Abstract
Malaria caused by Plasmodium falciparum is a major cause of global infant mortality, and no effective vaccine currently exists. Multiple potential vaccine targets have been identified, and immunoepidemiology studies have played a major part in assessing those candidates. When such studies are carried out in high-transmission settings, individuals are often superinfected with complex mixtures of genetically distinct P. falciparum types, making it impossible to directly correlate the genotype of the infecting antigen with the antibody response. In contrast, in regions of low transmission P. falciparum infections are often genetically simple, and direct comparison of infecting genotype and antigen-specific immune responses is possible. As a test of the utility of this approach, responses against several domains and allelic variants of the vaccine candidate P. falciparum merozoite surface protein 3 (PfMSP3) were tested in serum samples collected near Iquitos, Peru. Antibodies recognizing both the conserved C-terminal and the more variable N-terminal domain were identified, but anti-N-terminal responses were more prevalent, of higher titers, and primarily of cytophilic subclasses. Comparing antibody responses to different PfMSP3 variants with the PfMSP3 genotype present at the time of infection showed that anti-N-terminal responses were largely allele class specific, but there was some evidence for responses that cross-reacted across allele classes. Evidence for cross-reactive responses was much stronger when variants within one allele class were tested, which has implications for the rational development of genotype-transcending PfMSP3-based vaccines.
INTRODUCTION
The effort to develop a vaccine targeting blood stage Plasmodium falciparum parasites, which are responsible for virtually all malaria-related deaths worldwide, has been notably impacted by two recent phase IIb trials which did not result in detectable protection (21, 27). While disappointing, these results have had the beneficial impact of triggering extensive discussion of how vaccine candidates are selected and what data are necessary to rationally advance them along the vaccine development pipeline (3, 7, 10). These analyses clearly identify genetic diversity as one of the most significant problems in P. falciparum vaccine development. Blood stage vaccine candidates are of particular concern on this score, as they are exposed to the adaptive immune system, a strong selective pressure which can drive genetic diversity (36). Indeed, many blood stage antigens appear to be under balancing selective pressure, suggesting that immune responses to them are largely allele specific and that multiple allelic variants cocirculate within a given parasite population (18, 38).
Immunoepidemiology studies have been an extremely useful tool in the malaria vaccine development process. However, as the vaccine development process moves forward, there is an urgent need for these studies to tackle the question of genetic diversity and allele-specific immune responses head on. Allele-specific responses are frequently detected using antibody depletion experiments, where antibodies that recognize one antigen are depleted from a serum sample by multiple incubations with that antigen before the presence of antibodies that recognize a different antigen variant is detected. Such studies have been extremely successful, with important implications for vaccine candidates such as AMA1 (23, 24), MSP1 (14, 34), and MSP3 (4, 22, 25). However, they are not always possible in all studies because of sample volume limitations, particularly when multiple different variants of each antigen are used in separate competition experiments on the same serum sample.
An alternative and simple method to detect allele-specific immune responses would be to directly compare the immune response to multiple antigen genotypes with that of the antigen genotype present in the P. falciparum infection from which the sample has been taken. If an individual serum sample contained antibodies that recognized only the infecting allele type, this would be a strong argument for allele specificity. Such an approach is clearly difficult in hyperendemic transmission environments, where individuals are routinely infected with multiple overlapping P. falciparum genotypes. In contrast, in hypoendemic environments, where P. falciparum infections are genetically simple and often spaced by several months, the responses against both the infecting and noninfecting genotypes could be reasonably compared, and a direct correlation between genetic variation and the immune response could be inferred. In the context of a longitudinal study, where the infection history of each individual is known for an extended period of time and hence the length of time since they had been exposed to other allelic types is established, that comparison would be even more powerful.
To test the validity of this approach, we used samples from a longitudinal epidemiological cohort near Iquitos, Peru (1), and investigated responses against P. falciparum merozoite surface protein 3 (PfMSP3). PfMSP3 is encoded by one member of a multigene family (30), is expressed on the surface of merozoites (16, 19), and consists of two major domains, a polymorphic N-terminal domain and a relatively conserved C-terminal domain (11, 15). Genetic diversity within the N-terminal domain consists of sequence polymorphisms and multiple indel mutations, which define two allele classes termed 3D7 and K1 (6, 11). Antibodies targeting PfMSP3 are associated with long-term clinical protection, and full-length PfMSP3 provides strong protection against homologous challenge in an Aotus monkey model (8, 26). To date, PfMSP3 vaccine efforts have so far focused almost exclusively on the C-terminal domain, both because it is highly conserved and because specific subregions of the C-terminal domain can generate protective immune responses in vitro (5, 13, 17, 29, 32). However, anti-PfMSP3 N-terminal domain antibodies are also able to elicit protective responses (28, 29).
As a proof-of-concept study to test the approach of using sera from genotyped infections in a hypoendemic environment to identify allele-specific immune responses, we tested anti-PfMSP3 antibody responses in samples that had previously been genotyped for their infecting PfMSP3 allele. Responses against both variants of the PfMSP3 N-terminal domain in circulation at the study site were tested, as well as the more conserved C-terminal domain and two other N-terminal variants not found at the study site. The magnitude and isotype distribution of each response were measured, and correlations between responses to the infecting and noninfecting antigens were compared. The results confirm that the majority of anti-PfMSP3 responses are against the N terminus, rather than the C-terminal domain targeted by current vaccines. These responses were largely allele specific, as others have shown in hyperendemic samples (22, 25), and this validates the approach of using genotyped samples from hypoendemic transmission environments as an approach to identify allele-specific responses. The comparison of responses across multiple N-terminal variants, which has not previously been reported, provided no evidence for allele specificity within one allele class, which has important implications for the development of any PfMSP3-based vaccine that includes the highly immunogenic N-terminal domain.
