob. Chemother. 60:1258–1263 [DOI] [PubMed] [Google Scholar]
- 49. Wright J. S., III, Jin R., Novick R. P. 2005. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc. Natl. Acad. Sci. U. S. A. 102:1691–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.