Abstract
The cyclic-dimeric-GMP (c-di-GMP)-binding protein PilZ has been implicated in bacterial motility and pathogenesis. Although BB0733 (PlzA), the only PilZ domain-containing protein in Borrelia burgdorferi, was reported to bind c-di-GMP, neither its role in motility or virulence nor it's affinity for c-di-GMP has been reported. We determined that PlzA specifically binds c-di-GMP with high affinity (dissociation constant [Kd], 1.25 μM), consistent with Kd values reported for c-di-GMP-binding proteins from other bacteria. Inactivation of the monocistronically transcribed plzA resulted in an opaque/solid colony morphology, whereas the wild-type colonies were translucent. While the swimming pattern of mutant cells appeared normal, on swarm plates, mutant cells exhibited a significantly reduced swarm diameter, demonstrating a role of plzA in motility. Furthermore, the plzA mutant cells were significantly less infectious in experimental mice (as determined by 50% infectious dose [ID50]) relative to wild-type spirochetes. The mutant also had survival rates in fed ticks lower than those of the wild type. Consequently, plzA mutant cells failed to complete the mouse-tick-mouse infection cycle, indicating plzA is essential for the enzootic life cycle of B. burgdorferi. All of these defects were corrected when the mutant was complemented in cis. We propose that failure of plzA mutant cells to infect mice was due to altered motility; however, the possibility that an unidentified factor(s) contributed to interruption of the B. burgdorferi enzootic life cycle cannot yet be excluded.
INTRODUCTION
Cyclic dimeric-GMP (c-di-GMP) [bis(3′,5′)-cyclic diguanylic acid] is a ubiquitous bacterial second messenger. c-di-GMP is produced from 2 molecules of GTP by the activity of diguanylate cyclase and is degraded by phosphodiesterase enzymes. The activities of these cyclase and phosphodiesterase enzymes are often regulated by changing physiological conditions. c-di-GMP has been implicated in the regulation of diverse bacterial properties, including biosynthesis of adhesins and exopolysaccharide matrix for biofilm formation, different forms of motility, long-term survival, and response to environmental stresses, in addition to production of virulence factors and to virulence (14, 29, 39, 76, 88, 96). These cellular functions regulated by c-di-GMP often utilize multiple receptors and signaling mechanisms. Several types of c-di-GMP receptors have been reported, including PilZ, PelD, I-site effectors, FleQ, and Clp (29, 30, 40, 76). Although many of the downstream mechanisms remain to be identified, studies indicate that c-di-GMP can impact cellular signaling at multiple levels, including transcription, translation, posttranslation, protein activity, protein secretion, and protein stability (7, 15, 17, 30, 41, 48, 50, 57, 58, 70, 86, 93).
One c-di-GMP receptor, the PilZ protein, was originally identified in silico as a putative conserved sequence based on the binding of c-di-GMP to the BcsA1 protein, a component of the cellulose synthase complex of Acetobacter xylinus (2, 69, 94). The “PilZ domain,” named after the PilZ protein from Pseudomonas aeruginosa, is required for the assembly of functional type 4 pili (T4P) (1). Recently, proteins containing PilZ domains have been shown to bind c-di-GMP in several species of bacteria (see references 29 and 76 and references therein). Strong affinity of c-di-GMP for PilZ domains has been observed, with dissociation constants (Kd) in the range of 1 nM to 2 μM, and this strong affinity is consistent with the low concentration of c-di-GMP in cells (29, 37, 76, 78). Furthermore, PilZ proteins from a number of bacteria have been implicated in the regulation of specific cellular processes, such as motility, polysaccharide synthesis, biofilm formation, and virulence of animal and plant pathogens (5, 13, 15, 30, 41, 48, 60, 70). The roles of PilZ appear to be different in different bacterial species. For example, as mentioned above, PilZ of P. aeruginosa (PA2960) is necessary for T4P biogenesis and twitching motility. A pilZ knockout mutant is defective in the secretion of PilA polymers to the bacterial surface, though intracellular PilA pools are not affected (1). On the other hand, a PilZ ortholog knockout mutant of Neisseria meningitidis is piliated but is not able to produce bacterial aggregates (11); a Xanthomonas campestris pv. campestris PilZ knockout mutant has slightly reduced T4P-dependent motility on semisolid Eiken agar (47); and a pilZ mutation in Xanthomonas axonopodis pv. citri resulted in increased swarming motility relative to the parental wild-type cells (27). A PilZ domain-containing protein, Alg44, was shown to upregulate alginate production in P. aeruginosa (48). Furthermore, in Escherichia coli (and Salmonella enterica), the reduced motility of a phosphodiesterase mutant is alleviated by a pilZ mutation (58, 70), indicating a role of PilZ in bacterial motility. Specifically, the E. coli PilZ protein, YcgR, was found to control motility by directly interacting with and altering flagellar switch protein FliG, FliM, or motor protein MotA function in response to c-di-GMP (3, 7, 17, 57). In Vibrio cholerae, a mutation in genes encoding PilZ proteins also resulted in altered motility (60). These observations suggest that signaling by PilZ could be modulated differently in different species.
Although c-di-GMP may have different roles in different species of bacteria, little is known about this signaling system in Borrelia burgdorferi, the Lyme disease spirochete. The spirochete resides in disparate host environments, including Ixodes ticks and mammalian hosts. During transmission between hosts, B. burgdorferi modulates gene expression in response to environmental factors, such as nutrient availability, pH, CO2, temperature, host factors, and immune response (10, 55, 65, 74, 90, 91). c-di-GMP levels, which are controlled by a two-component system, are also believed to be involved in bacterial adaptation to different host environments (84, 88, 95). While many species of bacteria were reported to contain multiple enzymes responsible for controlling the levels of c-di-GMP (22), bioinformatics, as well as experimental analysis, indicates that B. burgdorferi utilizes only a limited number of enzymes to modulate c-di-GMP: a single diguanylate cyclase, two phosphodiesterases, and a single c-di-GMP-binding PilZ homolog designated PlzA (2, 21, 22, 66, 71, 87). B. burgdorferi diguanylate cyclase (BB0419/Rrp1) was shown to regulate a diverse set of genes, many of which were reported to be associated with B. burgdorferi virulence, implying a role of c-di-GMP in pathogenesis (66, 71). We recently demonstrated that BB0363 is a phosphodiesterase which specifically hydrolyzed c-di-GMP into pGpG (87). Furthermore, a bb0363/pdeA mutant exhibited impaired motility, and those mutant cells were unable to infect experimental mice, irrespective of the size or route of inoculum (needle or tick), demonstrating a role of c-di-GMP in B. burgdorferi motility and virulence (87).
Recently, PlzA was shown to specifically bind c-di-GMP, and the expression of plzA was shown to be modulated under different environmental conditions (21). While PilZ has different functions in different bacteria, a role for the lone c-di-GMP-binding protein PlzA has not been demonstrated in B. burgdorferi. Here we demonstrate that PlzA binds c-di-GMP in vitro with strong affinity. The deletion of plzA reduces motility and infectivity in experimental mice. The mutant cells exhibit lowered viability in ticks. Consequently, the plzA cells are unable to complete the mouse-tick-mouse infection cycle, consistent with a role for PilZ in bacterial pathogenesis. Furthermore, by constructing a plzA mutant in the pdeA mutant background, where the cellular c-di-GMP level is 2× higher than that in the wild-type cells, we demonstrate that PlzA regulates B. burgdorferi motility in a different manner from in E. coli and that elevated c-di-GMP in the B. burgdorferi pdeA mutant regulates motility by a mechanism independent of PlzA. A mechanism of altered motility and infectivity is proposed.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Low-passage, virulent B. burgdorferi strain B31-A3-68 Δbbe002 was used as the wild type throughout the study (a kind gift from R. Rego and P. Rosa, Rocky Mountain Laboratories, NIH) (63). This strain is a derivative of A3-68 (63), lacking circular plasmid 9 (cp9) and linear plasmid 56 (lp56), with the bbe002 gene inactivated with a PflgB-aadA cassette to increase transformation frequency (36, 63). B31-A3-68 Δbbe002 is a derivative of strain B31 (16). The genome of the virulent B31 strain has been sequenced and was found to contain a total of 21 plasmids with 12 linear and 9 circular plasmids, in addition to the 960-kb linear chromosome (12, 20). B. burgdorferi was cultured in liquid Barbour-Stoenner-Kelly (BSK-II) medium as described below; plating BSK medium was prepared with 0.6% agarose (52). Swarm plate assays were carried out with 0.35% agarose in plating BSK medium; approximately 1 × 106 cells in a volume of 5 μl were spotted onto plates containing plating BSK medium diluted 1:10 in Dulbecco's phosphate-buffered saline without divalent cations. Because B. burgdorferi is a slow-growing organism with an 8- to 12-h generation time, swarm plates were incubated for 5 to 7 days (52, 53). The paired Student's t test (Minitab, Inc.) was used to compare wild-type and plzA mutant cell swarm diameters.
