Abstract
Although parasite strain-restricted CD8 T cell responses have been described for several protozoa, the precise role of antigenic variability in immunity is poorly understood. The tick-borne protozoan parasite Theileria annulata infects leukocytes and causes an acute, often fatal lymphoproliferative disease in cattle. Building on previous evidence of strain-restricted CD8 T cell responses to T. annulata, this study set out to identify and characterize the variability of the target antigens. Three antigens were identified by screening expressed parasite cDNAs with specific CD8 T cell lines. In cattle expressing the A10 class I major histocompatibility complex haplotype, A10-restricted CD8 T cell responses were shown to be focused entirely on a single dominant epitope in one of these antigens (Ta9). Sequencing of the Ta9 gene from field isolates of T. annulata demonstrated extensive sequence divergence, resulting in amino acid polymorphism within the A10-restricted epitope and a second A14-restricted epitope. Statistical analysis of the allelic sequences revealed evidence of positive selection for amino acid substitutions within the region encoding the CD8 T cell epitopes. Sequence differences in the A10-restricted epitope were shown to result in differential recognition by individual CD8 T cell clones, while clones also differed in their ability to recognize different alleles. Moreover, the representation of these clonal specificities within the responding CD8 T cell populations differed between animals. As well as providing an explanation for incomplete protection observed after heterologous parasite challenge of vaccinated cattle, these results have important implications for the choice of antigens for the development of novel subunit vaccines.
INTRODUCTION
CD8 T cells have been shown to play a key role in immunity to a variety of intracellular pathogens (65), including a number of protozoan parasites (33, 36, 38, 55). A characteristic feature of CD8 T cell responses is that, in individual hosts, they are often directed against a few dominant epitopes (67). Consequently, mutations in sites encoding these epitopes can result in escape from immune recognition. This is well established as an important phenomenon in infections with some RNA viruses that exhibit a high rate of mutation, most notably, HIV-1 (18, 26, 39). Parasite strain-restricted CD8 T cell responses have also been reported for several protozoan infections, including human malaria and theileriosis in cattle (15, 17, 35). In the case of Theileria parva, variation between animals in the strain specificity of CD8 T cell responses has been shown to correlate with incomplete cross-protection between parasite strains (54). These observations suggest that polymorphism of the target parasite antigens may have arisen as a result of CD8 T cell imposed immune selection.
The bovine tick-borne parasites Theileria parva and T. annulata infect and transform leukocytes, causing acute lymphoproliferative diseases that result in high levels of mortality and heavy production losses (25). Although the two parasites infect different subsets of leukocytes (52) and are transmitted by different tick species, the biology of the host-parasite relationship and the resultant disease processes are essentially similar. After invasion of leukocytes by sporozoites, which involves entry into the cytosol, development to the schizont stage results in activation of a number of host cell signaling pathways (10, 21, 51), leading to activation and proliferation of the infected cells. Synchronous division of the parasite and host cell ensures that infection is retained in both daughter cells following cell division (23, 61), enabling parasite multiplication to occur by clonal expansion of the cells that are initially infected. These biological features enable parasitized cells to be cultured in vitro as continuously growing cell lines (22). Despite the intimate relationship of the parasite with the host cell, inoculation of animals with a few thousand allogeneic T. annulata-infected cells results in transfer of infection into cells of recipient animals (24), by a mechanism that is poorly understood. This property has allowed parasitized cell lines to be used for vaccination, but such cell lines need to be subjected to prolonged passage in vitro followed by testing in vivo to ensure that they are fully attenuated for virulence (40, 41). Although T. annulata-infected cells have been used with success to vaccinate cattle in the field, experimental vaccine trials have indicated that the stimulated immunity is less effective against challenge with heterologous parasite strains compared to the homologous strain (9, 41).
Cattle experimentally immunized with T. annulata-infected cell lines, generate strong parasite-specific CD8 T cell responses specific for the recipient animals' parasitized cells, coinciding with clearance of the immunizing infection (1, 8, 24, 27, 44). Given the evidence from studies of T. parva that such responses play a key role in immunity (33, 34), antigens recognized by T. annulata-specific CD8 T cells represent potential candidates for the development of alternative vaccines. We have previously reported that cattle carrying the A10 class I major histocompatibility complex (MHC) haplotype, immunized with the C9 clone of T. annulata, generate strong A10-restricted CD8 T cell responses that exhibit parasite strain specificity (27). These findings suggest that variability in CD8 T cell target antigens may facilitate escape from protective immune responses and hence have important implications for vaccination.
The primary aim of the present study was to identify parasite antigens recognized by CD8 T cells from T. annulata-immune cattle, focusing particularly on A10-restricted T cells, in order to investigate the nature and extent of antigenic variability. We report the identification of three CD8 T cell antigens and demonstrate that one of these antigens is a highly dominant target of the A10-restricted response. We also show that extensive polymorphism of this antigen results in differential recognition of variants by CD8 T cells and we present evidence that the region of the gene encoding the T cell epitopes has undergone positive selection for sequence diversity.
MATERIALS AND METHODS
T. annulata-immune animals.
Clinically normal castrated male Friesian/Holstein (Bos taurus) cattle were used in these studies. Animals carrying different combinations of defined MHC haplotypes, for which the expressed class I genes have been identified and cloned, were selected for the study. Animals were typed for class I MHC by using a combination of monoclonal antibodies and PCR/sequencing methods (12, 13). Selected animals were immunized with a cloned population of T. annulata (C9) derived from the Ankara isolate (37, 48) as described previously (34). Immunized animals bearing the A10, A14, A15, A18, and A31 MHC haplotypes served as donors of CD8 T cells for antigen screening.
Parasitized cells.
Cell lines infected with T. annulata were established from experimental animals by infection of peripheral blood mononuclear cells with sporozoites from cryopreserved stocks of T. annulata, as previously described (6). Cell lines were established with the Ankara and Gharb uncloned isolates and the cloned C9 derivative of Ankara. These isolates are genotypically distinct (63) and originate from geographically distant locations: T. annulata Ankara from Turkey and T. annulata Gharb from Morocco. Cloned infected cell lines were generated from the Ankara and Gharb isolates by limiting dilution cloning of cell lines infected with the respective isolates. All lines were maintained in RPMI 1640 medium containing 10 mM HEPES buffer, supplemented with 10% heat-inactivated fetal bovine serum, 5 × 10−5 M 2-mercaptoethanol, 2 mM l-glutamine, and 50 mg of penicillin-streptomycin (Pen-Strep)/ml (RPMI culture medium). Cell lines infected with the Muguga stock of T. parva were used as antigen-presenting cells in analyses of T cell specificity using synthetic peptides; these were prepared as described for T. annulata.
Parasitized cells from naturally infected cattle were also used to analyze sequence variation in the antigen encoding gene. These experiments used two T. annulata-infected cell line stocks isolated from infected cattle in Tunisia and samples of blood collected in EDTA from five infected cattle in Turkey. Previous multilocus genotyping demonstrated that the Turkish field isolates each represented multiple parasite genotypes, while the Tunisian cell lines each represented a single genotype (62, 63).
Parasite-specific CD8 T cell lines.
Methods used for generation and maintenance of parasite-specific CD8 T cell lines and clones used in the present study have been described previously (16). Cloned CD8 T cell lines obtained by limiting dilution were expanded and maintained in 48-well culture plates by stimulation at 10- to 14-day intervals with gamma-irradiated autologous parasitized cells in RPMI culture medium supplemented with 100 U of recombinant human interleukin-2 (rHuIL-2; Chiron, Ltd., United Kingdom)/ml. Their surface phenotype was determined by indirect immunofluorescence staining and analysis by flow cytometry on a FACSCalibur analyzer (Becton Dickinson). Immunofluorescence was performed with antibodies specific for CD3 (MM1A; IgG1) CD8 (IL-A51; IgG1), CD4 (IL-A12; IgG2a), and the bovine γδ T cell receptor (GB21A; IgG2b).
Assays of CD8 T cell specificity.
A 4-h indium oxine [111In] (Amersham Medical) release assay was used to assess the cytotoxicity of CD8 T cell clones as previously described (16). Briefly, for screening of clones, 100 μl of cells from expanded 48-well cultures were incubated with 5 × 103 111In-labeled T. annulata-infected target cells in V-bottom 96-well plates. In subsequent assays, defined numbers of effector cells were used at a range of effector-to-target ratios. Plates were incubated at 37°C for 4 h in a humidified atmosphere of 5% CO2 in air. Maximum 111In release was measured by incubating target cells in 0.1% Tween 20 in H2O for 4 h, and spontaneous (background) release was measured by incubating targets in RPMI growth medium without effectors. Radioactivity in supernatants was measured by using a gamma counter, and the percent cytotoxicity was calculated as 100 × (test release − spontaneous release)/(maximum release − spontaneous release).
