Abstract
BRENDA (BRaunschweig ENzyme DAtabase) represents a comprehensive collection of enzyme and metabolic information, based on primary literature. The database contains data from at least 83 000 different enzymes from 9800 different organisms, classified in ∼4200 EC numbers. BRENDA includes biochemical and molecular information on classification and nomenclature, reaction and specificity, functional parameters, occurrence, enzyme structure, application, engineering, stability, disease, isolation and preparation, links and literature references. The data are extracted and evaluated from ∼46 000 references, which are linked to PubMed as long as the reference is cited in PubMed. In the past year BRENDA has undergone major changes including a large increase in updating speed with >50% of all data updated in 2002 or in the first half of 2003, the development of a new EC-tree browser, a taxonomy-tree browser, a chemical substructure search engine for ligand structure, the development of controlled vocabulary, an ontology for some information fields and a thesaurus for ligand names. The database is accessible free of charge to the academic community at http://www.brenda.uni-koeln.de.
INTRODUCTION
The development of BRENDA was begun in 1987 at the German National Research Center for Biotechnology (GBF) and continues at the Cologne University Bioinformatics Centre. Initially, BRENDA was published as a series of books (1). Since 1998 all data have been presented in a relational database system with access free to the academic community at http://www.brenda.uni-koeln.de. Commercial users are required to purchase a licence.
Enzymes represent the largest and most diverse group of all proteins, catalysing all chemical reactions in the metabolism of all organisms. They play a key role in the regulation of metabolic steps within the cell. With the development and progress of projects of structural and functional genomics and metabolomics, the systematic collection, accessibility and processing of enzyme data becomes even more important in order to analyse and understand biological processes.
BRENDA, a protein function database (2) contains a huge amount of enzymic and metabolic data and is updated and evaluated by extracting information from the primary literature. Since 2002 the annotation speed has been tripled to 1000 EC numbers per year.
Major developments in the past few years were the ongoing conversion from an EC number/organism-specific to a protein-molecule-specific database. Furthermore, the presentation and the advanced search engine via the world wide web was improved. Tools like an EC browser, the taxonomy browser and a sequence-based search engine were integrated. BRENDA now provides the opportunity to search for substructures of ligands and a thesaurus of those chemical compounds that are involved in enzyme reactions. In terms of systematic access and analysis of data, a controlled vocabulary for organism-specific information, i.e. intracellular localization and enzyme source, was established.
CONTENT OF BRENDA
BRENDA represents a comprehensive relational database containing all enzymes classified according to the EC system of the Enzyme Nomenclature Committee (IUBMB) (3). This classification is based on the type of reaction (e.g. oxidation, reduction, hydrolysis, group transfer) catalysed by the enzyme.
In contrast to other databases, BRENDA is not limited to a specific aspect of the enzyme or to a specific organism. It covers organism-specific information on functional and molecular properties, enzyme names, catalysed reaction, occurrence, sequence, kinetics, substrates/products, inhibitors, cofactors, activators, structure and stability.
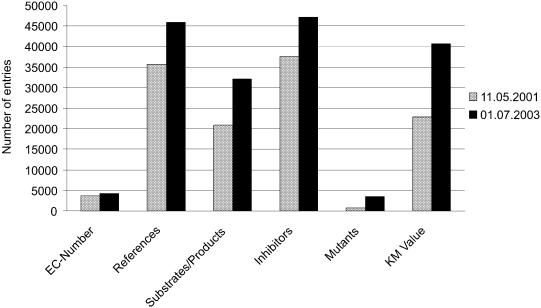
Presently, BRENDA holds information on 4200 EC numbers, which represent more than 83 000 different enzyme molecules. The data in BRENDA are continuously updated by manual extraction of relevant parameters from publications searched in the literature databases, i.e. PubMed (4) and SciFinder (5), and all entries are checked for internal inconsistencies. In addition, automated literature extraction is being developed to provide an almost complete overview of the literature. New information fields are continuously added as necessary. Figure 1 shows the quantitative increase of entries since 2001 in BRENDA exemplified by some significant information fields (EC number, literature references, substrates/products, inhibitors, mutants, Km value).
Figure 1.
Development of BRENDA data content 2001–2003 in some significant data fields.
SEARCH FEATURES
BRENDA is stored as a relational database, containing all data in 46 tables, enabling different queries.
The quick search mode provides easy access to the data of each information field individually. The search results are displayed in a comprehensive table format and also as a compact printable version. The table version includes links to a reference-specific view, pictures of molecules functioning as ligands, Gene Ontology (GO) definitions (6), PDB entries (7), amino acid sequences (8) and cross-references to other databases.
The advanced search mode allows one to combine 25 different query criteria. In addition to the possibility of restricting the query to a unique organism, the search can be extended to an upper level of a branch of the BRENDA taxonomy tree (TaxTree).
The BRENDA TaxTree is based on the tree published by the NCBI (4). The TaxTree search allows users to browse and search for organism names or nodes in the NCBI taxonomy browser. In addition, a search can be performed on organisms that are stored only in BRENDA, but do not occur in any sequence database. Special features show the availability of data for a specific organism class or organism in BRENDA.
The ECTree in BRENDA displays the enzyme classification as defined by the IUBMB (3).