BB0733 recombinant protein expression.
Recombinant BB0733 protein (PlzA) was expressed with an N-terminal 6×His (His6) fusion tag. To express BB0733 protein in E. coli, the coding sequence was amplified by PCR using the primers (5′→3′ throughout) R-BB0733-F (CTTTTATCTAGAAAAATAAGAG) and T-BB0733-R (CTTTGATTAATTGAAATAATC). The amplified DNA fragment was ligated into an E. coli expression vector pTrc His TOPO (Invitrogen, Inc.) by TA cloning. The vector was then digested and sequenced to confirm its integrity. E. coli cells containing pbb0733-TRC were induced with 0.25 mM isopropyl β-d-1-thiogalactopyranoside at 30°C for 4 h and Ni2+ affinity purified under native conditions. Purified recombinant BB0733 protein was resolved by SDS-PAGE to determine protein concentration and purity.
c-di-GMP-binding assays by equilibrium dialysis.
Equilibrium dialysis was performed as described previously (70). Briefly, DispoBiodialyzer cassettes (The Nest Group, Southborough, MA) contain 2 chambers separated by a 5-kDa-cutoff membrane. Sixty microliters of c-di-GMP (2 to 50 μM) was injected into one chamber, and 60 μl (10 μM) of test protein BB0733 or positive control E. coli c-di-GMP-binding PilZ receptor YcgR (70) was injected into the other chamber; the c-di-GMP and test proteins were contained in a mixture of 300 mM NaCl, 0.5 mM EDTA, 10% glycerol, and 50 mM sodium phosphate (pH 7.4). Following gentle rocking at room temperature for 24 h to reach equilibrium, samples from both sides of the cassettes were removed, boiled for 4 min, centrifuged, and filtered through a 0.22-μm-pore microfilter (Millipore Ultrafree MC). c-di-GMP concentrations in both samples were determined via reverse-phase high-performance liquid chromatography (HPLC) on a Supelcosil LC-18T column using the same buffer and gradient program described in reference 71, and c-di-GMP was quantified at 254 nm. The binding constant was determined with Graph Pad Prism 5 software.
RT-PCR.
Exponentially growing B. burgdorferi wild-type cells (2 × 107 cells ml−1) were treated with RNAprotect followed by total RNA isolation with the RNeasy minikit (Qiagen, Inc.). Contaminating DNA in the RNA samples was removed by RNase-free Turbo DNase I (Ambion, Inc.) digestion for 3 h at 37°C followed by RNeasy minipurification. For reverse transcription-PCR (RT-PCR), cDNA was prepared from 1 μg RNA with Superscript III reverse transcriptase according to the manufacturer's protocol (Invitrogen, Inc.). The RT-PCR primer sequences were a1 (TGTTAAATATATAAGCAGCTTATGCG), a2 (GCGGGCAATACTGTATCTTCTTGA), b1 (TTGAGATGAGAATAGGCTCTTATACC), b2 (GCGGGCAATACTGTATCTTCTTGA), c1 (TTGAGTGCTTCTGCTTATTCTTCT), and c2 (GATACGAAAATACTTATCACAAAAGC). A Bio-Rad iScript cDNA synthesis kit and an iCycler detection system were used to measure bb0733 transcript levels according to the manufacturer's instructions. The gene coding for B. burgdorferi enolase was used a reference gene (54, 87). The gene-specific primers were RT-enolase-F (TGGAGCGTACAAAGCCAACATT), RT-enolase-R (TGAAAAACCTCTGCTGCCATTC), Bb0733-rt-F (TTGAGATGAGAATAGGCTCTTATACC), and Bb0733-rt-R (GCGGGCAATACTGTATCTTCTTGA). The relative level of expression was calculated by the threshold cycle (2−ΔΔCT) method (46, 79, 87).
Construction of a plzA (bb0733) mutant.
Targeted inactivation of bb0733 (a 786-bp gene) was achieved by homologous recombination using a promoterless kanamycin (Pl-Kan) cassette (87). PCR amplification of bb0733, construction of the inactivation plasmids, and electroporation of linear bb0733-Pl-Kan DNA into competent cells were carried out as described previously (52, 53, 73, 87). Briefly, bb0733 DNA was PCR amplified with the GoTaq PCR system (Promega, Inc.) and primers BB0733-KO-F (AGTTTTGCATTTGGAAGCTGGA) and BB0733-KO-R (ATTCCCAAAAGTGCCCTCATG). The amplified DNA was ligated into the pGEM-T Easy vector (Promega, Inc.), yielding pbb0733-Easy. Pl-Kan was similarly PCR amplified from PflgB-Kan (8) with the primers PrLs-Kan AflII-F (CTTAAGTAGTTAAAAGCAAT) and PrLs-Kan AflII-R (CTTAAGTTAGAAAAACTCAT) (restriction sites are in boldface). The AflII-restricted Pl-Kan DNA was then inserted into the AflII sites within bb0733 (located 167 and 676 bp from the ATG start codon) of the pbb0733-Easy vector, yielding pbb0733-Pl-Kan. This deletion-insertion was in frame. Restriction mapping indicated that the direction of transcription of Pl-Kan was the same as that of bb0733. bb0733-Pl-Kan was digested and separated from pbb0733-Pl-Kan vector by NotI digestion, precipitated, and electroporated into the wild-type competent cells (52). Mutants were selected on solid growth medium containing 200 μg ml−1 kanamycin plus 100 μg ml−1 streptomycin. Resistant colonies were analyzed by PCR for the presence of the Pl-Kan cassette with the primers PrLs-Kan AflII-F and -R as well as primers BB0733-KO-F and -R. PCR-positive mutants were further examined for their plasmid contents by using 21 sets of primers to detect 21 linear and circular plasmids (100).
Complementation of the bb0733 (plzA) mutant.
To complement bb0733::Pl-Kan, the promoter of bb0733 (PplzA; 400 bp upstream from the ATG start codon of the bb0733/plzA gene) and the coding sequence was PCR amplified by PCR with primers 733 comp-F-SpeI (GCGCCACTAGTTGTGATAATAGAAAAGCTAACTGCGAAATA) and 733-comp-R-NotI (GCGCGCGGCCGCCAAATACAACCAAGAAGACTTAAAC). The PCR product was ligated into pGEM-T Easy. The bb0733 promoter-bb0733 (Pbb0733-bb0733/PplzA-plzA) DNA was then inserted into suicide vector pXLF14301 (99) by SpeI-NotI restriction digestion, yielding pXLFBB0733. Fifty micrograms of the resultant plasmid was electroporated into the bb0733::Pl-Kan competent cells. Transformants were selected in plating BSK medium containing 100 μg ml−1 streptomycin, 200 μg ml−1 kanamycin plus 40 μg ml−1 gentamicin. Resistant transformants were analyzed by PCR for the presence of kanamycin (Kan) and gentamicin (Gen) genes and an intact bb0733 gene. Positive clones were further examined for their plasmid content by PCR.
SDS-PAGE and Western blotting.
SDS-PAGE and Western blotting by an enhanced chemiluminescent detection method (GE Healthcare) were carried out as previously reported (23). B. burgdorferi strain B31-A3-68 Δbbe002 (wild-type), plzA, or plzA-pdeA cells were cultured to 5 × 107 cells ml−1. Cells were pelleted, resuspended in phosphate-buffered saline (PBS), and equalized to protein content with the Bradford assay reagent (Bio-Rad, Inc.). Samples were boiled, and approximately 5 μg of cell lysate was loaded per lane. Proteins were transferred to polyvinylidene difluoride (PVDF) membrane and Western blotted with monoclonal FlaB (A. Barbour), polyclonal MotB (J. Carroll), FliG1 and FliG2 (C. Li), FliM (D. Blair), CheY3, and FliL antisera. Specific monoclonal or polyclonal reactivity to B. burgdorferi FlaB, MotB, FliG1, FliG2, FliM, FliL, and CheY3 has been reported previously (42, 43, 51, 53, 54, 72; M. A. Motaleb et al., unpublished data).
Experimental mouse-tick-mouse infection model of B. burgdorferi.
Six-week-old female C3H/HeN mice were purchased from Charles River Laboratories, North Carolina, and housed in the East Carolina University animal facility at the Brody School of Medicine according to the institutional guidelines for the care and use of laboratory animals. For infection via needle, 5 × 102 to 5 × 106 in vitro-grown spirochetes were injected subcutaneously as described previously (16, 35, 64). The number of spirochetes was determined with a Petroff-Hausser chamber, and each clone was verified for its retention of the lp25, lp28-1, and lp36 plasmids. Mice were bled 2 weeks postinfection for immunoblot analysis of mouse sera against B. burgdorferi antigens to determine infectivity, as described previously (35, 80, 87). Reisolation of B. burgdorferi from mouse skin, bladder, and joint tissues was performed 6 weeks postinfection to assess the ability of spirochetes to infect mice as described previously (16, 26, 83, 87). Mouse tissues were placed in BSK-II growth medium and incubated for up to 35 days; the presence of spirochetes was determined by dark-field microscopy. The 50% infectious dose (ID50) was calculated as described below (35, 62).