MATERIALS AND METHODS
Study site, subjects, and sample collection.
A detailed description of the MIGIA cohort study has been previously described (1). Briefly, the study involves the residents of the Zungarococha community, a cluster of four villages located south of Iquitos in the Peruvian Amazon. The village residents have homogeneous housing construction, income levels, and access to health care, provided by the MIGIA cohort physicians at a community health post. Travel outside the community is rare, with the most frequent travel being to the city of Iquitos, where malaria transmission is nonexistent. The Zungarococha community was chosen as the focus of the MIGIA cohort because of the presence of continuing stable hypoendemic transmission of both P. vivax and P. falciparum, with frequency of infection rates for P. falciparum being <0.5 infection/person/year (35).
Cases of P. falciparum were detected by both active and passive means. Passive cases involved symptomatic individuals presenting at a local health post located in Zungarococha village, where they were tested for malaria parasites by Giemsa-stained microscopy and underwent a comprehensive medical evaluation. PCR verification of the microscopy result was subsequently performed using species-specific primers. Active case detection involved random sampling of villagers over the course of the malaria transmission season. All confirmed cases of Plasmodium infection were treated with appropriate antimalarial medication. Blood samples from confirmed P. falciparum infections were separated into serum and packed red blood cell (RBC) fractions by centrifugation, and each was cataloged and stored at −80°C until needed.
Antigen construction and purification.
Domain-specific PfMSP3 recombinant proteins were amplified from HB3 (HB3 N-terminal domain, C-terminal domain), Dd2 (K1 N-terminal domain), or 3D7 (3D7 and Nig80 N-terminal domains) genomic DNA. The forward and reverse primers used were 5′-CCGGCTCGAGGATTTTAGTGGTGGAGAATTTTCGTGGCC-3′ and 5′-CGGGATCCTTATTCCCAACCTAAAATATAATC-3′, 5′-CCGGCTCGAGTCTATGGAATTCGGAGGTTTTAC-3′ and 5′-CGGGATCCTTATTCCCAACCTAAAATATAATC-3′, and 5′-CCGGCTCGAGGATTATATTTTAGGTTGGGAATTTGGAGG-3′ and 5′-CGGGATCCTTAATGATTTTTAAAATATTTGGATAATTC-3′ for the HB3 and 3D7 N-terminal domains, K1 N-terminal domain, and conserved domain, respectively. The Nig80 N-terminal fragment was created using PCR overlap extension (9) with 5′-ATGCGGATCCGATTTTAGTGGTGGAGAATTTTTGTGGCCTGG-3′ and 5′-CTGCTTCTTTAGCAGCTTCTTCTGCCTCTTTAGAAGCATTTTCAGCATCTTCGGAAGC-3′ primers for the 5′ fragment, 5′-GCTGCTAATGATGCTGAAAATGCTTCAAAAGAGGC-3′ and 5′-AATTCTGCAGTTATTCCCAACCTAAAATATAATC-3′ for the 3′ fragment, and 5′-ATGCGGATCCGATTTTAGTGGTGGAGAATTTTTGTGGCCTGG-3′ and 5′-AATTCTGCAGTTATTCCCAACCTAAAATATAATC-3′ for the splicing reaction. Antigens were expressed as hexa-His-tagged fusion proteins from pET15b (Novagen) or pRSETA (Invitrogen) vectors in the Escherichia coli BL21(DE3)pLysS Rosetta strain (Novagen) and purified using affinity and anion-exchange chromatography, as previously described (2).
Circular dichroism.
To confirm correct folding of each recombinant antigen, circular dichroism (CD) spectra were obtained on an Aviv model 400 CD spectrometer (Lakewood, NJ). Data were collected from 260 nm to 185 nm in 0.5-nm increments with a 10-s average time per point. Spectra were baseline corrected and smoothed and converted to mean residue ellipticity for plotting and analysis. Secondary structural calculations were performed with PROSEC.
Enzyme-linked immunosorbent assay (ELISA) and competition assays.
Purified PfMSP3 antigen (50 ng) was used to coat each well, and a 1:100 dilution of patient serum was used for each assay. Bound antibodies were detected by the addition of horseradish peroxidase (HRP)-conjugated anti-IgG (Chemicon) at a dilution of 1:5,000 or HRP-conjugated IgG isotype-specific (Southern Biotech) and IgM-specific (Fisher Scientific) secondary antibodies at a dilution of 1:1,000. ChromoPure human IgG (Jackson ImmunoResearch Laboratories) was used to standardize total IgG antibody responses. IgG isotype antibody responses are presented as optical densities of 1:100 serum dilutions due to the unavailability of isotype-specific standards. All plates were read at 450 nm using a Uniread 800 ELISA plate reader (GeneMate, Kaysville, UT). Serially diluted positive pools were run concurrently with each set of ELISAs to ensure that all optical density at 450 nm (OD450) measurements remained within the linear range and to facilitate ELISA normalization. Responses were scored as ELISA positive if they were three standard deviations above the mean of a negative-control pool of serum obtained from Peruvian residents in Iquitos who have never been infected with P. falciparum. Serum samples from 11 individuals were used in a pool for all negative controls. Negative cutoff values for HB3 N-terminus, K1 N-terminus, and C-terminus antigens were 0.888 μg/ml, 1.026 μg/ml, and 0.979 μg/ml, respectively. Competition assays were performed as previously described (22). Titrating K1 or HB3 N-terminus antigens were added at a 1-ng, 100-ng, or 1,000-ng concentration to compete for anti-HB3 or anti-K1 antibody binding, respectively. As a control, homologous antigen was used to outcompete antibody binding to the ELISA plate.