SOURCE/TISSUE
The BRENDA team has created a hierarchical ontology for enzyme sources or tissue. The enzymes of multicellular organisms are either found in all parts of the body or are restricted to a single tissue. For practical purposes however, they are frequently isolated from cultured cells which are derived from normal or cancerous tissues. The Source/Tissue field of BRENDA comprises terms of tissues, cell lines and cell types from uni- and multicellular organisms. BRENDA has developed its own ontology corresponding to the rules and formats of the GO Consortium (6), thus providing the first (enzyme source) ontology for all organisms. The tissue tree is divided into four areas: animal, plant, fungi and other source, which are each separated into subtrees. The whole body of an animal, for example, is divided into 18 subtrees, i.e. the skeletal, hematopoietic and visceral systems. Each different cell type, cancer cell type or cell line is assigned to the tissue from which it has developed or to which it is related. The BRENDA Tissue Ontology (BTO) numbers are unique for each term. Most of the terms have definitions and synonyms included and different relationships are defined between terms, like ‘part of’, ‘develops from’, ‘is a’ or simply ‘is related to’.
LOCALIZATION
The data field localization contains the part of the cell where the enzyme is located. BRENDA now has a new controlled vocabulary in this data field. In cooperation with the GO Consortium (6), the BRENDA team is developing a common shared vocabulary for the Localization terms. The cellular component terms of GO are arranged in a concise ontology and the vocabulary of BRENDA is now consistent with these terms. It includes the GO-numbers which are unique for each term and a link leading to the AmiGO tree view of the term of interest.
LIGANDS
Another essential part of BRENDA is the information on ligands, which function as natural or in vitro substrates/products, inhibitors, activating compounds, cofactors, bound metals, etc. Now ∼500 000 enzyme–ligand relationships are stored with more than 46 000 different chemical compounds functioning as ligands.
BRENDA-LIGAND THESAURUS
In addition to the systematic name, which accurately describes a chemical compound, trivial names, abbreviations and synonyms are widely used. BRENDA-LIGAND now provides a search for all included synonyms of a given compound and, thus facilitates finding all enzyme–ligand-related information. This thesaurus is based on the generation of unique and chiral SMILES strings (9,10) for ligand structures in the database.
BRENDA-LIGAND SUBSTRUCTURE SEARCH
As most of the ligand structures have now been entered, chemical substructure searches are possible. BRENDA provides subgraph matching searches for most of the small molecules functioning as ligands, using a molecular editor (11).
In order to achieve rapidity and high precision, the search consists of two steps. The first step, used as a prefilter, is the very fast fingerprint scan (12) of all structures in the database. In the second step all found structures are tested for subgraph matching by a module for Maximum Common Substructure searches (12).
The substructure to search can be transferred from any ligand search or can be uploaded in several formats and edited by a structure sketch tool.
SUMMARY
BRENDA is a literature-based information system of functional enzyme data and metabolism. It provides various search modes for overall or organism-specific queries and now includes a tool for substructure searches of ligands. Additional new features are searches within the TaxTree and the inclusion of a controlled vocabulary for subcellular localization.
REFERENCES
- 1.Schomburg D. and Schomburg,I. (2001) Springer Handbook of Enzymes. 2nd edn. Springer, Heidelberg, Germany. [Google Scholar]
- 2.Schomburg I., Chang,A. and Schomburg,D. (2002) BRENDA, enzyme data and metabolic information. Nucleic Acids Res., 30, 47–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Webb E.C., NC-IUBMB. (1992) Enzyme Nomenclature: Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes. Academic Press, New York, NY. [Google Scholar]
- 4.Wheeler D.L., Church,D.M., Lash,A.E., Leipe,D.D., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Tatusova,T.A., Wagner,L. et al. (2001) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 29, 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ridley D.D. (2002) SciFinder and SciFinder Scholar. J. Wiley & Sons, New York, NY. [Google Scholar]
- 6.Ashburner C.A., Ball,J.A., Blake, D., Botstein,H., Butler,J.M., Cherry,A.P., Davis, K., Dolinski,S.S., Dwight,J.T., Eppig,M.A. et al. (2000) Gene Ontology: tool for the unification of biology. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berman H.M., Westbrook,J., Feng,Z., Gillilan,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O’Donovan,C., Phan,I. et al. (2003) The Swiss-Prot protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weininger D. (1988) SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci., 28, 31–36. [Google Scholar]
- 10.Weininger D., Weininger,A. and Weininger,J. (1989) SMILES. 2. Algorithm for generation of unique SMILES notation. J. Chem. Inf. Comput. Sci., 29, 97–101. [Google Scholar]
- 11.Csizmadia P. (2000) MarvinSketch and MarvinView: molecule applets for the World Wide Web. In Pombo-Villar,E., Neier,R. and Lin,S.K. (eds), Proceedings of ECSOC-3, The Third International Electronic Conference on Synthetic Organic Chemistry, September 1–30, 1999. MDPI, Basel, pp. 367–369. http://www.mdpi.org/ecsoc-3.htm. [Google Scholar]
- 12.Steinbeck C., Han,Y., Kuhn,S., Horlacher,O., Luttmann,E. and Willighagen,E. (2003) The Chemistry Development Kit (CDK): An open-source java library for chemo- and bioinformatics. J. Chem. Inf. Comput. Sci., 43, 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]