For tick infection studies, naïve Ixodes scapularis larvae were purchased from Oklahoma State University. Two independent experiments were performed with ticks. Ticks were kept at 23 to 24°C under a 14-h-light/10-h-dark photoperiod in a humidified chamber with 85 to 90% relative humidity. Approximately 100 larval ticks were fed to repletion on spirochete-infected mice for 5 to 7 days, allowed to fall off, and collected. One subset of larvae was dissected 7 days after repletion, and the isolated midguts were analyzed by indirect immunofluorescence assays (IFAs) for the presence of spirochetes (83, 87). A second subset of fed larvae was surface sterilized with 3% H2O2 followed by 70% ethanol, crushed in BSK-II medium, and plated to determine the number of CFU per tick. A third subset of larvae was crushed, and genomic DNA was extracted with the DNeasy blood and tissue kit (Qiagen, Inc.). Quantitative PCR (qPCR) was performed to quantitate the level of the tick actin gene by using primers ActinT-F (GATCATGTTCGAGACCTTCA) and ActinT-R (CGATACCCGTGGTACGA) versus the level of B. burgdorferi flaB by using primers FlaB qRT-F (TTGCTGATCAAGCTCAATATAACCA) and FlaB qRT-R (TTGAGACCCTGAAAGTGATGC), as described previously (97, 99). The remaining fed larvae were allowed to molt to nymphs. The infected nymphs were then fed to repletion on naïve 6-week-old female C3H/HeN mice, allowed to fall off, and collected. Subsets of nymphs were processed for IFA or plated to determine the number of CFU per tick as described above. Six weeks postinfection, mouse sera were collected for immunoblot analysis, and tissues were collected for reisolation of B. burgdorferi.
Determination of ID50 and statistical analysis.
The dose required to infect 50% of the mice inoculated was experimentally determined for the wild-type, plzA mutant, and complemented plzA+ strains as described previously (35, 62). The data from the ID50 infection experiment and the single-dose infection experiment for each strain were combined for the estimations of the 50% infectious dose. Comparison between strain ID50 values was made by using a generalized linear model with a probit link function. This method is also known as probit regression (35), and in it we assume identical slopes in the response/log-dose relationship but different intercepts for each strain. Graphically those assumptions manifest themselves as dose-response curves with lateral shifts corresponding to the changes in intercept. Additionally, an overdispersion parameter was fit in order to accommodate greater homogeneity in infection rates than would otherwise be permitted by the model. All calculations were carried out with JMP V9 software (SAS Institute, Inc., Cary, NC).
Artificial inoculation of ticks by immersion studies.
Tick immersion studies were performed as described previously (87). Briefly, approximately 150 tick larvae were artificially infected by immersion (in duplicate) in equal-density exponential-phase B. burgdorferi cultures (5 × 107 cells/ml), as described previously (4, 59, 83, 87). Ticks were fed to repletion on separate naïve mice for 5 to 7 days, allowed to fall off, and collected. Subsets of larvae were dissected 7 to 10 days after repletion, and the crushed ticks were analyzed by immunofluorescence assay (IFA) for the presence of spirochetes (83, 87). Fed larvae were treated as described above to determine the number of CFU per tick. Reisolation of spirochetes from mouse ears, joints, and bladders was performed as described above.
IFAs.
Ticks were dissected in 25 μl PBS-5 mM MgCl2 in Teflon-coated microscope slides, mixed by pipetting, and then air dried (4, 87). To avoid quenching by hemin in the blood, dissected tick contents were 10-fold serially diluted. Slides were blocked with 0.75% bovine serum albumin (BSA) in PBS-5 mM MgCl2 and washed with PBS-5 mM MgCl2. Spirochetes were detected with a 1:100 dilution of goat anti-B. burgdorferi antisera labeled with fluorescein isothiocyanate (Kirkegaard & Perry Laboratories). Images were captured with a Zeiss Axio Imager M1 microscope coupled with an AxioCam MRc digital camera (Carl Zeiss, Inc.).
RESULTS
BB0733 binds c-di-GMP with high affinity.
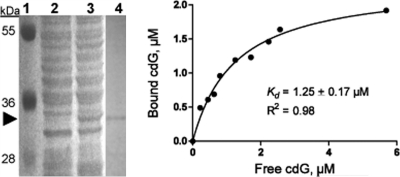
Bioinformatics BLAST analysis suggests that BB0733 is the only PilZ domain-containing protein in B. burgdorferi (PilZ domain; Pfam PF07238) (1, 2, 20). BB0733, which has been named “PlzA,” shares significant amino acid sequence similarity with orthologs in other Borrelia strains (21). Although PlzA shares poor homology with PilZ proteins of other species of bacteria, it contains the conserved residues of the “PilZ domain” (RXXXR; D/NZSXXG where “X” is any amino acid and “Z” is a hydrophobic residue) but lacks the so-called “PilZ-N”-terminal domain (N-terminal domain of E. coli PilZ protein YcgR [PF07317]) (2, 70) and therefore is considered a “stand-alone” c-di-GMP effector protein (21). Recently, using X-ray cross-linking studies, Freedman et al. confirmed that PlzA specifically binds c-di-GMP and that the conserved arginine (R) residues of 160RXXXR (number based on amino acid residues of BB0733 protein) (20) is critical for c-di-GMP binding (21). However, a dissociation constant (Kd) of PlzA for c-di-GMP was not determined (21). To determine the Kd of PlzA, synthetic c-di-GMP (38, 41) and recombinant, purified His6-PlzA (Fig. 1) were used in equilibrium dialysis, as described previously (70). An E. coli c-di-GMP-binding PilZ protein, YcgR, was used as a positive control (70). Figure 1 shows that His6-PlzA is highly purified and binds c-di-GMP; His6-PlzA did not bind other nucleotides tested (cyclic AMP [cAMP], GTP, and cyclic GMP [cGMP]; data not shown) confirming the findings reported by Freedman et al. (21). The dissociation constant of PlzA was determined to be 1.25 ± 0.17 μM, indicating high affinity for c-di-GMP, consistent with Kd values (1 nM to 2 μM) reported for c-di-GMP-binding proteins from other bacteria (29, 37).
Fig. 1.
(Left panel) SDS-PAGE analysis of expressed His6-BB0733/PlzA recombinant protein from E. coli. Lane 1, protein size marker with sizes in kDa; lanes 2 and 3, uninduced and induced E. coli lysates, respectively; lane 4, His6-PlzA protein after purification in an Ni2+-agarose column. An arrowhead indicates the His6-PlzA (∼32-kDa) protein band. (Right panel) Plot of equilibrium binding between PlzA and synthetic c-di-GMP (cdG). Equilibrium dialysis was performed as described above, and the binding constant was determined with GraphPad Prism 5 (one site, specific binding). Results with GTP, cAMP, or cGMP were not shown since no binding with His6-PlzA was detected.
Inactivation and complementation of plzA in B. burgdorferi.
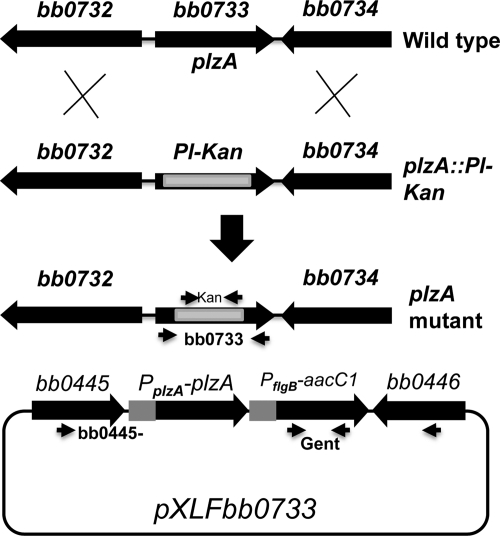
B. burgdorferi genome sequence analysis predicted that the genes upstream (bb0732) and downstream (bb0734) of bb0733 are transcribed in the opposite direction from bb0733 (Fig. 2). Using RT-PCR, we confirmed that bb0733 is a monocistronic mRNA that is expressed in growing B. burgdorferi cells and that the bb0732 and bb0734 genes are not cotranscribed (data not shown). bb0732 is annotated as a penicillin-binding protein that is predicted to be responsible for biosynthesis and degradation of murein sacculus and peptidoglycan of cell envelope, whereas bb0734 encodes a Sua5/YciO/YrdC/YwlC family protein of unknown function. In Shewanella oneidensis and the psychrophile Colwellia psychrerythraea 34H, the Sua5/YciO/YrdC/YwlC family protein is a putative translation factor involved in translation, as well as ribosomal structure and biogenesis (28, 49).