Statistical analysis.
Comparisons between proportions of ELISA antigens and allele infections were performed using the two-group chi-square test or Fisher's exact test when the assumptions of the chi-square were not tenable. Comparisons between mean IgG levels for ELISA antigens, separately for allele infections (see Fig. 3 to 5), were performed using the Kruskal-Wallis test, since IgG levels and antibody responses were determined to not be normally distributed. When a statistically significant overall result was obtained, the Dunn multiple comparison procedure was used to determine which specific pairs of means were significantly different. Spearman correlation analyses were performed to examine the relationships between IgG levels of the ELISA antigens (see Fig. 5C and D and Fig. 6A). All statistical tests were two sided and were performed using a 5% significance level (i.e., alpha = 0.05). SAS software (version 9.1.3; SAS Institute, Inc., Cary, NC) was used to perform all statistical analyses.
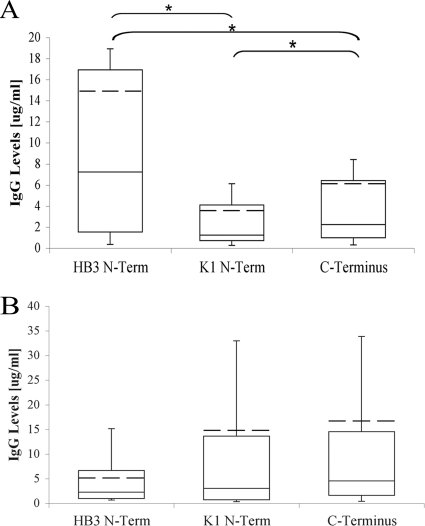
Fig. 3.
Anti-PfMSP3 responses are primarily anti-N-terminal domain allele-specific responses. IgG antibody levels (in micrograms per milliliter) against each PfMSP3 domain antigen in HB3-infected (Fig. 3A, n = 335) and K1-infected (Fig. 3B, n = 34) individuals. Whiskers represent upper and lower quartiles, and mean values are indicated by the hashed line.
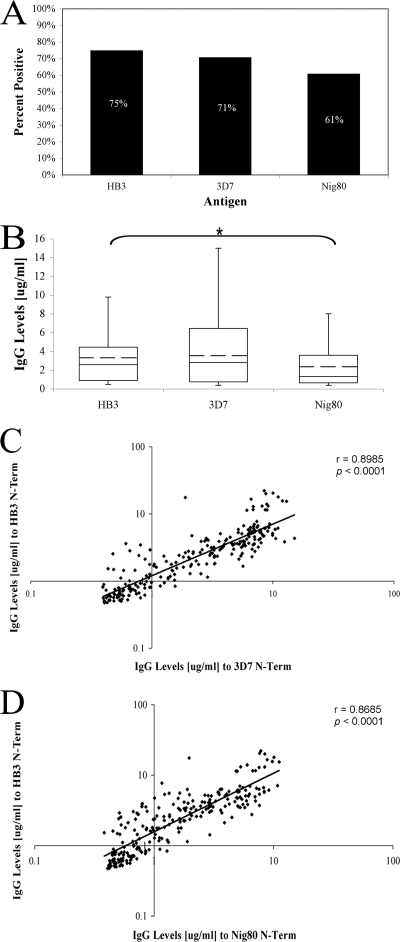
Fig. 5.
Anti-PfMSP3 N-terminal domain responses across variants within the 3D7 allele class. (A) Percent positive responses of ELISA-positive HB3-infected individuals (n = 242) against 3D7 class mutants 3D7 and Nig80. (B) IgG antibody levels (in micrograms per milliliter) against 3D7 class mutants. The whiskers denote the upper and lower quartiles. The mean is denoted by the hashed line. *, P = 0.001. (C and D) Pairwise correlation between anti-HB3 responses and either anti-3D7 (C) or anti-Nig80 (D) antibody responses; the black line indicates a best-fit exponential line.
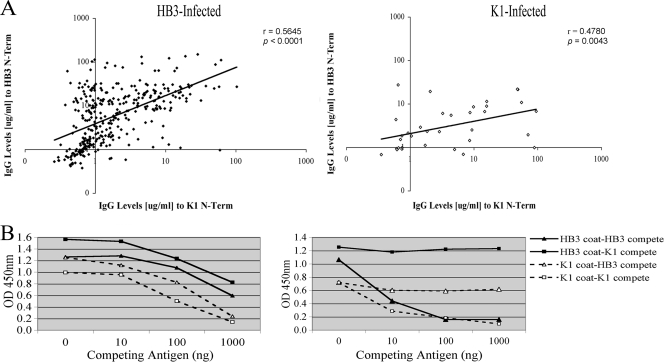
Fig. 6.
Anti PfMSP3 N-terminal (N-Term) domain responses across allele classes. (A) Pairwise correlation between anti-HB3 and anti-K1 antibody responses separated on the basis of their infecting PfMSP3 allele; the black line indicates a best-fit exponential line. n = 369. (B) Competition ELISAs were performed using titrating amounts of competing antigen (0 to 1,000 ng) to assess cross-reaction; some samples showed evidence of cross-reactivity (left panel), while others showed only allele-specific responses (right panel).
Ethics committee approval.
This study was approved by review boards of the University of Alabama at Birmingham, New York University, Universidad Peruana Cayetano Heredia, and the Peruvian Ministerio de Salud, Instituto Naccional de Salud. Written consent was obtained from all participants prior to study enrollment.