Measurement of gamma interferon (IFN-γ) secretion by CD8 T cells following antigen recognition was used to screen for candidate antigens and in additional analyses of antigenic specificity. A biological assay, based on the detection of IFN-γ-induced upregulation of class II MHC on bovine endothelial cells, was used as described previously (3). Briefly, 5 × 103 to 10 × 103 bovine CD8 T cells were incubated with 5 × 104 antigen-presenting cells in 200 μl of RPMI growth medium containing 100 U of rHuIL-2, in 96-well round bottom plates at 37°C for 24 to 48 h. Supernatants were collected, and 100 μl of each supernatant was added to bovine endothelial cells plated at 2 × 104 cells per well in 96-well flat-bottom plates. After incubation at 37°C for 24 to 48 h, the endothelial cells were harvested by the addition of 0.25% trypsin-EDTA (Invitrogen, United Kingdom), and the levels of MHC class II expression were assessed by staining with the anti-bovine class II DR monoclonal antibody IL-A21 (14) followed by fluorescein isothiocyanate-labeled anti-mouse immunoglobulin (Sigma, United Kingdom). The results are expressed as percent MHC class II-positive bovine endothelial cells.
Selection of parasite cDNAs for antigen screening.
A total of 70 T. annulata cDNAs containing 66 unique sequences were used for antigen screening (the genes are listed in Table S1 in the supplemental material). These comprised a set of 10 cDNAs representing T. annulata orthologues of T. parva genes previously identified as encoding antigens recognized by CD8 T cells in animals immune to T. parva (19) and a further set of 60 cDNAs originally selected for their potential involvement in host cell transformation (four genes were represented in both sets). The latter set of 60 cDNAs were identified as described previously (51), by using bioinformatic analyses of available Theileria genome sequences and a set of expression sequence tags obtained from a cDNA library representing purified T. annulata macroschizonts. Since expression by macroschizonts and possession of a predicted signal peptide sequence were two of the parameters used to select these 60 genes, the encoded proteins were also considered to be candidates for recognition by CD8 T cells. Full-length cDNAs were identified from a schizont library. Orthologues of the T. parva CD8 antigen genes were identified by similarity searches of the T. annulata genome database (http://old.genedb.org/genedb/annulata/) using the NCBI BLAST program. Full-length cDNAs were amplified by PCR using primers located at the putative ATG start and termination codons and cloned into the eukaryotic expression vector pCDNA3neo.
Antigen screening assay.
Five CD8 T cell lines, restricted by-products of the A10, A14, A15, A18, and A31 class I MHC haplotypes, were used for antigen screening. Each line comprised a pool of 10 to 20 cloned T cell lines, which were shown in initial screens to be specific for T. annulata and restricted by the respective class I MHC haplotype. The antigen screening assay involved measurement of IFN-γ release after incubation of the CD8 T cell line with COS cells that had been cotransfected with each T. annulata cDNA and cDNAs encoding the class I heavy chains expressed by the respective class I MHC haplotype: N*002019 and N*01201 for A10; N*02301, N*02401, and N*02501 for A14; N*00901, N*02401, and N*02501 for A15; N*01301 for A18; and N*02101 and N*02201 for A31 (5). After identification of a positive parasite cDNA, the restricting class I allele was identified by testing COS-7 cells cotransfected with the parasite cDNA and the individual class I cDNAs (20).
The screening method was performed essentially as described by Graham et al. (19), but with the following modifications. COS cells were plated at 2 × 104 cells per well in 96-well flat-bottom plates in Dulbecco modified Eagle medium (DMEM) with 10% fetal calf serum, 2 mM l-glutamine, and 50 μg of Pen-Strep/ml. The following day the cells were transfected with 100 ng of parasite DNA, 100 ng of bovine class I DNA, and 0.9 μl of Fugene (Roche Diagnostics) in 50 μl of DMEM without additives for 4 h at 37°C. The transfection medium was then replaced with 100 μl of DMEM, and the cells were incubated for 24 h at 37°C. Between 1 × 104 and 5 × 104 CD8 T cells in 150 μl of RPMI growth medium were then added to each well, and the plates were incubated for a further 24 to 48 h prior to removal of 100 μl of medium for IFN-γ detection.
Mapping of CD8 T cell epitopes.
A combination of two methods was used to locate and identify the epitope(s) recognized by immune CD8 T cells. First, cloned fragments of the positive cDNAs obtained by PCR amplification were screened for recognition, as described for the full-length cDNAs, in order to localize the epitope(s) to one discrete region. Overlapping synthetic peptides (17-mers overlapping by 12 residues; Pepscan, Ltd., Netherlands) representing the positive region were then screened to identify the epitope. Peptides were incubated at 1 μg/ml for 60 min at 37°C, together with an appropriate MHC-matched T. parva-infected cell line, and cells pulsed with each peptide were then used as target cells in an indium release cytotoxicity assay to test for recognition by the CD8 T cell line. The minimum peptide length recognized by the T cells was then determined by screening target cells pulsed with 10-fold dilutions of 12-mer, 11-mer, 10-mer, and 9-mer peptides covering the region identified by the 17-mer peptides in the initial screen.
Cloning and sequencing of Ta9 alleles.
Parasite DNA was prepared either from 300-μl EDTA blood samples by using a Wizard Genomic DNA purification system (Promega, United Kingdom) or from 107 infected cells harvested from cell lines using a Qiagen QIAamp DNA minikit.
Utilizing the published T. annulata genome (C9) (37), forward and reverse PCR primers for the Ta9 gene were designed in the signal peptide and 3′ downstream regions, respectively: forward, CACAATGAATCTCCTAACATCTGG (nucleotide positions 4 to 20); and reverse, GCTCGTCTAATTAAACTCTTCT (3′ untranslated region). An aliquot of each DNA preparation was PCR amplified in a total reaction volume of 20 μl under conditions previously described (29), using a Techne TC-512 thermocycler with the following settings: 94°C for 2 min; followed by 30 cycles of 94°C for 50 s, 50°C for 50 s, and 65°C for 90 s; with a final extension period of 15 min at 65°C. A mixture of Taq polymerase and a proofreading polymerase (Pfu) at a ratio of 15:1 was used to improve the fidelity of the reaction, and the PCR products were subsequently cloned into the sequencing vector pCR4-TOPO (Invitrogen, United Kingdom). Up to 20 μg of plasmid DNA was isolated from each culture with a proprietary kit (Qiagen, United Kingdom) using the technique described in the QIAprep miniprep handbook. Air-dried DNA (1 to 2 μg) was prepared for each sequencing reaction, which was undertaken by a commercial sequencing service (MWG Biotech, Germany). M13 universal (forward) and M13 reverse primer sites, present in the vector flanking sequence, were used to generate sequence reads to provide at least 2× coverage of every nucleotide. Sequence data were received electronically in standard chromatogram format (SCF). Corresponding FASTA files were generated and used to assemble complete consensus sequences with the ContigExpress feature of the software package Vector NTI Advance (Invitrogen). Wherever present, ambiguous nucleotide sequence was reexamined in SCF files before the decision was made to accept or reanalyze the sequence.
Sequence analysis.
DNA sequence polymorphism was evaluated by using the computer package DnaSP (47). The software was used to estimate several measures of DNA sequence variation within and between populations, including the McDonald-Kreitman test (31). To test whether the level of synonymous or nonsynonymous polymorphisms deviated from the neutral prediction of equal numbers either within T. annulata or between Theileria species, a Fisher exact test of significance was applied to the 2×2 matrix containing the results for each domain, with a low P value reflecting a departure from neutrality. The “neutrality index” odds ratio was also calculated for each locus (46) to indicate if there was an excess (ratio > 1) or deficiency (ratio < 1) of nonsynonymous substitutions within alleles from the same species, and this was used as a qualitative and quantitative indicator of the direction and degree of selection. To detect amino acid sites under positive selection and to determine the ratio of nonsynonymous to synonymous evolutionary changes (dN/dS) values across alleles, a maximum-likelihood method, using the HyPhy platform, was used (42, 43). This permitted the dN/dS ratio to be determined for each codon and, where dN was greater or less than dS, a P value was derived from a two-tailed binomial distribution to assess the significance. Amino acid position numbers referred to in the results relate to the reference C9 genome sequence.