Fig. 2.
Inactivation and complementation of bb0733 (plzA). (Top) In-frame insertion of the Pl-Kan cassette within the bb0733 gene by homologous recombination resulted in the creation of plzA mutant. Approximate locations of kan and bb0733 primer sets are shown by thin arrows that were used to confirm the bb0733 inactivation by PCR (Fig. 3). (Bottom) Complementation plasmid pXLFbb0733 containing the bb0733 promoter (PplzA) that drives the expression of the bb0733 (plzA) gene was used to complement the plzA mutant in cis by genomic integration in a region where genes bb0445 and bb0446 converge. Approximate locations of the primer sets (bb0445 and bb0446 and gentamicin [Gent]) used to confirm the integration of PplzA-plzA-PflgB-aacC1 are shown by thin horizontal arrows. PCR primer products of each of these primer set are shown in Fig. 3.
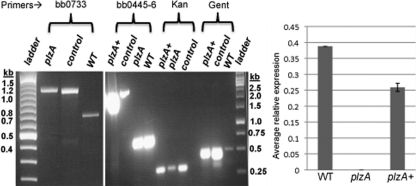
The expression of B. burgdorferi PlzA was reported to be maintained throughout the enzootic cycle; however, its expression was significantly increased in ticks after a blood meal (21). A function of plzA has not been demonstrated. To investigate the function of PlzA during the enzootic life cycle of B. burgdorferi, we inactivated plzA with a promoterless kanamycin cassette (Pl-Kan) by allelic exchange, as described previously (87). Electroporation of the plzA::Pl-Kan linear DNA (Fig. 2) into the wild-type competent cells yielded ∼10 kanamycin-resistant clones. PCR analysis confirmed that a 509 bp internal fragment of plzA is replaced by the 872-bp Pl-Kan cassette in all clones, confirming inactivation of plzA (Fig. 3, left panel). Characterization of several positive clones confirmed identical genotypes and phenotypes; one randomly chosen clone was used for subsequent analysis.
Fig. 3.
Confirmation of plzA inactivation, complementation in cis, and restoration of plzA transcript synthesis. (Left panel) PCR was used to confirm the integration of Pl-Kan within the bb0733 (plzA) gene. The locations of PCR primers are shown in Fig. 2. An 892-bp DNA containing the Pl-Kan cassette was inserted after deletion of a 509-bp fragment from the plzA gene. The size of PCR products for lanes containing plzA and the positive “control” DNA is 1,132 bp; the size for the wild type (WT) is 783 bp. A 100-bp ladder was used (Fermentas, Inc.). (Middle panel) Confirmation of the integration of PplzA-plzA-PflgB-aacC1 between genes bb0445 and bb0446. The PCR primers used to amplify a specific region are indicated at the top of the agarose gels. The sizes of all PCR products were amplified as expected. Controls in both gels were the plasmid DNAs that were used to inactivate plzA or complement the plzA mutant. A 1-kb DNA ladder (Fermentas, Inc.) was used in the middle panel. (Right panel) Real-time PCR detecting bb0733 mRNA from the indicated strains. The bar represents the mean + standard error of the mean (SEM) from 2 independent experiments. Average relative expression of bb0733 from each strain was calculated by the 2−ΔΔCT method. Enolase was used as an internal control (54, 87).
To restore the synthesis (and phenotype [see below]) of plzA at the wild-type level, we complemented the plzA mutant by in cis genomic integration where genes bb0445 and bb0446 converge (Fig. 2, bottom). The insertion of PplzA-plzA was downstream of both the bb0445 and bb0446 genes and less likely to affect their expression, as described previously (44, 99). We chose to complement the mutant in cis, since the shuttle vectors developed so far to complement B. burgdorferi mutants are multicopy plasmids (89) that result in overexpression of the target gene (43, 53, 77, 89). Accordingly, we introduced suicide vector pXLFbb0733 (Fig. 2, bottom) containing an intact plzA gene under its own promoter (PplzA-plzA) into the mutant cells. Positive clones were characterized by PCR by using kanamycin, gentamicin, and bb0445-bb0446 gene-specific primers (Fig. 3, middle panel). PCR analysis confirmed that PplzA-plzA integrated within the bb0445-bb0446 region, as expected (Fig. 2 and 3). To detect the expression of plzA in the wild-type, plzA mutant, and complemented plzA (plzA+) strains, we employed qPCR using gene-specific primers. Real-time PCR detected the plzA transcripts in wild-type and the complemented plzA+ cells but not in the plzA mutant cells (Fig. 3, right panel). Furthermore, the level of plzA transcripts synthesized in the complemented plzA+ cells was approximately 70% the level of wild-type cells, indicating the restoration of plzA synthesis in the complemented cells.
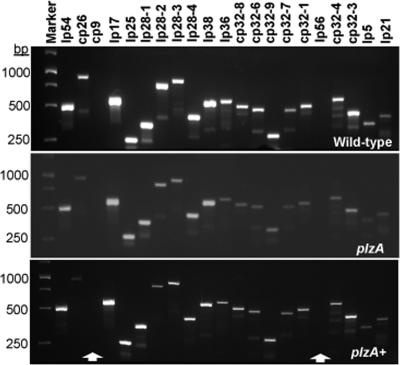
To confirm retention of B. burgdorferi endogenous plasmids required for infectivity (56, 61) in the plzA mutant and complemented strains, PCR-based plasmid profiling using each of the plasmid-specific primers was employed (61, 100). As shown in Fig. 4, the plasmid profiles of the wild-type, plzA mutant, and complemented plzA+ strains were the same, confirming retention of all plasmids required for infectivity (see below).
Fig. 4.
PCR analysis confirmed that the wild-type, plzA mutant, and the complemented plzA+ cells retained endogenous plasmids required for infection of mice and ticks. Twenty-one sets of PCR primers were used to detect 12 linear (lp) and 9 circular (cp) B. burgdorferi plasmids (12, 100). Arrows indicate the cp9 and lp56 plasmids were lost from all strains.
plzA mutant is deficient in swarming motility.
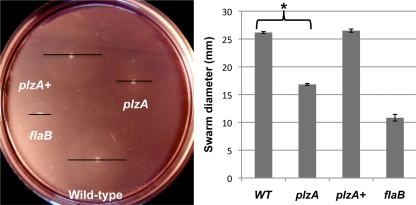
Mutation of a gene encoding PilZ has been reported to generate various phenotypes: rough, rugose, opaque, or translucent bacterial colony morphology and altered motility (2, 13, 27, 58, 60, 98). To determine if a mutation in plzA altered the B. burgdorferi swimming pattern or swarming motility (42, 53), the mutant cells were analyzed by microscopy and swarm plate motility assays in 0.35% agarose (52). Although cellular morphology and the swimming pattern (run, pause/flex, and reverse) (42, 53) of the plzA mutant cells were indistinguishable from those of the wild-type cells, the swarm diameter of plzA mutant cells was significantly reduced (P = 0.001) compared to that of the wild-type cells when the plates were incubated for 7 days (Fig. 5). Additionally, while the colony morphology of the wild-type cells was translucent, the mutant cells colonies were solid/opaque (which the figure was not clear enough to show). Both the swarming motility and colony morphology phenotypes were restored to wild type upon complementation (Fig. 5 and data not shown), indicating that the phenotypes associated with plzA cells are solely due to the mutation and not a secondary alteration elsewhere. Noticeable growth defects were not observed in plzA mutant cells, indicating the decreased-swarming phenotype was not due to a growth defect (not shown). These results indicate that plzA is essential for normal B. burgdorferi motility and colony morphology.
Fig. 5.
Swarm plate motility assays confirmed that the plzA mutant was defective in motility. A representative swarm assay with plzA mutant cells incubated for 7 days is shown. Since the swarm diameters were not clearly visible, a black bar was placed on each swarm, which represents the size of their swarm diameters (left panel). Swarm diameters from 4 assays were measured in millimeters ± standard deviations (right panel). A flagellinless, nonmotile flaB mutant was used as a control (51). *, significant difference (P = 0.001) between wild-type (WT) and plzA mutant cells.