RESULTS
Production of PfMSP3 domain antigens.
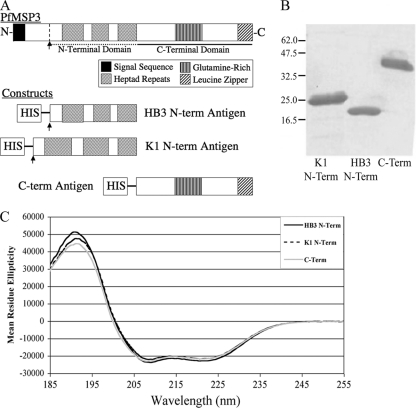
Our previous genotyping study established that both PfMSP3 allele classes, 3D7-like and K1-like, were in circulation at the MIGIA study site and were identical in sequence to the previously published HB3 and K1 sequences, respectively (12). Three recombinant antigens were generated to represent the PfMSP3 domains in circulation at the study site; two N-terminal domain antigens, HB3 and K1, and one C-terminal domain antigen corresponding to the C-terminal sequence shared by both alleles (Fig. 1A). Purification of these antigens yielded milligram quantities of >95% pure protein (Fig. 1B), and far-UV circular dichroism (CD) spectroscopy readings (Fig. 1C) were consistent with previous biophysical studies (2), indicating that all three antigens have native secondary structures.
Fig. 1.
Purity and folding of PfMSP3 domain antigens. (A) Three PfMSP3 constructs were generated to test antibody responses by ELISA: HB3 and K1 allele N-terminal domains and the conserved C-terminal domain. The N-terminal domains began at the proteolytic cleavage site and included all three heptad repeats. The C-terminal domain began just C terminal to the last heptad repeat. (B) Coomassie blue-stained SDS-PAGE demonstrates PfMSP3 construct antigens are >95% pure as measured by scanning densitometry. (C) Circular dichroism spectroscopy reveals signature peaks at 215 nm and 190 nm indicative of constructs being comprised largely of alpha-helical structures, consistent with published data for full-length PfMSP3.
Comparison of antibody responses to infecting and noninfecting antigens.
The three antigens were used to detect anti-PfMSP3 responses in time-of-infection serum samples from 369 distinct P. falciparum infections collected between 2003 and 2006, all of which had previously been genotyped for the infecting PfMSP3 allele (12). More than 90% of P. falciparum infections at this study site are of the PfMSP3 3D7-like allele class, so samples were selected to reflect these observed allele frequencies: 335 from HB3 infections and 34 from K1 infections. No samples were from individuals coinfected with more than one allele, which happens in less than 0.5% of infections because of the low transmission rates (12). Serum samples were tested against all three antigens, and ELISA results were normalized and converted into absolute concentrations of anti-PfMSP3 IgG.
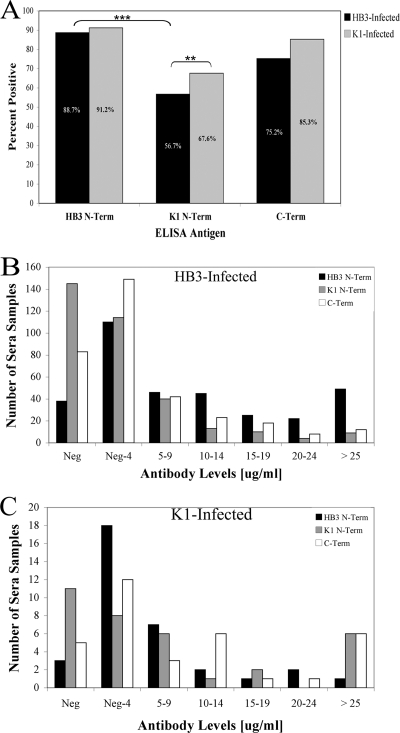
Approximately 90% of individuals had positive responses against the HB3 N-terminal antigen, compared to 75 to 85% who had positive responses against the C-terminal antigens (Fig. 2A). When responses were compared with those to the infecting PfMSP3 genotype, there was evidence of a skew toward a response to the infecting allele: a significantly higher percentage of HB3-infected individuals had positive responses against the HB3 N-terminal domain than against either the K1 N-terminal domain or the C-terminal domain (Fig. 2A, P < 0.0001), and there was a significant increase in anti-K1 responses in K1-infected individuals relative to HB3-infected individuals (P = 0.016). Interestingly, although anti-N-terminal domain responses were skewed toward the infecting genotype, many individuals responded to both the infecting and noninfecting genotypes; 57% of HB3-infected individuals generated positive responses against the noninfecting K1 N-terminal domain. When the distribution of antibody responses is taken into consideration, more than twice as many HB3-infected individuals had anti-HB3 N-terminal domain antibody levels greater than 10 μg/ml compared to C-terminal domain antibody responses, and more than three times as many had anti-HB3 antibody levels above 20 μg/ml (P < 0.0001) (Fig. 2B). We did not detect a significant difference comparing N-terminal and C-terminal responses in K1-infected individuals, which is likely due to the small sample size of K1-infected individuals (Fig. 2C).
Fig. 2.