RESULTS
Identification of T. annulata antigens and epitopes recognized by CD8 T cells.
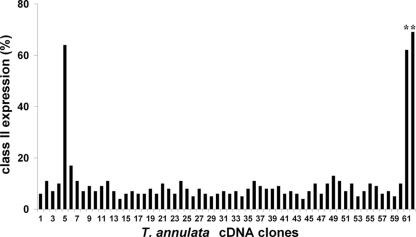
Five CD8 T cell lines restricted by different MHC haplotypes (A10, A14, A15, A18, and A31) were used to screen the expressed products of 70 cDNAs derived from the schizont stage of T. annulata, using an IFN-γ release assay to detect antigen recognition. Screens with three of the five CD8 T cell lines identified positive cDNAs; an example of the results obtained from one of these lines is shown in Fig. 1. The CD8 T cell lines restricted by A10 and A31 each recognized a single parasite cDNA (annotated in the genome as TA15705 and TA17545, respectively); the A14-restricted line also recognized TA15705 and an additional cDNA (TA14970). The characteristics of these genes and the antigens they encode are summarized in Table 1. The two antigens encoded by TA14970 and TA15705 represent the orthologues of the T. parva antigens, Tp5 and Tp9, recognized by immune CD8 T cells, and have been named Ta5 and Ta9, respectively. The third antigen, not previously identified in T. parva, has been named Ta11. Further screening of cells transfected with the positive parasite cDNAs along with cDNA encoding the individual bovine MHC class I heavy chains expressed by the respective MHC haplotypes demonstrated that in each case the antigens were presented by a single class I, with Ta9 and Ta5 both presented by the same class I product on the A14 haplotype (Table 1).
Fig. 1.
Identification of a CD8 T cell target antigen by screening with T. annulata-specific T cells. COS-7 cells cotransfected with the N*02101 and N*02201 MHC class I heavy-chain cDNAs, along with 60 individual T. annulata cDNAs, were screened with an MHC A31-restricted CD8+ T cell line. Supernatants harvested after incubation of the T cells with the transfected cells for 24 h were screened for IFN-γ release by assaying induction of class II MHC expression on a bovine endothelial cell line. Levels of class II expression, measured by flow cytometry using a DR-specific monoclonal antibody (IL-A21), are expressed as percentages of positive cells based on gates set with unstimulated endothelial cells stained with the same antibody. A positive result was obtained with cDNA number 5 (TA17545). Two IFN-γ-positive control wells were included (*). A further 10 parasite cDNAs encoding orthologues of defined T. parva CD8 T cell antigens, screened separately, did not yield any positive results.
Table 1.
Characteristics of T. annulata antigens identified by screening cells transfected with parasite cDNAs using CD8+ T cell lines
| T cell MHC restriction |
Parasite genea (chromosome no.) | Protein |
||||||
|---|---|---|---|---|---|---|---|---|
| MHC haplotype | Restricting class I | Name | Size (no. of amino acids) | Signal peptide | Epitope | Epitope positionb | Identity | |
| A10 | N*00201 | TA15705 (2) | Ta9 | 335 | Yes | QRSPMFEGTL | 40–49 | Hypothetical protein |
| A14 | N*02301 | TA15705 (2) | Ta9 | 335 | Yes | SKFPKMRMG | 64–72 | Hypothetical protein |
| N*02301 | TA14970 (2) | Ta5 | 155 | No | SKADVIAKY | 86–94 | Putative translation initiation factor eIF-1AF | |
| A31 | N*02201 | TA17545 (4) | Ta11 | 557 | Yes | KRKTEGYVF | 395–403 | Subtelomere-encoded variable secreted protein |
That is, the gene identifier used in the T. annulata genome database (http://old.genedb.org/genedb/annulata/).
The epitope positions refer to the amino acid residues in the respective proteins encoded by the reference cloned isolate of T. annulata (C9) for which the complete genome sequence is available.
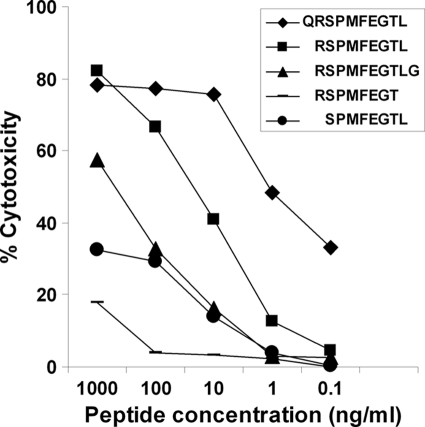
CD8 T cell epitopes within the antigens were identified by first screening cDNA subclones encoding fragments of the proteins and then localizing the epitopes by testing cells pulsed with overlapping synthetic peptides. As an example, use of an A10-restricted T cell line to screen COS-7 cells transfected with two overlapping PCR fragments covering the full length of the Ta9 cDNA, showed that the larger fragment (extending from 130 to 1,008 bp) was not recognized, demonstrating that the epitope(s) is encoded within the 5′ 130 bp (Fig. 2). Screening of a series of overlapping peptides covering this region identified two 17-mer contiguous peptides, encompassing amino acid residues 34 to 57, which gave levels of cytotoxicity of >40%. By testing overlapping peptides of different sizes (8 to 12 amino acids) covering this region, a minimum-length peptide of 10 amino acids QRSPMFEGTL (residues 40 to 49 of Ta9) that retained strong recognition by the A10-restricted, Ta9-specific T cell line was identified (Fig. 3). Using a similar approach, a second 9-mer epitope restricted by A14 (Ta964-72) was identified in Ta9, and single 9-mer epitopes were also identified in Ta5 (Ta586-94) and Ta11 (Tp11395-403). The sequences and locations of the epitopes are shown in Table 1.
Fig. 2.
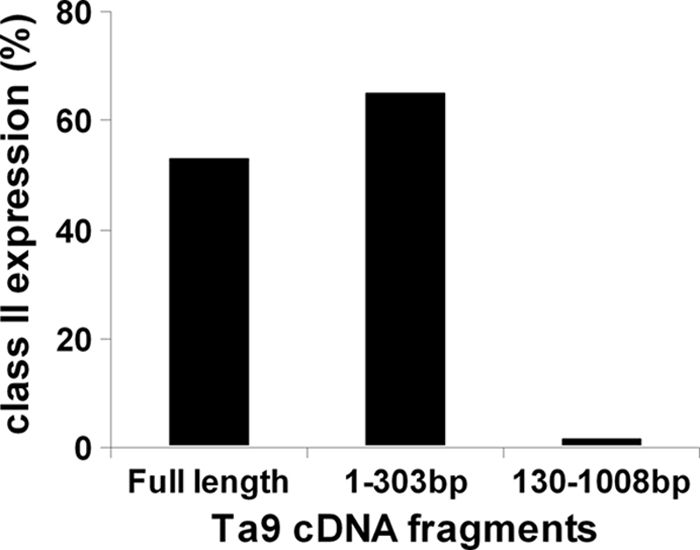
Localization of the Ta9 epitope-encoding region to a 130bp fragment. COS cells cotransfected with full-length cDNA or two overlapping cDNA fragments of TA15705 (Ta9), along with the N*01201 class I heavy chain cDNA, were screened for recognition by an A10-restricted CD8+ T cell line from animal 744, using an IFN-γ release assay. The results indicate that the epitope(s) is encoded within the 5′-terminal 130 bp.
Fig. 3.
Identification of the A10-restricted Ta9 epitope. A minimal length peptide representing the A10-restricted Ta9 epitope presented by the N*00201 class I gene product was identified by screening A10+ T. parva-infected cells pulsed with a range of concentrations of different length peptides, using an uncloned Tp9-specific CD8 T cell line in an 111In release cytotoxicity assay.
The majority of the A10-restricted CD8 T cell response is directed to Ta9.
In previous studies, we had shown that MHC class I A10-heterozygous animals immunized with T. annulata generated strong A10-restricted cytotoxic CD8 T cell responses displaying restricted recognition of different parasite strains, suggesting that A10 class I products present one or more dominant polymorphic antigens (27). Parasite-specific CD8 T cell lines derived from three of these A10+ animals were used to investigate responses to the Ta9 antigen. In order to determine the proportion of the response directed against Ta9, panels of 90 CD8+ T cell clones from each animal were tested for cytotoxicity on autologous and MHC haplotype-matched T. annulata-infected target cells and on autologous T. parva-infected target cells pulsed with peptide representing Ta940-49. A summary of the results is shown in Table 2.