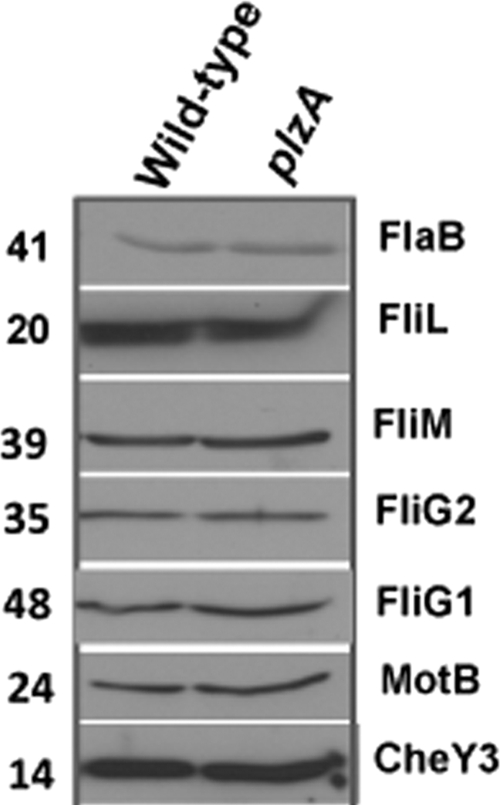
To determine if the deficient motility exhibited by plzA cells resulted from alteration of motility/chemotaxis protein synthesis, we performed Western blotting using specific antisera against the periplasmic flagellar protein FlaB; motor switch proteins FliM, FliG1, FliG2, and MotB; flagellar protein FliL; or the chemotaxis protein CheY3. As shown in Fig. 6, the levels of proteins detected by specific antisera in the plzA mutant cells were not altered compared with those of the wild-type cells. These results indicate that the reduced swarming observed in the plzA mutant cells was not due to a reduction in levels of FlaB, FliL, FligG1, FliG2, FliM, MotB, or CheY3 proteins.
Fig. 6.
The levels of prominent motility or chemotaxis proteins were not altered in the plzA mutant. Western blots using 5 μg of cell lysates from the indicated strains were probed with antisera against the indicated motility and chemotaxis proteins. Specific antisera against the proteins used are indicated on the right side of each blot. The values on the left are molecular sizes in kilodaltons.
plzA mutant cells are significantly attenuated in infecting mice.
In some bacteria, pilZ was shown to be required for the host colonization/disease processes (47, 60). In order to evaluate the infection potential of the plzA mutant, groups of C3H/HeN mice were challenged with 10-fold-increasing doses of wild-type, plzA::Pl-Kan, or isogenic complemented plzA+ cells to determine the 50% infectious dose (ID50). Two weeks postinoculation, the mice were bled and their sera were assessed for reactivity with B. burgdorferi antigen membrane protein A, also known as P39 (35, 80). Furthermore, to confirm the serology results, mice were sacrificed 6 weeks postinoculation, and spirochetes were reisolated from ear, bladder, and joint tissues. Serology results indicated that plzA mutant cells were attenuated in establishing an infection in mice (data not shown) and correlated well with reisolation of spirochetes from the tissues examined (Table 1). Whereas the plzA+ cells did not show an increase in the ID50 compared to the parental wild-type cells, the ID50 for the plzA mutant was reproducibly more than 1 logarithm higher than that of the wild type (or complemented plzA+ strain), indicating a significant attenuation in virulence (P = 0.0031) (Table 1).
Table 1.
plzA mutant cells are significantly attenuated in C3H/HeN mouse infection
| Type of strain | No. of mice infected/total at dose of: |
ID50 | ||||
|---|---|---|---|---|---|---|
| 5 × 102 | 5 × 103 | 5 × 104 | 5 × 105 | 5 × 106 | ||
| Wild type | 0/8 | 3/8 | 10/10 | 8 × 103 | ||
| plzA mutant | 0/8 | 1/8 | 6/12 | 6/10 | 10/10 | 9 × 104 |
| Complemented plzA+ | 1/8 | 4/8 | 10/10 | 4 × 103 | ||
The B. burgdorferi plzA mutant strain is unable to complete the mouse-tick-mouse infection cycle.
Because infection of and survival within the mammalian host represent only one facet of the B. burgdorferi enzootic life cycle, a more comprehensive evaluation of the behavior of these mutants in the tick vector was warranted. Naïve Ixodes scapularis larval ticks were allowed to feed on mice that were infected with mutant plzA (dose, 5 × 106/mouse), wild-type, or complemented strains (dose, 5 × 104/mouse). These doses were chosen to infect 100% of mice with wild-type, plzA, or complemented plzA+ cells (Table 1). Nine days after feeding on infected mice, a subsample of ticks were analyzed by immunofluorescence assays (IFAs) to determine the percentage of ticks infected with the different cells (Fig. 7 and data not shown). Spirochete loads in those fed ticks were determined by qPCR as well as by crushing ticks and plating on B. burgdorferi growth plates to determine the number of viable CFU per tick (Table 2). The majority of the fed larvae were allowed to molt to nymphs. As shown in Table 2, while almost 100% of naïve ticks that fed on wild-type- or the complemented plzA+ strain-infected mice became infected, only 55% of the ticks that fed on plzA mutant-infected mice were positive. Furthermore, the spirochete burden in the plzA mutant-infected ticks was >150-fold lower than that in the wild-type or isogenic complemented strains (Table 2). These results indicate that plzA mutant cells are deficient in ability to survive in ticks. Moreover, when the plzA strain-infected nymphs were fed on naïve mice, none of the mice became infected, whereas all naïve mice were infected with nymphs containing wild-type or the complemented plzA+ cells (Table 2). The failure of the plzA ticks to transmit the infection to naïve mice was not due to the number of nymphal ticks allowed to feed on mice. Since only 55% of plzA mutant ticks were positive (versus ∼100% for the wild-type or the complemented plzA+ strain), we allowed 30 plzA mutant nymphs to feed on a naïve mouse versus only 10 nymphs for the wild-type or complemented plzA+ cells (Table 2). Together, these results clearly demonstrate that B. burgdorferi PilZ protein PlzA appears essential to complete the mouse-tick-mouse infection cycle.
Fig. 7.
Immunofluorescence assays (IFAs) of immersed larval ticks 7 days postfeeding. Spirochetes were detected in the wild-type, plzA mutant, and complemented plzA+ strains; however, this is the only tick in which several B. burgdorferi plzA mutant cells were detected. In nearly all ticks infected with plzA mutant spirochetes by immersion, no spirochetes were detected by IFA 7 days postfeeding (n = 15).
Table 2.
Inability of plzA mutant cells to complete mouse-tick-mouse infection cycle
| Type of strain | Dose | % of: |
No. of viable spirochetes/tick | No. of nymphs/mouse | No. of mice infected/total | |
|---|---|---|---|---|---|---|
| Mice infected | Ticks positive | |||||
| Wild type | 5 × 104 | 100 | 95 | 3,947 ± 725 | 10 | 3/3 |
| plzA mutant | 5 × 106 | 100 | 55 | 20a | 30 | 0/5 |
| Complemented plzA+ | 5 × 104 | 100 | 100 | 3,006 ± 537 | 10 | 5/5 |
plzA-infected ticks had ≤20 spirochetes/tick (n = 10).
plzA mutant cells are deficient in surviving in artificially infected ticks.
To better determine why plzA cells in infected mice are deficient in the ability to transmit the infection to naïve ticks, we artificially infected naïve larval ticks with wild-type, plzA mutant, and complemented plzA+ cells by immersion (Table 3). Tick immersion studies (83, 85, 87) allow for direct artificial tick infection and serve two purposes: (i) to optimally infect naïve ticks with the wild-type and mutant spirochetes and determine their colonization and survivability within the tick vector and (ii) to examine the spirochete's potential to migrate from the arthropod host vector to the mammalian host. While ticks inoculated with wild-type or plzA+ cells were readily transmitted to and infected naïve mice, the plzA strain-infected ticks failed to transmit the infection to naïve mice (n = 2) (Table 3). We found that the plzA cells were severely defective in surviving in ticks that fed on the naïve mouse, although the tick's infection rates were similar (80%) between the wild-type and mutant cells (Table 3). The number of viable plzA spirochetes was very low compared to the number of viable wild-type or complemented plzA+ spirochetes (Table 3). These results indicated that a mutation in plzA resulted in cells that are deficient in surviving in fed ticks and, consequently, failed to transmit the infection from infected ticks to naïve mice (Tables 2 and 3).
Table 3.
Defective ability of plzA mutant to survive in ticks and to be transmitted to naïve mice based on tick immersion studies
| Type of strain | % of fed ticks positive for B. burgdorferi | No. of spirochetes/tick | No. of mice infected/total |
|---|---|---|---|
| Wild type | 80 | 3,100 ± 1,125 | 2/2 |
| plzA mutant | 80 | 330 ± 183 | 0/2 |
| Complemented plzA+ | 41 | 5,600 ± 4,869 | 2/2 |
DISCUSSION
c-di-GMP controls diverse cellular processes, including the motility and virulence of some bacteria (47, 60, 96). B. burgdorferi is an ideal organism with which to study c-di-GMP effects, in that there is no repetition of the genes that synthesize or hydrolyze c-di-GMP. The B. burgdorferi genome encodes a single protein for the GGDEF, EAL, or HD-GYP domains (22, 66, 71, 87); however, relatively little is known about how these proteins function mechanistically in different cellular process. Recently, we have shown that the EAL domain-containing phosphodiesterase BB0363 plays a role in motility and virulence of B. burgdorferi (87).