Distribution of anti-PfMSP3 responses in HB3- and K1-infected individuals. (A) Percent positive responses, by infected allele, either HB3 (N = 335) or K1 (N = 34), for each PfMSP3 domain antigen. Responses were scored positive if they were three standard deviations above the mean of a negative-control pool of serum from individuals who had never been infected with P. falciparum. Negative cutoff values for each antigen are listed in Materials and Methods. The double and triple asterisks denote P = 0.016 and P < 0.0001, respectively. (B and C) Number of individuals who exhibit given IgG antibody levels (in micrograms per milliliter) against each PfMSP3 domain antigen in HB3- (B) and K1-infected (C) individuals. Neg shows number of samples below the negative cutoff values, which are listed in Materials and Methods. Neg-4 are samples less than 5 μg/ml but greater than the negative cutoff, 5–9 are samples from 5 to 9 μg/ml above the negative cutoff, and so on.
Anti-PfMSP3 responses are primarily anti-N-terminal domain allele-specific responses.
Direct comparison of the antibody response against each antigen (in micrograms per milliliter) within each infecting allele class was then performed to characterize the strength of anti-PfMSP3 responses. In HB3-infected individuals, mean antibody levels reacting with the HB3 N-terminal domain were 2.2-fold higher than mean antibody levels that reacted with the C-terminal domain (P < 0.05) and also 3.1-fold higher than mean antibody levels that reacted with the K1 N-terminal domain (Fig. 3A). In K1-infected individuals, there was no significant difference in levels of antibody to the K1 N-terminal domain and C-terminal domain antibodies, but mean levels recognizing the K1 N-terminal domain were 2.8-fold higher than levels recognizing the HB3 N-terminal domain (this difference did not reach statistical significance, presumably because of the low numbers of K1-infected individuals) (Fig. 3B). The N-terminal domain therefore appears to be the immunodominant PfMSP3 domain in this study site and is largely targeted by allele-specific antibodies, in keeping with previous results from the Gambia (22, 25).
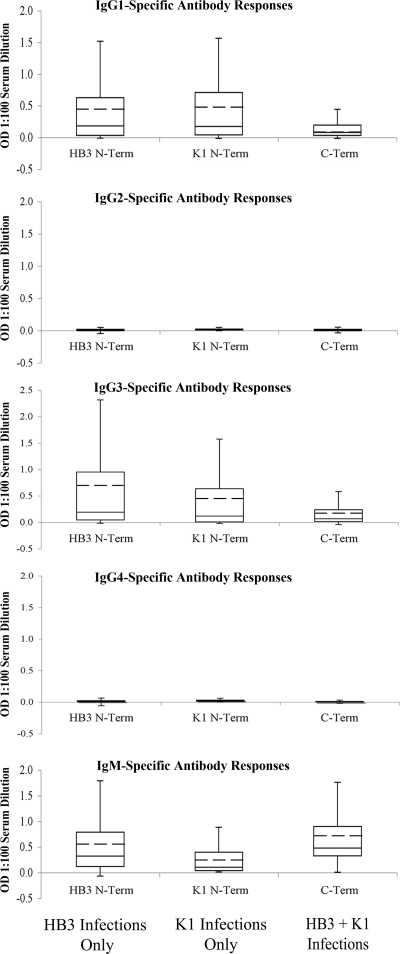
Anti-PfMSP3 N-terminal domain antibodies are primarily cytophilic subclasses.
Because cytophilic IgG subclasses IgG1 and IgG3 have been previously associated with protection against malaria (20, 33, 37), each antiserum sample that scored as positive by total IgG ELISA was tested for isotype class distribution using isotype-specific secondary antibodies and presented in terms of its optical density at a 1:100 dilution, because a lack of isotype-specific standards for normalization precluded conversion to absolute isotype-specific antibody levels. Because of sample volumes, isotype distribution was established only for the antigens present in the infection, namely, the infecting N-terminal domain and the conserved C-terminal domain. A total of 34 serum samples that were typed for antigen specificity (Fig. 3) had insufficient volume remaining to carry out isotype experiments, leaving 304 samples from HB3-infected individuals and 21 samples from K1-infected individuals to be typed. Anti-N-terminal domain antibodies were primarily of the IgG1 and IgG3 cytophilic subclasses; in HB3-infected individuals mean anti-HB3 N-terminal domain IgG3 levels were higher than mean IgG1 levels (P < 0.05), whereas in K1-infected individuals, mean IgG1 and IgG3 responses against the K1 N-terminal domain were identical (Fig. 4). In contrast, antibodies recognizing the C-terminal domain were significantly enriched for IgM responses regardless of the infecting allele (P < 0.05).
Fig. 4.
Responses against the PfMSP3 N-terminal domain are largely of cytophilic subclasses. PfMSP3 isotype antibody levels (OD450, 1:100 serum dilution) for IgG1-4 and IgM responses against N-terminal (N-Term) and C-terminal (C-Term) domains. Only homologous N-terminal domain responses were assessed. The whiskers denote the upper and lower quartiles. The mean is denoted by the hashed line.
Anti-PfMSP3 N-terminal domain responses across variants within the 3D7 allele class.
Although these data and others have shown that anti-N-terminal domain responses are primarily allele specific, no previous study has tested reactivity across variants within a given allele class. To test for allele specificity across such variants, N-terminal domain antigens for two other naturally occurring 3D7 class alleles were generated. One antigen was identical to the 3D7 allele, which differs from the HB3 sequence used in the initial studies at only a single nonsynonymous single nucleotide polymorphism (SNP). This SNP is the most prevalent polymorphism in the 3D7 class alleles globally (6) and has been detected at the MIGIA study site, being found in 10/630 previously genotyped 3D7 class alleles (12). None of these infections were included in this study, but, although it is unlikely, it is possible that some individuals in this study had previously been infected with this allele. As a more extreme difference, a previously published 3D7 class PfMSP3 sequence from Nigeria (Nig80), which possesses a heptad indel mutation in addition to the 3D7 SNP (25), was chosen. No similar Nig80 sequence has been reported across more than 600 infections at this study site or elsewhere in South America, making it extremely unlikely that any of the individuals at the MIGIA study site have been previously exposed to this allele.