Table 2.
MHC restriction and antigenic specificity of CD8 T cell clones generated from three MHC class I A10+ animals immunized with the C9 clone of T. annulata
| Animal (MHC type) | No. of T cell clones tested | No. of clones showing cytotoxic activitya on target cells (% killing [range]) |
|||
|---|---|---|---|---|---|
|
T. annulata C9 infectedb |
Ta9 peptide pulsedc |
||||
| Autologous | A10 matched | Unpulsed | Pulsed | ||
| 744 (A10/A20) | 90 | 81 | 81 (12–80) | 0 | 81 (69–100) |
| 1147 (A10/A15) | 90 | 65 | 65 (5–73) | 0 | 67 (73–93) |
| 219 (A10/A19) | 90 | 51 | 5 (58–81) | 0 | 5 (71–87) |
Clones were tested using a 4-h 111indium release cytotoxicity assay. Levels of killing of ≥5% were considered positive. Levels of killing on negative control targets were in all cases <3%.
MHC restriction specificity was determined by testing T. annulata-infected target cells derived from animals that shared either of the MHC haplotypes expressed by the respective animal.
Peptide-pulsed target cells consisted of T. parva-infected cells incubated with a 17-mer peptide containing the Ta940-49 epitope at a concentration of 1 μg/ml. In all three animals, all clones that killed the A10-matched T. annulata-infected cells also killed Ta9 peptide-pulsed target cells.
In two of the animals, all of the CD8 T cell clones that showed detectable specific killing of T. annulata-infected targets (81/90 for animal 744 and 65/90 for animal 1147) were found to be A10-restricted. In contrast, only a minority of the clones (5/51) that showed specific cytotoxicity from the third animal (219) were restricted by the A10 haplotype. All of the A10-restricted T cell clones from the three animals demonstrated specific killing of Ta940-49 peptide-pulsed targets but no killing of unpulsed targets. The levels of killing of pulsed targets were substantially higher (69 to 100% for all clones) than those obtained with T. annulata-infected target cells (5 to 80%) and two additional T cell clones that did not kill the parasitized cells demonstrated significant killing of peptide pulsed targets (Table 2).
These results provide clear evidence that Ta9 is a highly dominant antigen in A10-restricted CD8 T cell responses to T. annulata.
Characterization of the Ta9 gene family and interspecies diversity.
The Ta9 gene is predicted to encode a polypeptide of 335 amino acids, including a signal peptide at the N terminus. Analysis of available genomic sequences showed that the Ta9 gene is one member of a predicted five-gene family (cluster tribe_TA_TP:0028) located on chromosome 2 of T. annulata (37), with an orthologous family present in the T. parva genome.
To investigate the nature and extent of sequence diversity and whether the identified epitopes may be conserved between family members, Ta9 gene family sequences were characterized further and compared to those of the T. parva orthologues. All T. annulata family members have a single exon and analyses of expressed sequence tag data indicate that four of the genes, including Ta9, are relatively abundantly expressed in the macroschizont stage of the parasite (see Fig. S1 in the supplemental material). Predicted Ta9 family proteins are characterized by a conserved signal sequence, two loosely conserved motifs of 50 and 15 residues toward the C terminus, and a low-complexity central region (except the shorter TA15695) rich in proline and glutamine, part of which constitutes a predicted PEST domain. Considerable sequence variation exists among family members, with Ta9 possessing low levels of amino acid identity with paralogous proteins: 25% with TA15685, 27% with TA15695, 33% with TA15710, and 40% with TA15690. Neither of the two defined epitopes in Ta9 was found to be conserved among other family members. Only four residues in the Ta9 A10 epitope were identical to the corresponding region in one paralogue, TA15710, and just a single residue in the A14 epitope region was common to other family members. Using the amino acid sequence of Ta9 as a query, BLASTP searches of the nonredundant protein sequence database at NCBI did not identify any similar proteins of known function in other (non-Theileria) parasites or mammalian species, and no conserved domains were identified.
Comparison of the Ta9 family with its orthologous family in T. parva using dN/dS revealed a mean family interspecies dN/dS ratio of 0.2703, which is far greater than the average genome-wide figure of 0.1220 (37). This value is in line with other characterized antigen genes (64) and, together with the polymorphism identified across the paralogous genes, provides good evidence that these gene families are evolving relatively rapidly within their respective species.
Sequence analysis reveals a high level of polymorphism among alleles of the Ta9 antigen.
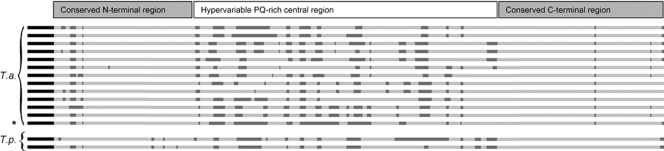
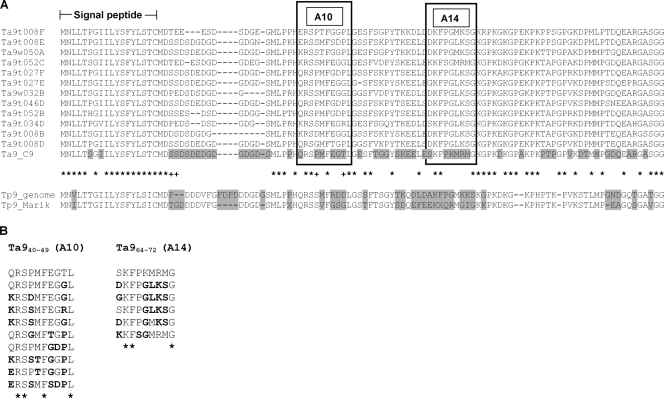
Amplicons corresponding to full-length Ta9 sequences were generated using each of five peripheral blood Turkish isolates and two cloned Tunisian cell lines as templates for PCR amplification. All full-length amplicons were cloned and sequenced. Ten distinct alleles of Ta9 were identified from 16 sequences obtained from Turkish samples examined and together with a further two distinct alleles from the Tunisian samples, a set of 12 discrete allelic sequences was obtained, all of which differed from the reference genome sequence (clone C9, Ankara isolate). As identified for the C9 sequence, all allelic sequences encoded a conserved predicted signal peptide and a central PQ-rich, low-complexity region. Encoded proteins varied widely in size from 335 to 391 amino acid residues, with length polymorphism principally due to insertions and deletions (indels) in the central PQ-rich region, as illustrated by the sequence alignment in Fig. 4. Similarly, two previously sequenced T. parva alleles of Tp9 (N. D. MacHugh and W. I. Morrison, unpublished data) exhibited several indels in this region, the remainder of the sequence aligning with T. annulata alleles at the N and C termini. An alignment of predicted allelic amino acid sequences corresponding to the N-terminal region of Ta9, along with the two T. parva orthologous sequences, is shown in Fig. 5. Considerable variation can be seen among T. annulata alleles, with only 54 of the 112 codons in the alignment conserved among all sequences. In marked contrast, the signal peptide is relatively well conserved across alleles, with 16 of the 18 residues invariant. The two CD8 T cell epitopes (positions shown in Fig. 5) display extensive sequence variation. Only 4 of the 10 residues of the Ta940-49 epitope and 3 of the 9 residues in the Ta964-72 epitope were conserved among all alleles; the 12 Ta9 sequences contained 9 variants of the Ta940-49 epitope, differing from the C9 genome sequence by 1 to 5 amino acids, and 5 variants of the Ta964-72 epitope, differing from C9 by 3 to 5 amino acids.
Fig. 4.
Alignment of nucleotide sequences of alleles of Ta9 and Tp9. Comparison of aligned sequences of the genes encoding Ta9 and Tp9 reveals that the signal peptide sequence (black) is conserved among all sequences while gaps in the alignment (dark gray) were identified throughout the hypervariable central region.
Fig. 5.
Ta9 alleles display extensive polymorphism in amino acid sequence. (A) Amino acid alignment of the conserved N-terminal region of the genes encoding Ta9 and Tp9 showing the signal peptide, polymorphic sites within each species (gray), and alignment gaps (−). Sites conserved among T. annulata alleles are marked with an asterisk (*), and those with evidence of positive selection are marked with a plus symbol (+). The A10 and A14 epitopes are indicated in boxes. (B) Allelic variants of the two CD8 T cell epitopes. Residues differing from the C9 sequence are indicated in boldface.