The c-di-GMP-binding PilZ domain proteins are important for mediating c-di-GMP signaling that controls bacterial motility and colony morphology and affects virulence. PlzA/BB0733 is the only recognizable PilZ domain-containing protein in B. burgdorferi (2), and recently it has been shown to specifically bind to c-di-GMP (21). The Kd value we determined for B. burgdorferi PilZ was 1.25 μM, which is consistent with the Kd values of other c-di-GMP-binding PilZ proteins (29, 37) and indicates strong affinity for c-di-GMP (Fig. 1). Cellular c-di-GMP concentrations, together with the effector components' affinities for c-di-GMP, are crucial for triggering many c-di-GMP-dependent outputs (29).
Analysis of the B. burgdorferi genome as well as our RT-PCR results indicates that plzA is transcribed monocistronically and is separated from divergently transcribed upstream and downstream genes by 309 and 205 bp, respectively. Although genetic manipulations in B. burgdorferi virulent, low-passage strains are challenging and often result in failure (33, 66, 68), we constructed a plzA mutant from the virulent wild-type strain B31-A3-68 Δbbe002 that retained all of the endogenous plasmids (Fig. 4) required for mouse and tick infections (Tables 1 to 3) (63). pilZ mutants of different bacteria exhibit different and diverse phenotypes, including but not limited to the translucent, “rdar” (rough, dry, and rugose colony morphology) morphotype and altered motility (60, 67, 70, 98). We discovered for the first time that the B. burgdorferi plzA mutant exhibited a opaque colony morphology consistent with other bacteria. The colony morphology is specific to the plzA mutation since complementation in cis resulted in restoration of the wild-type translucent colony morphology. Phase variation in other bacteria has been described to arise from programmed genetic rearrangements or variable expression patterns in an isogenic population (92). Molecular mechanisms underlying the translucent colony morphology exhibited by the B. burgdorferi plzA mutant are currently unknown and require further investigation. The plzA mutant cells' run-pause-reverse swimming pattern was indistinguishable from that of the wild-type cells (42, 53). However, the swarming motility on 0.35% agarose was markedly reduced compared to that of the isogenic complemented or the parental wild-type cells when plates were incubated for 7 days (Fig. 5). (B. burgdorferi cells do not swarm on 0.5% agarose [our unpublished results].) We note that when swarm plates were incubated for 5 days, the swarm diameters of the mutant strain were only slightly smaller than those of the wild-type cells; these results suggest that PilZ has a positive effect on B. burgdorferi motility.
In other species of bacteria, PilZ was reported to control motility in response to changes in the level of c-di-GMP (13, 58, 70). Specifically, in Caulobacter crescentus, the c-di-GMP-binding PilZ protein, DgrA, when complexed with c-di-GMP, diminished FliL synthesis, altering motility (13). However, this is likely not to be the case in B. burgdorferi: the levels of motility and chemotaxis proteins FliL, FliG1, FliG2, FliM, MotB, FlaB, or CheY3 were not altered in the plzA, pdeA, or plzA pdeA double mutant, as shown below (Fig. 6 and data not shown). Furthermore, in E. coli (and S. enterica), a mutation in the phosphodiesterase yhjH gene reduces motility, while a ycgR (encoding a PilZ protein) mutant exhibits normal motility. Furthermore, the motility defect of the yhjH mutant was rescued by deleting ycgR (24, 58, 70), indicating that YcgR mediated the reduction in motility due to elevated c-di-GMP (yhjH mutant). In this case, the PilZ protein YcgR controls motility posttranslationally: in response to elevated c-di-GMP, YcgR was reported to interact with motor switch proteins FliM, FliG, or MotA, interfering with the electrostatic interaction between the motor and switch protein controlling motility (3, 7, 17, 57). To determine if PlzA can function in response to c-di-GMP to regulate B. burgdorferi motility, we inactivated plzA in a phosphodiesterase (pdeA/bb0363) mutant background (87) where the cellular c-di-GMP level is 2× higher than that in the wild-type cells (our unpublished observation). We have demonstrated that bb0363 (pdeA) mutants exhibit reduced motility and that those mutant cells failed to reverse, whereas the wild-type cells ran, paused, and reversed (87). B. burgdorferi cells do not swim well in their BSK-II growth medium, and analysis of their swimming pattern requires highly viscous medium, such as methylcellulose (25, 43, 52, 87). The plzA mutant constructed in the pdeA mutant background (elevated c-di-GMP) did not rescue the pdeA cells' swimming behavior: the plzA pdeA double mutant cells' swimming pattern was indistinguishable from the pdeA cells' constant run-pause motility phenotype (87). These results suggest that PlzA regulates B. burgdorferi motility in a different manner than it does in E. coli and that elevated c-di-GMP in the B. burgdorferi pdeA mutant alters motility by a mechanism independent of PlzA. It is tempting to speculate that B. burgdorferi may possess an additional c-di-GMP-binding protein(s) that responds to elevated c-di-GMP in a manner similar to that reported in E. coli or C. crescentus to control motility (7, 13, 57, 87). Furthermore, our findings are consistent with the observations that PilZ proteins lacking the N-terminal YcgR domain are likely unable to interact with the flagellar switch FliG protein to control bacterial motility (17).
On the other hand, in some bacteria PilZ can act independent of c-di-GMP. For example, V. cholerae contains five PilZ domain proteins (60), but only PlzC and PlzD bind c-di-GMP (60). However, both plzB and plzC mutant cells were reported to exhibit decreased motility, and a plzD mutant displayed an increased swarming motility relative to wild-type V. cholerae (45, 60). In addition, a V. cholerae strain that produced elevated levels of c-di-GMP exhibited reduced motility due to diminished transcription of motility and chemotaxis genes (6, 60). However, as described above, the expression of B. burgdorferi motility or chemotaxis genes was not affected in pdeA, plzA, or pdeA plzA mutant cells.
While the motility (Fig. 5) of plzA mutant cells was attenuated relative to wild-type cells, the swimming pattern of the plzA cells was indistinguishable from that of the wild-type cells, and expression of motility and chemotaxis genes was also unaltered. These findings may suggest that assembly of the periplasmic flagella was not affected due to a mutation in plzA (96). We speculate that PlzA or a PlzA-regulated factor(s) may modulate the function of a motor protein, perhaps by modulating the MotB proton channel. A recent study with E. coli reported that amino acid residues K53 to T64 of MotB act as a plug to prevent proton flow before the MotA-MotB complex associates with the flagellar structure (31, 32). Because in plzA cells the swarming motility is attenuated but the swimming pattern was not altered, it appears likely that PlzA acts as a positive regulator in B. burgdorferi motility or chemotaxis.
Several studies have shown that a mutation in the gene encoding PilZ protein affects virulence of some bacteria. For example, a mutation in the plzD gene of V. cholerae results in cells that are 10-fold less infectious than the parental cells in a murine model (60). A mutation in the plant pathogen Xanthomonas campestris pv. campestris pilZ (xc3221) gene also resulted in cells that are significantly less virulent in the Chinese radish than their parental wild-type cells (47). Therefore, the >1-log-increased ID50 observed with the B. burgdorferi plzA mutant cells during mammalian infections is consistent with these other reports (Table 1). Although why plzA mutant cells are significantly (P = 0.0031) less infectious than the wild-type cells is unknown, we propose that motility is likely to be a factor and that wild-type motility is critical for B. burgdorferi infectivity (9, 43, 87; our unpublished observation). Future studies will test this hypothesis. However, we were surprised that the plzA cells are defective in transmitting the infection from infected mouse to naïve tick to mouse (Table 2). The defect in the transmission between tick and mammalian hosts was not due to the number of ticks allowed to feed on naïve mice (30 plzA mutant ticks versus 10 ticks per mouse for wild-type or plzA+ cells). Using mouse-tick-mouse infection cycle and tick-immersion studies (Tables 2 and 3), we determined that plzA cells were defective in surviving in fed ticks, although the mechanism is currently unknown (see below). Reduced motility is likely not responsible for the reduced survival of spirochetes in ticks since motility- or chemotaxis-defective mutants are able to survive and multiply before or after a blood meal from mice (87; our unpublished observation). We speculate that PlzA itself is required for survivability within ticks or PlzA is a regulator of a virulence determinant(s) impacting survivability in ticks and/or attenuation in mice. It is also possible that the opaque colony morphology of the plzA mutant cells may reflect an altered cell surface membrane structure, which may be related to the impaired ability of plzA cells to infect mice and/or decreased survivability within ticks. Tick saliva contains bioactive molecules and a plethora of proteolytic enzymes (19, 75). Whether tick-derived elements, host-derived products acquired from the tick's blood meal, or salivary gland factors (18, 19, 34, 75, 81, 82) were responsible for the reduced survival of the plzA mutant in the ticks remains to be determined. We and others have shown that B. burgdorferi uses different mechanisms to survive in different hosts (ticks/mammalian). Future studies are warranted to decipher the wide spectrum of PilZ effects on B. burgdorferi.