A total of 242 antiserum samples from HB3-infected individuals with sufficient serum volume remaining were used to test responses to both the 3D7 and Nig80 N-terminal antigens. Of these individuals, 71% had positive responses to the 3D7 antigen and 61% to the Nig80 antigen, compared to 75% to the HB3 antigen (Fig. 5A). There was no significant difference in mean IgG levels between the 3D7 and HB3 responders, but there was a 0.3-fold reduction in responses against the Nig80 antigen (P = 0.001) (Fig. 5B). To compare responses at an individual rather than a population level, the response to either the 3D7 or Nig80 antigen was compared with the response to the HB3 antigen in the same individual. Responses to the HB3 antigen correlated strongly with responses to both the 3D7 (Fig. 5C; Spearman's r = 0.8985, P < 0.0001) and Nig80 (Fig. 5D; Spearman's r = 0.8685, P < 0.0001) antigens. There is therefore little evidence of allele-specific responses between these three variants of the 3D7 allele class.
Anti-PfMSP3 N-terminal domain responses across allele classes.
Although responses against the PfMSP3 N-terminal domain were primarily against the infecting allele class, suggesting allele specificity (Fig. 2 and 3), it was notable that detectable responses against the K1 N-terminal domain antigen were present in 57% of HB3-infected individuals, suggesting that some anti-N-terminal domain responses are not allele specific. To investigate this more closely, responses against the infecting and noninfecting allele classes were compared for each individual infection. The strength of responses was clearly skewed toward the infecting N-terminal allele (Fig. 6A), but there was a statistically significant positive correlation between the strength of response against both the infecting and noninfecting N-terminal domains (Spearman's r = 0.5645, P < 0.0001 for HB3 infection; Spearman's r = 0.4780, P = 0.0043 for K1 infection).
To establish whether these represent true interallele cross-reactive responses, competition ELISAs were performed on the few serum samples where sufficient volume was present for such tests. Of the samples that had both strongly positive anti-HB3 and anti-K1 responses (upper right quadrant of Fig. 6A, above 10 μg/ml for both antigens), 11 samples had a sufficient volume of serum for the multiple rounds of depletion with different concentrations of antigens needed for competition assays. Of these 11 samples, in 6 cases responses against one antigen could be competed by preincubating with the heterologous antigen, denoting clear cross-reactive responses (Fig. 6B, left panel). In the other 5 cases, responses to both antigens were present but could be competed only with the homologous antigen, suggesting the simultaneous presence of allele-specific responses to both antigens (Fig. 6B, right panel). Both correlation and competition ELISA analyses therefore suggest that some antibody responses generated by the PfMSP3 N-terminal domain can cross-react across allele classes.
DISCUSSION
This report investigated the antibody responses against a leading vaccine candidate, P. falciparum merozoite surface protein 3 (PfMSP3), in a hypoendemic transmission environment, in order to test the approach of using genotyped samples from such environments to rapidly identify allele-specific and allele-transcending antibody responses. As seen in other studies, responses to PfMSP3 were widespread, with approximately 90% of individuals responding positively against the HB3 N-terminal antigen and 75% responding to the C-terminal antigen. The PfMSP3 N-terminal domain was significantly more immunogenic than the C-terminal domain, eliciting, in HB3 infected individuals, a 2.3-fold increase in mean IgG antibody levels and a 3.6-fold increase in “high-responder” individuals—those who exhibited IgG antibody levels greater than 20 μg/ml. These results are in line with data from the Gambia, which demonstrated significantly higher N-terminal domain antibody responses, especially in young children (22, 25). Additionally, anti-N-terminal domain antibodies were largely comprised of cytophilic IgG1 and IgG3 subclasses, which have previously been associated with protection against malaria (20, 33, 37).
The fact that the samples in this study had previously been genotyped for infecting PfMSP3 allele type, and came from individuals who had on average not had a previous P. falciparum infection for at least 1 year, allowed the extent of allele specificity to be tested directly. When comparing responses within the 3D7 allele class, there was little evidence for allele specificity, with responses against two noninfecting 3D7 class allele mutants exhibiting strong pairwise correlation with responses to the infecting HB3 N-terminal domain (P < 0.0001). These antigens differed at only a few amino acids, so presumably the majority of responses are recognizing epitopes that are conserved across the variants, perhaps the residues responsible for stabilizing the coiled-coil domains that are almost exclusively conserved within each allele class. The existence of responses against such conserved regions is not necessarily surprising but is important and has not been previously shown.
Comparing responses across N-terminal domain allele classes confirms previous findings that demonstrated that anti-N-terminal domain immune responses are weighted toward being allele specific (22, 25). However, this study also provides clear evidence that some anti-N-terminal domain responses target epitopes shared across allele classes. At a population level, 57% of HB3-infected individuals elicited positive responses against the K1 N-terminal domain, and there was a significant positive correlation between individual responses to the 3D7 and K1 class domains, although not as strong as the correlation between responses to variants within the 3D7 allele class. Competition ELISAs confirmed that at least some of these responses were truly cross-reactive, as they could be competed away with both the HB3 and K1 N-terminal domain antigens. In theory, these apparent cross-reactive responses could be explained by recent infections with a K1 class allele that were undetected in the cohort study. However, using available frequency-of-infection and allele frequency data for this study site, there is only a 0.84% probability of an undetected K1 infection occurring in any one individual within the past 2 years, clearly not enough to explain 57% of HB3-infected individuals having antiserum that can cross-react with K1 antigens. The most parsimonious explanation of the data is therefore that infection with HB3 allele PfMSP3 antigen generates a response against epitopes that are conserved across both allele classes, although at the expense of significant immunogenicity.