Evidence for positive selection of amino acid diversity in the Ta9 epitope.
To test for evidence of positive selection, dN/dS analysis was performed separately on the N and C termini of all sequences available after alignment positions with gaps were removed. The large number of alignment gaps made analysis of the central region unreliable. The results of this analysis are shown in Table 3. dN/dS ratios of 0.8661 and 0.8553 were obtained for the N and C termini, both of which far exceed the interspecies dN/dS ratio of 0.3166, as measured across the entire gene. In a codon-by-codon analysis, four positively selected sites (codons) were identified at the N terminus (Fig. 5), while in the slightly longer C terminus none were identified (P < 0.1). Two of these sites are within the A10 T-cell epitope (Ta940-49) identified for the T. annulata antigen, and statistical analysis confirmed that, compared to the rest of the gene, the epitope region was enriched for positively selected codons (P = 0.016, Fisher exact test). In addition, three of the sites have amino acid substitutions in the two T. parva alleles. To test for evidence of within-species positive selection using T. parva as an outspecies, the McDonald-Kreitman test was performed using all 13 T. annulata and both T. parva alleles at the N and C termini. Interestingly, the N terminus showed an excess of nonsynonymous (amino acid altering) substitutions when we compared within-species to between-species substitutions (43 versus 12, Table 3), while a similar number of synonymous (silent) substitutions was observed within and between the species for this region (15 versus 14). This was reflected in an elevated neutrality index value of 3.344 and a significant Fisher exact test result (P = 0.0241), indicating departure from neutrality of the N-terminal sequence. In contrast, there was no evidence for within-species positive selection at the C terminus as the number of nonsynonymous substitutions within the species was in line with the number of synonymous substitutions (neutrality index = 1.350, P = 0.5186).
Table 3.
Analysis for evidence of selection on the N- and C-terminal regions of the gene encoding Ta9
| Domain | Size C9 (no. of amino acids) | Codon-by-codon dN/dS (HyPhy) |
Calculated dN/dS |
McDonald-Kreitman testa |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Positively selected sites (P < 0.1) | Negatively selected sites (P < 0.1) | T. annulata allelic (HyPhy) | T. annulata vs T. parva (PAML) | Polymorphic changes within species |
Fixed differences between species |
Neutrality index | Pb | ||||
| Syn | Non | Syn | Non | ||||||||
| N terminus | 89 | 4 | 5 | 0.8661 | 0.3166 | 15 | 43 | 14 | 12 | 3.344 | 0.0241 |
| C terminus | 111 | 0 | 2 | 0.8553 | 0.3166 | 20 | 54 | 13 | 26 | 1.350 | 0.5186 |
Thirteen allelic sequences of T. annulata compared to two T. parva allelic sequences. Syn, synonymous; Non, nonsynonymous.
Calculated using the Fisher exact test.
Differential recognition of Ta9 alleles by distinct T cell clones.
The data presented above indicate that amino acid diversity in the Ta940-49 epitope may allow escape from an immune response. Previously, we have shown that A10-restricted CD8 T cell lines from the three A10+ animals examined in the current study exhibit variable killing when tested on infected cloned cell lines derived from the Ankara and Gharb isolates of T. annulata, and preliminary data indicated that cloned CD8 T cell lines exhibit different patterns of killing of these targets (34). Although we proposed that these T cells recognize different target antigens, the results of the present study have demonstrated that all of the A10-restricted CD8 T cell clones from these animals recognize the same epitope in Ta9 (see Table 2). In order to further investigate this finding, 20 A10-restricted CD8 T cell clones, which were successfully passaged and retained strong cytotoxic activity, were tested for their ability to recognize a panel of six cloned parasitized cell lines, three derived from the Ankara isolate and three from the Gharb isolate. As shown in Table 4, three patterns of specificity were observed: six of the clones did not recognize any of the parasitized cell lines, a further six clones recognized only one of the lines (TaG.5) and the remaining eight clones recognized two of the lines (TaG.5 and TaG.3). Sequencing of Ta9 cDNA representing four of the cloned parasitized cell lines used in these experiments revealed that they each expressed a unique allele of the Ta9 gene, all of which differed from the reference C9 sequence in the region encoding the Ta940-49 epitope. The predicted sequences of the Ta940-49 epitope in each parasite, in relation to their recognition by the CD8 T cell clones, are shown in Table 5. The sequences contained between one and four amino acid substitutions, affecting 6 of the 10 residues in the reference C9 sequence, including the two residues for which evidence of positive selection was obtained. Two variants (from cell lines TaA.1 and TaG.4), which were not recognized by any of the T cell clones, each contained four substitutions. A third variant from cell line TaG5, which was recognized by 14 of the 20 T cell clones, had a single substitution, whereas the fourth variant (TaG.3), also recognized by a subset of these T cell clones, had three substitutions. The results confirm that amino acid substitutions in the epitope are associated with differential recognition by Ta940-49-specific T cell clones. They also demonstrate that CD8 T cell clones differ in their tolerance of amino acid substitutions in the Ta940-49 epitope, allowing recognition of some alleles (with up to 3 amino acid substitutions) by a component of the CD8 T cell response.
Table 4.
Recognition of different cloned parasites by Ta940-49-specific CD8 T cell clones from cattle immunized with T. annulata C9
| Animal | T cell clone | Specific cytotoxicity (%)a |
||||||
|---|---|---|---|---|---|---|---|---|
| Ankara-derived parasites |
Gharb-derived parasites |
|||||||
| C9 | TaA.1 | TaA.3 | TaA.4 | TaG.4 | TaG.3 | TaG.5 | ||
| 744 | 1 | 65 | 0 | 0 | 0 | 1 | 1 | 4 |
| 4 | 63 | 1 | 0 | 1 | 0 | 1 | 1 | |
| 5 | 60 | 0 | 1 | 1 | 2 | 0 | 1 | |
| 6 | 59 | 0 | 0 | 1 | 1 | 1 | 1 | |
| 33 | 53 | 3 | 0 | 0 | 1 | 1 | 1 | |
| 34 | 34 | 0 | 0 | 2 | 2 | 0 | 1 | |
| 2 | 53 | 1 | 0 | 1 | 4 | 78 | 85 | |
| 1147 | 2 | 75 | 1 | 0 | 1 | 1 | 3 | 100 |
| 11 | 63 | 0 | 0 | 0 | 0 | 1 | 78 | |
| 24 | 50 | 0 | 0 | 0 | 0 | 0 | 47 | |
| 29 | 68 | 0 | 0 | 0 | 0 | 1 | 50 | |
| 40 | 76 | 1 | 0 | 0 | 0 | 0 | 75 | |
| 77 | 75 | 1 | 0 | 0 | 0 | 1 | 97 | |
| 1 | 73 | 1 | 0 | 0 | 5 | 74 | 66 | |
| 6 | 42 | 2 | 0 | 0 | 3 | 72 | 56 | |
| 14 | 80 | 1 | 1 | 0 | 3 | 81 | 88 | |
| 92 | 59 | 0 | 0 | 1 | 3 | 59 | 64 | |
| 219 | 6 | 66 | 2 | 1 | 1 | 2 | 55 | 66 |
| 11 | 80 | 1 | 1 | 1 | 5 | 67 | 78 | |
| 13 | 82 | 2 | 1 | 2 | 6 | 86 | 87 | |
CD8 T cell clones were tested in a 4-h 111indium release cytotoxicity assay using an effector/target ratio of approximately 8:1. Target cells designated TaA and TaG represent cloned parasitized cells derived from lines infected with the Ankara and Gharb isolates of T. annulata, respectively. C9 is the cloned parasite against which the T cell clones were generated and is also derived from the Ankara isolate. Positive results are indicated in boldface.
Table 5.
Sequence of Ta940-49 epitope variants from clones of T. annulata differentially recognized by CD8 T cell clones
| Cloned parasitea | Ta940-49 amino acid sequenceb | Recognition by T cell clones |
|
|---|---|---|---|
| No. of clones tested | No. positive | ||
| C9 | QRSPMFEGTL | 20 | 20 |
| TaG.5 | QRSPMFEGGL | 20 | 14 |
| TaG.3 | ERSPMFEEGL | 20 | 8 |
| TaG.4 | KRSSMFEEGL | 20 | 0 |
| TaA.1 | ERSPTFGGPL | 20 | 0 |
Origin of parasites and T cell clones described in Table 4.
Residues that differ from the C9 sequence are indicated in boldface. Residues showing evidence of positive selection are underlined.