ACKNOWLEDGMENTS
We thank Rita Tamayo and Philip Stewart for comments on the manuscript. We thank R. Rego, P. Rosa, C. Li, J. Benach, A. Barbour, J. Carroll, M. Gomelsky, D. Blair, U. Pal, E. Fikrig, and S. Samuels for providing reagents. We appreciate technical discussions from P. Policastro and A. Bestor, RML, NIH.
The research was sponsored by a National Institute of Arthritis, Musculoskeletal, and Skin Diseases, NIH, grant (R03AR054582) and an award of an East Carolina University Research and Development “start-up” fund to M.A.M.
Footnotes
Published ahead of print on 28 February 2011.
REFERENCES
- 1. Alm R. A., Bodero A. J., Free P. D., Mattick J. S. 1996. Identification of a novel gene, pilZ, essential for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 178:46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amikam D., Galperin M. Y. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6 [DOI] [PubMed] [Google Scholar]
- 3. Armitage J. P., Berry R. M. 2010. Time for bacteria to slow down. Cell 141:24–26 [DOI] [PubMed] [Google Scholar]
- 4. Battisti J. M., et al. 2008. Outer surface protein A protects Lyme disease spirochetes from acquired host immunity in the tick vector. Infect. Immun. 76:5228–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Benach J., et al. 2007. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 26:5153–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beyhan S., Tischler A. D., Camilli A., Yildiz F. H. 2006. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J. Bacteriol. 188:3600–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boehm A., et al. 2010. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141:107–116 [DOI] [PubMed] [Google Scholar]
- 8. Bono J. L., et al. 2000. Efficient targeted mutagenesis in Borrelia burgdorferi. J. Bacteriol. 182:2445–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Botkin D. J., et al. 2006. Identification of potential virulence determinants by Himar1 transposition of infectious Borrelia burgdorferi B31. Infect. Immun. 74:6690–6699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brooks C. S., Hefty P. S., Jolliff S. E., Akins D. R. 2003. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect. Immun. 71:3371–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carbonnelle E., Helaine S., Prouvensier L., Nassif X., Pelicic V. 2005. Type IV pilus biogenesis in Neisseria meningitidis: PilW is involved in a step occurring after pilus assembly, essential for fibre stability and function. Mol. Microbiol. 55:54–64 [DOI] [PubMed] [Google Scholar]
- 12. Casjens S., et al. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490–516 [DOI] [PubMed] [Google Scholar]
- 13. Christen M., et al. 2007. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 104:4112–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cotter P. A., Stibitz S. 2007. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr. Opin. Microbiol. 10:17–23 [DOI] [PubMed] [Google Scholar]
- 15. Duerig A., et al. 2009. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 23:93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Elias A. F., et al. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 70:2139–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fang X., Gomelsky M. 2010. A posttranslational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol. Microbiol. 76:1295–1305 [DOI] [PubMed] [Google Scholar]
- 18. Fogaca A. C., et al. 1999. Antimicrobial activity of a bovine hemoglobin fragment in the tick Boophilus microplus. J. Biol. Chem. 274:25330–25334 [DOI] [PubMed] [Google Scholar]
- 19. Francischetti I. M., Sa-Nunes A., Mans B. J., Santos I. M., Ribeiro J. M. 2009. The role of saliva in tick feeding. Front. Biosci. 14:2051–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fraser C. M., et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586 [DOI] [PubMed] [Google Scholar]
- 21. Freedman J. C., et al. 2010. Identification and molecular characterization of a cyclic-di-GMP effector protein, PlzA (BB0733): additional evidence for the existence of a functional cyclic-di-GMP regulatory network in the Lyme disease spirochete, Borrelia burgdorferi. FEMS Immunol. Med. Microbiol. 58:285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Galperin M. Y., Nikolskaya A. N., Koonin E. V. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11–21 [DOI] [PubMed] [Google Scholar]
- 23. Ge Y., Li C., Corum L., Slaughter C. A., Charon N. W. 1998. Structure and expression of the FlaA periplasmic flagellar protein of Borrelia burgdorferi. J. Bacteriol. 180:2418–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Girgis H. S., Liu Y., Ryu W. S., Tavazoie S. 2007. A comprehensive genetic characterization of bacterial motility. PLoS Genet. 3:1644–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goldstein S. F., Charon N. W., Kreiling J. A. 1994. Borrelia burgdorferi swims with a planar waveform similar to that of eukaryotic flagella. Proc. Natl. Acad. Sci. U. S. A. 91:3433–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grimm D., Elias A. F., Tilly K., Rosa P. A. 2003. Plasmid stability during in vitro propagation of Borrelia burgdorferi assessed at a clonal level. Infect. Immun. 71:3138–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guzzo C. R., Salinas R. K., Andrade M. O., Farah C. S. 2009. PILZ protein structure and interactions with PILB and the FIMX EAL domain: implications for control of type IV pilus biogenesis. J. Mol. Biol. 393:848–866 [DOI] [PubMed] [Google Scholar]
- 28. Heidelberg J. F., et al. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118–1123 [DOI] [PubMed] [Google Scholar]
- 29. Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263–273 [DOI] [PubMed] [Google Scholar]
- 30. Hickman J. W., Harwood C. S. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69:376–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hosking E. R., Manson M. D. 2008. Clusters of charged residues at the C terminus of MotA and N terminus of MotB are important for function of the Escherichia coli flagellar motor. J. Bacteriol. 190:5517–5521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hosking E. R., Vogt C., Bakker E. P., Manson M. D. 2006. The Escherichia coli MotAB proton channel unplugged. J. Mol. Biol. 364:921–937 [DOI] [PubMed] [Google Scholar]
- 33. Hyde J. A., Shaw D. K., Smith R., III, Trzeciakowski J. P., Skare J. T. 2010. Characterization of a conditional bosR mutant in Borrelia burgdorferi. Infect. Immun. 78:265–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hynes W. L., Ceraul S. M., Todd S. M., Seguin K. C., Sonenshine D. E. 2005. A defensin-like gene expressed in the black-legged tick, Ixodes scapularis. Med. Vet. Entomol. 19:339–344 [DOI] [PubMed] [Google Scholar]
- 35. Jewett M. W., et al. 2007. The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi. Mol. Microbiol. 64:1358–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kawabata H., Norris S. J., Watanabe H. 2004. BBE02 disruption mutants of Borrelia burgdorferi B31 have a highly transformable, infectious phenotype. Infect. Immun. 72:7147–7154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ko J., et al. 2010. Structure of PP4397 reveals the molecular basis for different c-di-GMP binding modes by Pilz domain proteins. J. Mol. Biol. 398:97–110 [DOI] [PubMed] [Google Scholar]
- 38. Kulasakara H., et al. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. U. S. A. 103:2839–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kumar M., Chatterji D. 2008. Cyclic di-GMP: a second messenger required for long-term survival, but not for biofilm formation, in Mycobacterium smegmatis. Microbiology 154:2942–2955 [DOI] [PubMed] [Google Scholar]
- 40. Leduc J. L., Roberts G. P. 2009. Cyclic di-GMP allosterically inhibits the CRP-like protein (Clp) of Xanthomonas axonopodis pv. citri. J. Bacteriol. 191:7121–7122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee V. T., et al. 2007. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 65:1474–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li C., et al. 2002. Asymmetrical flagellar rotation in Borrelia burgdorferi nonchemotactic mutants. Proc. Natl. Acad. Sci. U. S. A. 99:6169–6174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li C., Xu H., Zhang K., Liang F. T. 2010. Inactivation of a putative flagellar motor switch protein FliG1 prevents Borrelia burgdorferi from swimming in highly viscous media and blocks its infectivity. Mol. Microbiol. 75:1563–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li X., et al. 2007. The Lyme disease agent Borrelia burgdorferi requires BB0690, a Dps homologue, to persist within ticks. Mol. Microbiol. 63:694–710 [DOI] [PubMed] [Google Scholar]
- 45. Liu X., Beyhan S., Lim B., Linington R. G., Yildiz F. H. 2010. Identification and characterization of a phosphodiesterase that inversely regulates motility and biofilm formation in Vibrio cholerae. J. Bacteriol. 192:4541–4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 47. McCarthy Y., et al. 2008. The role of PilZ domain proteins in the virulence of Xanthomonas campestris pv. campestris. Mol. Plant Pathol. 9:819–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Merighi M., Lee V. T., Hyodo M., Hayakawa Y., Lory S. 2007. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65:876–895 [DOI] [PubMed] [Google Scholar]
- 49. Methe B. A., et al. 2005. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc. Natl. Acad. Sci. U. S. A. 102:10913–10918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Monds R. D., Newell P. D., Gross R. H., O'Toole G. A. 2007. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 63:656–679 [DOI] [PubMed] [Google Scholar]