The P. falciparum genome presents a large number of potential vaccine candidates, and it is therefore critical that go/no-go decisions based on solid evidence be applied to limit the number of candidates entering expensive later-stage vaccine trials (3, 7, 10). The PfMSP3 N-terminal domain had previously been largely eliminated from consideration based on concerns about sequence diversity and because the C-terminal domain has been extensively studied in the context of antibody-dependent cellular immunity assays (ADCI), the mechanism by which PfMSP3 is thought to induce protection in vivo (19, 29, 39). Long peptides based on the C-terminal domain can induce potent antibody responses in vivo that are capable of killing P. falciparum parasites, have good safety profiles in initial trials, and clearly warrant further investigation as candidate antigens (5, 13, 17, 26, 31, 32). However, the PfMSP3 N-terminal domain is also capable of inducing ADCI-mediated killing in vitro (29), data from both Peru (this paper) and the Gambia (25) suggest that the PfMSP3 N-terminal domain is a more potent immunogen than the C-terminal domain in vivo, and in the Gambia, responses against the N-terminal domain correlated with protection against clinical malaria (25). The totality of these data suggests that the PfMSP3 N-terminal domain could be reconsidered as a potential vaccine antigen if the problems of genetic diversity can be addressed (28). The findings reported here that anti-N-terminal domain responses include responses to epitopes conserved within an allele class, and also to a lesser extent to epitopes conserved across allele classes, raise the possibility that regions of the N-terminal domain could be identified that may overcome this problem. Mapping those conserved epitopes within the N-terminal domain, as well as testing for functional cross-protection in in vitro assays, is therefore a high priority for further research.
ACKNOWLEDGMENTS
We thank Patrick Sutton and Eva Clark for help with sample processing and Mike Jablonsky for his help with the circular dichroism. We thank all residents in the Zungarococha community who participate so willingly in the MIGIA cohort study. We thank all the staff of the MIGIA project, which is a strong collaboration with the Universidad Nacional de la Amazonia Peruana. We thank all for sample collection, clinic visits and management, and laboratory sample processing and care.
This work was supported by National Institutes of Health grants R21 AI072421 and R01 AI064849, and the UAB Sparkman Center for Global Health.
We declare no conflict of interest.
Footnotes
Published ahead of print on 7 March 2011.
REFERENCES
- 1. Branch O., et al. 2005. Clustered local transmission and asymptomatic Plasmodium falciparum and Plasmodium vivax malaria infections in a recently emerged, hypoendemic Peruvian Amazon community. Malar. J. 4:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burgess B. R., Schuck P., Garboczi D. N. 2005. Dissection of merozoite surface protein 3, a representative of a family of Plasmodium falciparum surface proteins, reveals an oligomeric and highly elongated molecule. J. Biol. Chem. 280:37236–37245 [DOI] [PubMed] [Google Scholar]
- 3. Coppel R. L. 2009. Vaccinating with the genome: a Sisyphean task? Trends Parasitol. 25:205–212 [DOI] [PubMed] [Google Scholar]
- 4. Demanga C. G., et al. 2010. Toward the rational design of a malaria vaccine construct using the MSP3 family as an example: contribution of antigenicity studies in humans. Infect. Immun. 78:486–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Druilhe P., et al. 2005. A malaria vaccine that elicits in humans antibodies able to kill Plasmodium falciparum. PLoS Med. 2:e344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Escalante A. A., Lal A. A., Ayala F. J. 1998. Genetic polymorphism and natural selection in the malaria parasite Plasmodium falciparum. Genetics 149:189–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Greenwood B., Targett G. 2009. Do we still need a malaria vaccine? Parasite Immunol. 31:582–586 [DOI] [PubMed] [Google Scholar]
- 8. Hisaeda H., et al. 2002. Merozoite surface protein 3 and protection against malaria in Aotus nancymai monkeys. J. Infect. Dis. 185:657–664 [DOI] [PubMed] [Google Scholar]
- 9. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59 [DOI] [PubMed] [Google Scholar]
- 10. Holder A. A. 2009. Malaria vaccines: where next? PLoS Pathog. 5:e1000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huber W., et al. 1997. Limited sequence polymorphism in the Plasmodium falciparum merozoite surface protein 3. Mol. Biochem. Parasitol. 87:231–234 [DOI] [PubMed] [Google Scholar]
- 12. Jordan S. J., Branch O. H., Castro J. C., Oster R. A., Rayner J. C. 2009. Genetic diversity of the malaria vaccine candidate Plasmodium falciparum merozoite surface protein-3 in a hypoendemic transmission environment. Am. J. Trop. Med. Hyg. 80:479–486 [PMC free article] [PubMed] [Google Scholar]
- 13. Lusingu J. P., et al. 2009. Satisfactory safety and immunogenicity of MSP3 malaria vaccine candidate in Tanzanian children aged 12-24 months. Malar. J. 8:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mamillapalli A., et al. 2007. Polymorphism and epitope sharing between the alleles of merozoite surface protein-1 of Plasmodium falciparum among Indian isolates. Malar. J. 6:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McColl D. J., Anders R. F. 1997. Conservation of structural motifs and antigenic diversity in the Plasmodium falciparum merozoite surface protein-3 (MSP-3). Mol. Biochem. Parasitol. 90:21–31 [DOI] [PubMed] [Google Scholar]