The data shown in Table 4 suggested that the representation of CD8 T cell clones capable of recognizing the TaG.3 and TaG.5 parasites differed between the three animals, but the numbers of clones examined were not sufficient to draw clear conclusions. Therefore, to address this question, larger panels of CD8 T cell clones were derived from animals 744 and 1147, in which the response is largely focused on Ta9, and tested for their ability to recognize the TaG.3 and TaG.5 parasitized lines. The results, summarized in Table 6, confirm differences between the animals' responses in the representation of clones reactive against cell lines expressing these Ta9 variants. Thus, ca. 10% of clones from animal 1147 that killed C9-infected cells also recognized TaG.3, whereas none of the 81 C9-reactive clones from animal 744 recognized this cell line. Similarly clones that recognized TaG.5 were more abundant in animal 1147 than in animal 744 (51% versus 10%). These results indicate that heterogeneity in the composition of fine antigenic specificities among T cells responding to this dominant epitope can result in variation between animals in the overall parasite strain specificity of the CD8 T cell response.
Table 6.
Difference in frequency of CD8 T cells reactive with variants of the Ta940-49 epitope in two animals immunized with the C9 clone of T. annulata
| Animal | No. of clones tested | No. of clones reactive witha: |
||
|---|---|---|---|---|
| C9 | TaG.5 | TaG.3 | ||
| 744 | 90 | 81 | 8 | 0 |
| 1147 | 90 | 67 | 34 | 7 |
CD8 T cell clones were tested for recognition of cell lines infected with cloned parasites using a 4-h 111indium release assay. All of the clones that recognized C9 were shown in a separate assay to recognize Ta940-49.
DISCUSSION
The present study has identified three T. annulata antigens recognized by CD8 T cells and demonstrated that one of these antigens (Ta9) is a highly dominant target in A10-restricted responses of Friesian/Holstein cattle. Sequencing of the Ta9 gene from field isolates of T. annulata identified a high level of sequence divergence, resulting in coding changes that conferred amino acid substitutions within defined CD8 T cell epitopes. Comparative analysis of the allelic sequences revealed high dN/dS ratios and evidence for positive selection of amino acid changes within the region encoding the CD8 T cell epitopes. Functional studies confirmed that sequence differences resulted in differential recognition by CD8 T cell clones specific for the A10-restricted epitope, but T cell clones differed in their ability to recognize allelic variants of the epitope, indicating potential for variability between animals in cross-reactivity of the CD8 T cell response.
Three of the five CD8 T cell lines used for antigen screening identified one or, in one case, two antigens among the 70 expressed T. annulata cDNAs used in the screens. T cells from animals of different MHC genotypes tended to recognize different antigens, suggesting differences in the dominance of the antigens dependent on the presenting MHC. Because our previous studies had provided evidence of parasite strain specificity of the CD8 T cell response in several MHC class I A10+ animals (27), further analyses focused on the Ta9 antigen identified by screening with an A10-restricted CD8 T cell line. All A10-restricted CD8 T cell clones obtained from the three animals studied (151 clones in total) were found to be specific for a single epitope in Ta9, demonstrating that Ta9 is a highly dominant antigen in the context of this MHC haplotype. Moreover, in two of the animals, all of the CD8 T cells that exhibited parasite-specific cytotoxic activity were shown to be specific for Ta9, whereas in the third animal the response was dominated by T cells restricted by the other MHC haplotype. In previous studies of similar CD8 T cell lines generated from T. parva-immune cattle, we showed that their T cell receptor (TCR) Vβ gene expression profile closely resembled that of CD8 T cells harvested ex vivo from the same animal undergoing parasite challenge, indicating that the composition of such lines is representative of the in vivo CD8 T cell memory population (50). A CD8 T cell line from an A14+ animal was also found to react with Ta9 and again a single epitope was identified, but the relative dominance of this epitope on the A14 MHC background was not determined. The failure to detect recognition of Ta9 by CD8 T cell lines restricted by three other class I MHC haplotypes (A15, A18, and A31) confirms that dominance of this antigen is dependent on host MHC genotype. MHC related hierarchy in dominance of epitopes is consistent with findings for T. parva in cattle (28) and is a well-recognized feature of CD8 T cell responses to viruses in humans and mice (2, 56, 66).
The gene encoding Ta9 belongs to a family of five genes, which have a conserved leader sequence and two loosely conserved motifs of unknown function in the C-terminal region, but the remaining portion of the genes, although structurally similar, exhibit divergent sequences. DNA and protein database searches did not reveal any orthologues of Ta9 in parasite genera other than Theileria or in any mammalian species. The general features of the central region of the Ta9 protein, namely, its PQ-rich nature and variation in sequence and length due to multiple insertions and deletions, are found in other Theileria proteins, including the polymorphic immunodominant antigen in T. parva (PIM), the Tasp protein of T. annulata, and the subtelomere-encoded variable secreted protein (SVSP) family in T. parva and T. annulata (49, 50, 57). The presence of a predicted signal peptide indicates that Ta9 is likely to be secreted by the parasite and hence be available for cytosolic antigen processing. This is supported by the recent finding that antiserum raised against a Ta9 fusion protein locates the polypeptide to the parasite and the cytoplasm of macroschizont-infected cells and detects a protein of the expected size in the cytosolic fraction of infected cell extracts (W. Weir and B. R. Shiels, unpublished data).
Studies of CD8 T cell responses to Trypanosoma cruzi in mice and humans have identified a large family of trans-sialidase proteins as dominant targets of the CD8 T cell response and demonstrated that the epitopes are conserved among multiple members of this family (30, 59). On the basis of these findings, these authors proposed that epitope abundance due to sharing of the epitope sequence among family members may account for immunodominance of the CD8 T cell response (30). However, no such sharing of epitope sequences is observed between members of the Ta9 gene family.
Sequencing of the Ta9 gene from parasites isolated from naturally infected cattle demonstrated a high level of sequence polymorphism, with 12 distinct alleles identified among a small panel of parasite isolates. In addition to variation due to multiple insertions and deletions in the central region of the gene, the N- and C-flanking regions showed extensive sequence variation such that less than 50% of the codons were conserved among all alleles. This included variation among the T cell epitope sequences, which exhibited up to 5 amino acid substitutions compared to the reference C9 sequence. Sequence diversity in the epitopes was reflected by differential recognition of the allelic variants by specific CD8 T cells, indicating that some variants have the ability to escape the T cell response induced against the C9 parasite genotype.
Interrogation of Ta9 sequences by dN/dS analysis and the McDonald-Kreitman test revealed convincing evidence of positive selection at the N-terminal region (following the signal sequence). Two such positively selected sites at the N-terminal of Ta9 occur within an identified T cell epitope region, suggesting that immune selection may be acting on this site, driving the population to diversify and leading to a failure of specific CD8 T cells to recognize particular divergent epitopes. Evidence for CD8 T cell-imposed immune selection for sequence variation has been obtained for several RNA viruses, notably human immunodeficiency virus and influenza A virus (26, 60). Because such viruses undergo a rapid rate of mutation, it has been possible to monitor the emergence of the mutations in host populations in which the immune responses to the viruses have been defined. Immune selection of T cell epitopes has not been widely documented in protozoan parasite populations and, to date, the only other evidence of this phenomenon is in the circumsporozoite protein of Plasmodium falciparum (45). Eukaryotic genomes of apicomplexan parasites evolve relatively slowly and, while factors such as sexual recombination promote diversity within a population, the underlying mutation rate is much lower than that of viral genomes (7). Thus, it is likely that the positive selection observed among Ta9 alleles has developed over millennia during the course of many parasite generations against a predominantly Bos indicus/buffalo genetic background. This is consistent with the suggestion that selective pressure on parasite T cell antigens may be relatively weak at the population level (32) because, in natural host-parasite populations, variation in the specificity of the CD8 T cell response combined with reassortment in the parasite genome acts to dilute the selective pressure.