- 51. Motaleb M. A., et al. 2000. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc. Natl. Acad. Sci. U. S. A. 97:10899–10904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Motaleb M. A., Miller M. R., Bakker R. G., Li C., Charon N. W. 2007. Isolation and characterization of chemotaxis mutants of the Lyme disease spirochete Borrelia burgdorferi using allelic exchange mutagenesis, flow cytometry, and cell tracking. Methods Enzymol. 422:421–437 [DOI] [PubMed] [Google Scholar]
- 53. Motaleb M. A., et al. 2005. CheX is a phosphorylated CheY phosphatase essential for Borrelia burgdorferi chemotaxis. J. Bacteriol. 187:7963–7969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Motaleb M. A., Sal M. S., Charon N. W. 2004. The decrease in FlaA observed in a flaB mutant of Borrelia burgdorferi occurs posttranscriptionally. J. Bacteriol. 186:3703–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ojaimi C., et al. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 71:1689–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ouyang Z., et al. 2009. BosR (BB0647) governs virulence expression in Borrelia burgdorferi. Mol. Microbiol. 74:1331–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Paul K., Nieto V., Carlquist W. C., Blair D. F., Harshey R. M. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol. Cell 38:128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pesavento C., et al. 2008. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 22:2434–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Policastro P. F., Schwan T. G. 2003. Experimental infection of Ixodes scapularis larvae (Acari: Ixodidae) by immersion in low passage cultures of Borrelia burgdorferi. J. Med. Entomol. 40:364–370 [DOI] [PubMed] [Google Scholar]
- 60. Pratt J. T., Tamayo R., Tischler A. D., Camilli A. 2007. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 282:12860–12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Purser J. E., Norris S. J. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc. Natl. Acad. Sci. U. S. A. 97:13865–13870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Reed L. J., Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493–497 [Google Scholar]
- 63. Rego R. O., Bestor A., Rosa P. A. 2011. Defining the plasmid-encoded restriction-modification systems of the Lyme disease spirochete Borrelia burgdorferi. J. Bacteriol. 193:1161–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Revel A. T., et al. 2005. bptA (bbe16) is essential for the persistence of the Lyme disease spirochete, Borrelia burgdorferi, in its natural tick vector. Proc. Natl. Acad. Sci. U. S. A. 102:6972–6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Revel A. T., Talaat A. M., Norgard M. V. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. U. S. A. 99:1562–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rogers E. A., et al. 2009. Rrp1, a cyclic-di-GMP-producing response regulator, is an important regulator of Borrelia burgdorferi core cellular functions. Mol. Microbiol. 71:1551–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Romling U. 2005. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell. Mol. Life Sci. 62:1234–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rosa P. A., Tilly K., Stewart P. E. 2005. The burgeoning molecular genetics of the Lyme disease spirochaete. Nat. Rev. Microbiol. 3:129–143 [DOI] [PubMed] [Google Scholar]
- 69. Ross P., et al. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279–281 [DOI] [PubMed] [Google Scholar]
- 70. Ryjenkov D. A., Simm R., Romling U., Gomelsky M. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 281:30310–30314 [DOI] [PubMed] [Google Scholar]
- 71. Ryjenkov D. A., Tarutina M., Moskvin O. V., Gomelsky M. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187:1792–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sal M. S., et al. 2008. Borrelia burgdorferi uniquely regulates its motility genes and has an intricate flagellar hook-basal body structure. J. Bacteriol. 190:1912–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Samuels D. S., Mach K. E., Garon C. F. 1994. Genetic transformation of the Lyme disease agent Borrelia burgdorferi with coumarin-resistant gyrB. J. Bacteriol. 176:6045–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Samuels D. S., Radolf J. D. 2009. Who is the BosR around here anyway? Mol. Microbiol. 74:1295–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sauer J. R., McSwain J. L., Bowman A. S., Essenberg R. C. 1995. Tick salivary gland physiology. Annu. Rev. Entomol. 40:245–267 [DOI] [PubMed] [Google Scholar]
- 76. Schirmer T., Jenal U. 2009. Structural and mechanistic determinants of c-di-GMP signalling. Nat. Rev. Microbiol. 7:724–735 [DOI] [PubMed] [Google Scholar]
- 77. Seshu J., et al. 2006. Inactivation of the fibronectin-binding adhesin gene bbk32 significantly attenuates the infectivity potential of Borrelia burgdorferi. Mol. Microbiol. 59:1591–1601 [DOI] [PubMed] [Google Scholar]
- 78. Simm R., Morr M., Remminghorst U., Andersson M., Romling U. 2009. Quantitative determination of cyclic diguanosine monophosphate concentrations in nucleotide extracts of bacteria by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry. Anal. Biochem. 386:53–58 [DOI] [PubMed] [Google Scholar]
- 79. Simm R., Remminghorst U., Ahmad I., Zakikhany K., Romling U. 2009. A role for the EAL-like protein STM1344 in regulation of CsgD expression and motility in Salmonella enterica serovar Typhimurium. J. Bacteriol. 191:3928–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Simpson W. J., Burgdorfer W., Schrumpf M. E., Karstens R. H., Schwan T. G. 1991. Antibody to a 39-kilodalton Borrelia burgdorferi antigen (P39) as a marker for infection in experimentally and naturally inoculated animals. J. Clin. Microbiol. 29:236–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sonenshine D. E., Ceraul S. M., Hynes W. E., Macaluso K. R., Azad A. F. 2002. Expression of defensin-like peptides in tick hemolymph and midgut in response to challenge with Borrelia burgdorferi, Escherichia coli and Bacillus subtilis. Exp. Appl. Acarol. 28:127–134 [DOI] [PubMed] [Google Scholar]
- 82. Sonenshine D. E., Hynes W. L. 2008. Molecular characterization and related aspects of the innate immune response in ticks. Front. Biosci. 13:7046–7063 [DOI] [PubMed] [Google Scholar]
- 83. Stewart P. E., Bestor A., Cullen J. N., Rosa P. A. 2008. A tightly regulated surface protein of Borrelia burgdorferi is not essential to the mouse-tick infectious cycle. Infect. Immun. 76:1970–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Stock A. M., Robinson V. L., Goudreau P. N. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183–215 [DOI] [PubMed] [Google Scholar]
- 85. Strother K. O., de Silva A. 2005. Role of Borrelia burgdorferi linear plasmid 25 in infection of Ixodes scapularis ticks. J. Bacteriol. 187:5776–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sudarsan N., et al. 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321:411–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sultan S. Z., Pitzer J. E., Miller M. R., Motaleb M. A. 2010. Analysis of a Borrelia burgdorferi phosphodiesterase demonstrates a role for cyclic-di-guanosine monophosphate in motility and virulence. Mol. Microbiol. 77:128–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tamayo R., Pratt J. T., Camilli A. 2007. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 61:131–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Tilly K., et al. 2006. Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect. Immun. 74:3554–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Tilly K., Rosa P. A., Stewart P. E. 2008. Biology of infection with Borrelia burgdorferi. Infect. Dis. Clin. North Am. 22:217–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Tokarz R., Anderton J. M., Katona L. I., Benach J. L. 2004. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect. Immun. 72:5419–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. van der Woude M. W., Baumler A. J. 2004. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17:581–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Weber H., Pesavento C., Possling A., Tischendorf G., Hengge R. 2006. Cyclic-di-GMP-mediated signalling within the sigma network of Escherichia coli. Mol. Microbiol. 62:1014–1034 [DOI] [PubMed] [Google Scholar]
- 94. Weinhouse H., et al. 1997. c-di-GMP-binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. FEBS Lett. 416:207–211 [DOI] [PubMed] [Google Scholar]
- 95. West A. H., Stock A. M. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369–376 [DOI] [PubMed] [Google Scholar]
- 96. Wolfe A. J., Visick K. L. 2008. Get the message out: cyclic-Di-GMP regulates multiple levels of flagellum-based motility. J. Bacteriol. 190:463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Yang X. F., Pal U., Alani S. M., Fikrig E., Norgard M. V. 2004. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J. Exp. Med. 199:641–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yildiz F. H., Visick K. L. 2009. Vibrio biofilms: so much the same yet so different. Trends Microbiol. 17:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhang X., Yang X., Kumar M., Pal U. 2009. BB0323 function is essential for Borrelia burgdorferi virulence and persistence through tick-rodent transmission cycle. J. Infect. Dis. 200:1318–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhou X., Miller M. R., Motaleb M., Charon N. W., He P. 2008. Spent culture medium from virulent Borrelia burgdorferi increases permeability of individually perfused microvessels of rat mesentery. PLoS One 3:e4101. [DOI] [PMC free article] [PubMed] [Google Scholar]