- 16. McColl D. J., et al. 1994. Molecular variation in a novel polymorphic antigen associated with Plasmodium falciparum merozoites. Mol. Biochem. Parasitol. 68:53–67 [DOI] [PubMed] [Google Scholar]
- 17. Nebie I., et al. 2009. Humoral and cell-mediated immunity to MSP3 peptides in adults immunized with MSP3 in malaria endemic area, Burkina Faso. Parasite Immunol. 31:474–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ochola L. I., et al. 2010. Allele frequency-based and polymorphism-versus-divergence indices of balancing selection in a new filtered set of polymorphic genes in Plasmodium falciparum. Mol. Biol. Evol. 27:2344–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oeuvray C., et al. 1994. A novel merozoite surface antigen of Plasmodium falciparum (MSP-3) identified by cellular-antibody cooperative mechanism antigenicity and biological activity of antibodies. Mem. Inst. Oswaldo Cruz 89(Suppl. 2):77–80 [DOI] [PubMed] [Google Scholar]
- 20. Oeuvray C., et al. 2000. Cytophilic immunoglobulin responses to Plasmodium falciparum glutamate-rich protein are correlated with protection against clinical malaria in Dielmo, Senegal. Infect. Immun. 68:2617–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ogutu B. R., et al. 2009. Blood stage malaria vaccine eliciting high antigen-specific antibody concentrations confers no protection to young children in Western Kenya. PLoS One 4:e4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Osier F. H., et al. 2007. Naturally acquired antibodies to polymorphic and conserved epitopes of Plasmodium falciparum merozoite surface protein 3. Parasite Immunol. 29:387–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Osier F. H., et al. 2010. Allelic diversity and naturally acquired allele-specific antibody responses to Plasmodium falciparum apical membrane antigen 1 in Kenya. Infect. Immun. 78:4625–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Polley S. D., et al. 2004. Human antibodies to recombinant protein constructs of Plasmodium falciparum apical membrane antigen 1 (AMA1) and their associations with protection from malaria. Vaccine 23:718–728 [DOI] [PubMed] [Google Scholar]
- 25. Polley S. D., et al. 2007. Plasmodium falciparum merozoite surface protein 3 is a target of allele-specific immunity and alleles are maintained by natural selection. J. Infect. Dis. 195:279–287 [DOI] [PubMed] [Google Scholar]
- 26. Roussilhon C., et al. 2007. Long-term clinical protection from falciparum malaria is strongly associated with IgG3 antibodies to merozoite surface protein 3. PLoS Med. 4:e320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sagara I., et al. 2009. A randomized controlled phase 2 trial of the blood stage AMA1-C1/Alhydrogel malaria vaccine in children in Mali. Vaccine 27:3090–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saul A. 2007. Malaria vaccines based on the Plasmodium falciparum merozoite surface protein 3—should we avoid amino acid sequence polymorphisms or embrace them? J. Infect. Dis. 195:171–173 [DOI] [PubMed] [Google Scholar]
- 29. Singh S., et al. 2004. Identification of a conserved region of Plasmodium falciparum MSP3 targeted by biologically active antibodies to improve vaccine design. J. Infect. Dis. 190:1010–1018 [DOI] [PubMed] [Google Scholar]
- 30. Singh S., et al. 2009. A conserved multi-gene family induces cross-reactive antibodies effective in defense against Plasmodium falciparum. PLoS One 4:e5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sirima S. B., et al. 2007. Safety and immunogenicity of the Plasmodium falciparum merozoite surface protein-3 long synthetic peptide (MSP3-LSP) malaria vaccine in healthy, semi-immune adult males in Burkina Faso, West Africa. Vaccine 25:2723–2732 [DOI] [PubMed] [Google Scholar]
- 32. Sirima S. B., et al. 2009. Safety and immunogenicity of the malaria vaccine candidate MSP3 long synthetic peptide in 12-24 months-old Burkinabe children. PLoS One 4:e7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stanisic D. I., et al. 2009. Immunoglobulin G subclass-specific responses against Plasmodium falciparum merozoite antigens are associated with control of parasitemia and protection from symptomatic illness. Infect. Immun. 77:1165–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sutton P. L., Clark E. H., Silva C., Branch O. H. 2010. The Plasmodium falciparum merozoite surface protein-1 19 KD antibody response in the Peruvian Amazon predominantly targets the non-allele specific, shared sites of this antigen. Malar. J. 9:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sutton P. L., Neyra V., Hernandez J. N., Branch O. H. 2009. Plasmodium falciparum and Plasmodium vivax infections in the Peruvian Amazon: propagation of complex, multiple allele-type infections without super-infection. Am. J. Trop. Med. Hyg. 81:950–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Takala S. L., Plowe C. V. 2009. Genetic diversity and malaria vaccine design, testing and efficacy: preventing and overcoming “vaccine resistant malaria.” Parasite Immunol. 31:560–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Taylor R. R., Allen S. J., Greenwood B. M., Riley E. M. 1998. IgG3 antibodies to Plasmodium falciparum merozoite surface protein 2 (MSP2): increasing prevalence with age and association with clinical immunity to malaria. Am. J. Trop. Med. Hyg. 58:406–413 [DOI] [PubMed] [Google Scholar]
- 38. Tetteh K. K., et al. 2009. Prospective identification of malaria parasite genes under balancing selection. PLoS One 4:e5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Trucco C., et al. 2001. The merozoite surface protein 6 gene codes for a 36 kDa protein associated with the Plasmodium falciparum merozoite surface protein-1 complex. Mol. Biochem. Parasitol. 112:91–101 [DOI] [PubMed] [Google Scholar]