A striking feature of the Ta940-49-specific CD8 T cell clones examined in the present study was their differential recognition of allelic variants of the Ta9 epitope. The data show that some clones will tolerate up to three amino acid substitutions in the epitope, while others fail to recognize a variant with a single substitution. In each case, the levels of killing against cells expressing the variant epitopes were similar to those obtained with the reference C9 epitope. These findings indicate that at least those allelic variants that are recognized by some of the T cell clones retain the ability to bind to the N*00201 class I MHC and show that allelic polymorphism per se is not necessarily sufficient to avoid CD8 T cell recognition. Moreover, comparison of the composition of the CD8 T cell response in two of the animals revealed marked differences in the representation of T cell clones reactive with some of the variant epitopes. The composition of expressed TCR gene rearrangements has been shown to influence the fine specificity of human virus epitope-specific CD8 T cell clones, such that different TCRs specific for the same epitope may interact differentially with exposed amino acid residues in the epitope (11, 58). Hence, the different patterns of cross-reactivity of the Ta9-specific T cell clones, and also the difference between animals in the representation of these specificities in the response, almost certainly reflect the expressed TCR gene repertoire of the epitope-specific T cells. Although this work needs to be extended to further animals, these findings clearly demonstrate that the overall strain specificity of the CD8 T cell response with respect to the Ta940-49 epitope can vary between animals. Moreover, they point to the possibility of biasing the composition of the CD8 T cell response toward more cross-reactive T cells, by simultaneous immunization with parasites or vaccine constructs expressing two or more Ta9 allelic variants.
The results of the present study have important implications for understanding cross-protection between parasite strains and for vaccine development. Unlike the current experimental study, which utilized a cloned population of T. annulata for immunization (9), available evidence indicates that cattle in the field are often infected with genetically heterogeneous mixtures of parasites (4, 63), which is likely to broaden the antigenic specificity of the CD8 T cell response and reduce the likelihood of breakthrough infections upon subsequent challenge. However, the capacity to escape immune recognition may be of benefit to the parasite in field locations where parasite challenge is low and hence individual animals may be more likely to be infected with oligoclonal parasite populations. The generation of dominant responses against polymorphic antigens is of particular relevance for vaccination. The prolonged passage of T. annulata-infected cell lines, which is required to achieve attenuation of live vaccines, results in selection of clonal or near-clonal populations in the cell lines used for vaccination (9, 53). Hence, induction of highly focused immune responses against polymorphic antigens by cell line vaccines may account for the reported poorer protection obtained against heterologous parasite challenge compared to homologous challenge (9, 41). Moreover, the use of such antigens for the development of a subunit vaccine would present a substantial challenge. More extensive studies of the antigenic specificity of CD8 T cell responses to T. annulata in animals of different MHC genotypes are required to determine the extent of polymorphism in other target antigens (both dominant and subdominant) and to assess the potential for using a selected mixture of antigens to achieve herd immunity by vaccination.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by Wellcome Trust grant WT075820.
We thank Shirley Ellis, Institute for Animal Health, Compton, United Kingdom, for providing cloned cDNAs encoding defined bovine class I MHC heavy chains.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 7 February 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Ahmed J. S., Hartwig H., Schein E. 1999. Generation of Theileria annulata-specific cytotoxic T lymphocytes coincides with the control of tropical theileriosis. Parasitol. Res. 85:870–872 [DOI] [PubMed] [Google Scholar]
- 2. Altfeld M., et al. 2006. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8+ T cell response against HIV-1. PLoS Med. 3:e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ballingall K. T., et al. 2000. A highly sensitive, non-radioactive assay for T cell activation in cattle: applications in screening for antigens recognized by CD4+ and CD8+ T cells. J. Immunol. Methods 239:85–93 [DOI] [PubMed] [Google Scholar]
- 4. Ben Miled L., et al. 1994. Genomic and phenotypic diversity of Tunisian Theileria annulata isolates. Parasitology 108(Pt. 1):51–60 [DOI] [PubMed] [Google Scholar]
- 5. Birch J., Murphy L., MacHugh N. D., Ellis S. A. 2006. Generation and maintenance of diversity in the cattle MHC class I region. Immunogenetics 58:670–679 [DOI] [PubMed] [Google Scholar]
- 6. Brown C. G., Stagg D. A., Purnell R. E., Kanhai G. K., Payne R. C. 1973. Infection and transformation of bovine lymphoid cells in vitro by infective particles of Theileria parva. Nature 245:101–103(Letter.) [DOI] [PubMed] [Google Scholar]
- 7. Buonagurio D. A., et al. 1986. Evolution of human influenza A viruses over 50 years: rapid, uniform rate of change in NS gene. Science 232:980–982 [DOI] [PubMed] [Google Scholar]
- 8. Conze G., et al. 1998. Evidence for strain specificity in cytotoxic T-lymphocyte-mediated, major histocompatibility complex class I-dependent killing of Theileria annulata-infected cells. Parasitol. Res. 84:593–595 [DOI] [PubMed] [Google Scholar]
- 9. Darghouth M. A., et al. 1996. A preliminary study on the attenuation of Tunisian schizont-infected cell lines of Theileria annulata. Parasitol. Res. 82:647–655 [DOI] [PubMed] [Google Scholar]
- 10. Dobbelaere D. A., Kuenzi P. 2004. The strategies of the Theileria parasite: a new twist in host-pathogen interactions. Curr. Opin. Immunol. 16:524–530 [DOI] [PubMed] [Google Scholar]
- 11. Douek D. C., et al. 2002. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J. Immunol. 168:3099–3104 [DOI] [PubMed] [Google Scholar]
- 12. Ellis S. A., et al. 2005. Serological and molecular diversity in the cattle MHC class I region. Immunogenetics 57:601–606 [DOI] [PubMed] [Google Scholar]
- 13. Ellis S. A., Staines K. A., Stear M. J., Hensen E. J., Morrison W. I. 1998. DNA typing for BoLA class I using sequence-specific primers (PCR-SSP). Eur. J. Immunogenet. 25:365–370 [DOI] [PubMed] [Google Scholar]
- 14. Fraser D. C., et al. 1996. Functional expression of a cattle MHC class II DR-like antigen on mouse L cells. Immunogenetics 43:296–303 [DOI] [PubMed] [Google Scholar]
- 15. Gilbert S. C., et al. 1998. Association of malaria parasite population structure, HLA, and immunological antagonism. Science 279:1173–1177 [DOI] [PubMed] [Google Scholar]
- 16. Goddeeris B. M., Morrison W. I. 1988. Techniques for the generation, cloning and characterization of bovine cytotoxic T cells specific for the protozoan Theileria parva. J. Tissue Culture Methods 11:101–110 [Google Scholar]
- 17. Goddeeris B. M., Morrison W. I., Teale A. J., Bensaid A., Baldwin C. L. 1986. Bovine cytotoxic T-cell clones specific for cells infected with the protozoan parasite Theileria parva: parasite strain specificity and class I major histocompatibility complex restriction. Proc. Natl. Acad. Sci. U. S. A. 83:5238–5242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goulder P. J., et al. 1997. Patterns of immunodominance in HIV-1-specific cytotoxic T lymphocyte responses in two human histocompatibility leukocyte antigens (HLA)-identical siblings with HLA-A*0201 are influenced by epitope mutation. J. Exp. Med. 185:1423–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Graham S. P., et al. 2006. Theileria parva candidate vaccine antigens recognized by immune bovine cytotoxic T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 103:3286–3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Graham S. P., et al. 2008. Characterization of the fine specificity of bovine CD8 T-cell responses to defined antigens from the protozoan parasite Theileria parva. Infect. Immun. 76:685–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heussler V. T., et al. 2002. Hijacking of host cell IKK signalosomes by the transforming parasite Theileria. Science 298:1033–1036 [DOI] [PubMed] [Google Scholar]
- 22. Hulliger L. 1965. Cultivation of three species of Theileria in lymphoid cells in vitro. J. Protozool. 12:649–655 [DOI] [PubMed] [Google Scholar]
- 23. Hulliger L., Wilde K. H., Brown C. G., Turner L. 1964. Mode of multiplication of Theileria in cultures of bovine lymphocytic cells. Nature 203:728–730 [DOI] [PubMed] [Google Scholar]
- 24. Innes E. A., Millar P., Brown C. G., Spooner R. L. 1989. The development and specificity of cytotoxic cells in cattle immunized with autologous or allogeneic Theileria annulata-infected lymphoblastoid cell lines. Parasite Immunol. 11:57–68 [DOI] [PubMed] [Google Scholar]
- 25. Irvin A. D., Morrison W. I. 1987. Immunopathology, immunology, and immunoprophylaxis of Theileria infections, p. 223–274 In Soulsby E. J. L. (ed.), Immune responses in parasitic infections: immunology, immunopathology and immunoprophylaxis of Theileria infections. CRC Press, Inc., Baton Rouge, FL [Google Scholar]
- 26. Jones N. A., et al. 2004. Determinants of human immunodeficiency virus type 1 escape from the primary CD8+ cytotoxic T lymphocyte response. J. Exp. Med. 200:1243–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. MacHugh N. D., Burrells A. C., Morrison W. I. 2008. Demonstration of strain-specific CD8 T cell responses to Theileria annulata. Parasite Immunol. 30:385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. MacHugh N. D., et al. 2009. CD8+ T-cell responses to Theileria parva are preferentially directed to a single dominant antigen: implications for parasite strain-specific immunity. Eur. J. Immunol. 39:2459–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. MacLeod A., et al. 2000. Minisatellite marker analysis of Trypanosoma brucei: reconciliation of clonal, panmictic, and epidemic population genetic structures. Proc. Natl. Acad. Sci. U. S. A. 97:13442–13447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martin D. L., et al. 2006. CD8+ T-cell responses to Trypanosoma cruzi are highly focused on strain-variant trans-sialidase epitopes. PLoS Pathog. 2:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McDonald J. H., Kreitman M. 1991. Adaptive protein evolution at the adh locus in Drosophila. Nature 351:652–654 [DOI] [PubMed] [Google Scholar]
- 32. McKeever D. J. 2009. Bovine immunity: a driver for diversity in Theileria parasites? Trends Parasitol. 25:269–276 [DOI] [PubMed] [Google Scholar]
- 33. McKeever D. J., et al. 1994. Adoptive transfer of immunity to Theileria parva in the CD8+ fraction of responding efferent lymph. Proc. Natl. Acad. Sci. U. S. A. 91:1959–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morrison W. I. 2007. The biological and practical significance of antigenic variability in protective T cell responses against Theileria parva. Vet. Parasitol. 148:21–30 [DOI] [PubMed] [Google Scholar]
- 35. Morrison W. I., et al. 1987. Cytotoxic T cells elicited in cattle challenged with Theileria parva (Muguga): evidence for restriction by class I MHC determinants and parasite strain specificity. Parasite Immunol. 9:563–578 [DOI] [PubMed] [Google Scholar]
- 36. Morrot A., Zavala F. 2004. Effector and memory CD8+ T cells as seen in immunity to malaria. Immunol. Rev. 201:291–303 [DOI] [PubMed] [Google Scholar]
- 37. Pain A., et al. 2005. Genome of the host-cell transforming parasite Theileria annulata compared with T. parva. Science 309:131–133 [DOI] [PubMed] [Google Scholar]
- 38. Parker S. J., Roberts C. W., Alexander J. 1991. CD8+ T cells are the major lymphocyte subpopulation involved in the protective immune response to Toxoplasma gondii in mice. Clin. Exp. Immunol. 84:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Phillips R. E., et al. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354:453–459 [DOI] [PubMed] [Google Scholar]
- 40. Pipano E. 1981. Schizonts and tick stage in immunization against Theileria annulata infection, p. 242–252 In Irvin A. D., Cunningham M. P., Young A. (ed.), Advances in the control of theileriosis. Martinus Nijoff Publishers, The Hague, Netherlands [Google Scholar]
- 41. Pipano E. 1989. Vaccination against Theileria annulata theileriosis, p. 203–234 In Wright I. G. (ed.), Veterinary protozoan and hemoparasite vaccines. CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 42. Pond S. L., Frost S. D. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22:1208–1222 [DOI] [PubMed] [Google Scholar]
- 43. Pond S. L., Frost S. D., Muse S. V. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679 [DOI] [PubMed] [Google Scholar]
- 44. Preston P. M., Brown C. G., Spooner R. L. 1983. Cell-mediated cytotoxicity in Theileria annulata infection of cattle with evidence for BoLA restriction. Clin. Exp. Immunol. 53:88–100 [PMC free article] [PubMed] [Google Scholar]
- 45. Putaporntip C., Jongwutiwes S., Hughes A. L. 2009. Natural selection maintains a stable polymorphism at the circumsporozoite protein locus of Plasmodium falciparum in a low endemic area. Infect. Genet. Evol. 9:567–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rand D. M., Kann L. M. 1996. Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol. Biol. Evol. 13:735–748 [DOI] [PubMed] [Google Scholar]
- 47. Rozas J., Rozas R. 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175 [DOI] [PubMed] [Google Scholar]
- 48. Schein E. 1975. On the life cycle of Theileria annulata (Dschunkowsky and Luhs, 1904) in the midgut and hemolymph of Hyalomma anatolicum excavatum (Koch, 1844). Z. Parasitenkd. 47:165–167 [DOI] [PubMed] [Google Scholar]
- 49. Schmuckli-Maurer J., et al. 2009. Expression analysis of the Theileria parva subtelomere-encoded variable secreted protein gene family. PLoS One 4:e4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schnittger L., et al. 2002. Characterization of a polymorphic Theileria annulata surface protein (TaSP) closely related to PIM of Theileria parva: implications for use in diagnostic tests and subunit vaccines. Mol. Biochem. Parasitol. 120:247–256 [DOI] [PubMed] [Google Scholar]
- 51. Shiels B., et al. 2006. Alteration of host cell phenotype by Theileria annulata and Theileria parva: mining for manipulators in the parasite genomes. Int. J. Parasitol. 36:9–21 [DOI] [PubMed] [Google Scholar]
- 52. Spooner R. L., Innes E. A., Glass E. J., Brown C. G. 1989. Theileria annulata and T. parva infect and transform different bovine mononuclear cells. Immunology 66:284–288 [PMC free article] [PubMed] [Google Scholar]
- 53. Sutherland I. A., et al. 1996. Theileria annulata: altered gene expression and clonal selection during continuous in vitro culture. Exp. Parasitol. 83:125–133 [DOI] [PubMed] [Google Scholar]
- 54. Taracha E. L., Goddeeris B. M., Teale A. J., Kemp S. J., Morrison W. I. 1995. Parasite strain specificity of bovine cytotoxic T cell responses to Theileria parva is determined primarily by immunodominance. J. Immunol. 155:4854–4860 [PubMed] [Google Scholar]
- 55. Tarleton R. L., Koller B. H., Latour A., Postan M. 1992. Susceptibility of β2-microglobulin-deficient mice to Trypanosoma cruzi infection. Nature 356:338–340 [DOI] [PubMed] [Google Scholar]
- 56. Townsend A. R., McMichael A. J. 1985. Specificity of cytotoxic T lymphocytes stimulated with influenza virus. Studies in mice and humans. Prog. Allergy 36:10–43 [DOI] [PubMed] [Google Scholar]
- 57. Toye P. G., et al. 1995. Characterization of the gene encoding the polymorphic immunodominant molecule, a neutralizing antigen of Theileria parva. J. Immunol. 155:1370–1381 [PubMed] [Google Scholar]
- 58. Turnbull E. L., et al. 2006. HIV-1 epitope-specific CD8+ T cell responses strongly associated with delayed disease progression cross-recognize epitope variants efficiently. J. Immunol. 176:6130–6146 [DOI] [PubMed] [Google Scholar]
- 59. Tzelepis F., et al. 2008. Infection with Trypanosoma cruzi restricts the repertoire of parasite-specific CD8+ T cells leading to immunodominance. J. Immunol. 180:1737–1748 [DOI] [PubMed] [Google Scholar]
- 60. Voeten J. T., et al. 2000. Antigenic drift in the influenza A virus (H3N2) nucleoprotein and escape from recognition by cytotoxic T lymphocytes. J. Virol. 74:6800–6807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. von Schubert C., et al. 2010. The transforming parasite Theileria co-opts host cell mitotic and central spindles to persist in continuously dividing cells. PLoS Biol. 8:e100498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Weir W. 2006. Genomic and population genetic studies on Theileria annulata. Ph.D. thesis University of Glasgow, Glasgow, Scotland [Google Scholar]
- 63. Weir W., et al. 2007. Genetic exchange and sub-structuring in Theileria annulata populations. Mol. Biochem. Parasitol. 154:170–180 [DOI] [PubMed] [Google Scholar]
- 64. Weir W., Karagenc T., Baird M., Tait A., Shiels B. R. 2010. Evolution and diversity of secretome genes in the apicomplexan parasite Theileria annulata. BMC Genomics 11:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wong P., Pamer E. G. 2003. CD8 T cell responses to infectious pathogens. Annu. Rev. Immunol. 21:29–70 [DOI] [PubMed] [Google Scholar]
- 66. Yewdell J. W. 2006. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity 25:533–543 [DOI] [PubMed] [Google Scholar]
- 67. Yewdell J. W., Bennink J. R. 1999. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu. Rev. Immunol. 17:51–88 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.