Abstract
Release 4.0 of ProTherm, thermodynamic database for proteins and mutants, contains ∼14 500 numerical data (∼450% of the first version) of several thermodynamic parameters along with experimental methods and conditions, and structural, functional and literature information. The sequence and structural information of proteins is connected with thermodynamic data through links between entries in Protein Data Bank, Protein Information Resource and SWISS-PROT and the data in ProTherm. We have separated the Gibbs free energy change obtained at extrapolated temperature from the data on denaturation temperature measured by the thermal denaturation method. We have added the statistics of amino acid replacements and links to homologous structures to each protein. Further, we have improved the search and display options to enhance search capability through the web interface. ProTherm is freely available at http://gibk26.bse.kyutech.ac.jp/jouhou/Protherm/protherm.html.
INTRODUCTION
Thermodynamic data for proteins are essential for understanding the mechanism of protein folding and stability, and for designing stable mutants. Due to the advancement in molecular biology and biochemistry, a large number of proteins have been characterized. Accordingly, the accumulation of thermodynamic data has been steadily increasing (1,2). The compilation of thermodynamic data along with the sequence and structural information would be a valuable resource for developing algorithms/methods to elucidate the mechanism of protein folding and stability and to predict the stability change upon mutations. We have designed an electronically accessible database, ProTherm (3–5), including several thermodynamic parameters along with sequence and structural information, experimental methods and conditions, and literature information. Furthermore, we have provided a web interface to facilitate searching the database, sorting and visualizing the results. The thermodynamic data available in ProTherm have been effectively used in understanding the mechanism of protein stability upon mutations and for predicting the stability of protein mutants (6–13).
CURRENT DEVELOPMENTS
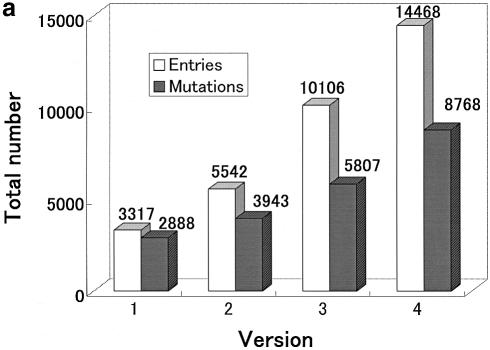
(i) Release 4.0 contains 14 468 entries including the latest data, ∼436% of release 1.0 (3317 entries).
(ii) A table is included showing the number of mutations for each type of amino acid replacement (380 possibilities), deposited in ProTherm. Furthermore, the most commonly occurring mutations are highlighted with different colors.
(iii) Previously, we assigned unique PDB codes to proteins from different sources and mentioned only the appropriate codes. Currently, we provide a link to all homologous PDB codes with a sequence identity of >95%.
(iv) The sequence and structure of proteins have been connected with thermodynamic data through 3DinSight (14), with cross-links between entries in PDB (15), PIR (16) and Swiss-Prot (17) and ProTherm. These cross-link tables can also be used to create pointers from these databases to ProTherm.
(v) Search options are provided for molecular weight, number of states and reversibility. Molecular weight and amino acid length are added in Display Option.
(vi) Temperature units for T, Tm and ΔTm can be selected as C (Celsius) or K (Kelvin). The energy unit of ΔH, ΔG, ΔΔG, ΔGH20, ΔΔGH20 and ΔCp can be selected from kcal and kJ before a search.
(vii) So far, we have included Tm data and ΔG/ΔΔG/ΔH data in the same entry. However, the ΔG/ΔΔG/ΔH data are often extrapolated at different temperatures. We had placed all the temperatures at Tm field without distinguishing T from Tm for thermal denaturation experiments. In order to avoid confusion for users, we have separated these data and placed the extrapolated temperature in a new T field.
(viii) Each article usually contains multiple data and they are entered in different entries. Hence, we added a new field ‘RELATED_ENTRIES’, which provides the list of entries that contain data from the same paper.
DATABASE STATISTICS
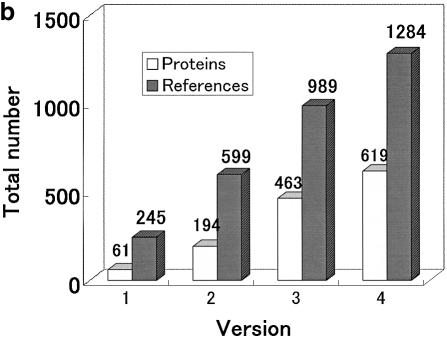
The progressive growth of ProTherm is depicted in Figure 1. Figure 1a and b show the increase in the total numbers of entries and mutations, and proteins and research articles, respectively, in each release of ProTherm. We observed a substantial increase in data in all versions. Currently, the number of data for wild-type proteins, single, double and multiple mutants are, respectively, 5700, 7217, 1050 and 501. Based on solvent accessibility, 3917 are buried, 2163 are partially buried and 2323 are exposed. In terms of secondary structure, 3392 are in helix, 2220 in strand, 920 in turn and 2198 in coil regions. The majority of data are obtained from CD (5777) and DSC (4721) experiments followed by fluorescence (2856). Further, 8748 data are obtained by thermal denaturation (8748), and 3320 and 2296 data from GdnHCl and urea denaturation, respectively.
Figure 1.
Growth of ProTherm: (a) total numbers of entries and mutations; (b) numbers of all proteins and references.
DATA SCRUTINY AND REFINEMENT
To improve the checking process, before uploading the data to the website directly, the data are put in a temporary test site for preliminary checking by database members. After their approval, the data will be made available to the users and in the meantime we send emails to the corresponding authors of each paper informing them of the inclusion and correctness of their data. We have received feedback from authors, which helps us to maintain the data quality. We also receive suggestions and comments on ProTherm, and refine the functionality of ProTherm according to them.
CITATION OF ProTherm
The users of ProTherm are asked to cite this article in their publication including the URL http://gibk26.bse.kyutech.ac.jp/jouhou/protherm/protherm.html. Suggestions and other materials for inclusion in the database are welcome and should be sent to protherm@rtcmain.bse.kyutech.ac.jp.
Acknowledgments
ACKNOWLEDGEMENTS
The development of ProTherm is partially supported by a Grant-in-Aid for Publication Scientific Research Results from the Japan Society for the Promotion of Sciences (JSPS).
REFERENCES
- 1.Pfeil W. (1998) Protein Stability and Folding: A Collection of Thermodynamic Data. Springer, New York, NY. [Google Scholar]
- 2.Pfeil W. (2001) Protein Stability and Folding, Supplement 1: A Collection of Thermodynamic Data. Springer, New York, NY. [Google Scholar]
- 3.Gromiha M.M., An,J., Kono,H., Oobatake, M, Uedaira,H. and Sarai,A. (1999) ProTherm: thermodynamic database for proteins and mutants. Nucleic Acids Res., 27, 286–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gromiha M.M., Uedaira,H., An,J., Selvaraj,S., Prabakaran,P. and Sarai,A. (2002) ProTherm, thermodynamic database for proteins and mutants: developments in version 3.0. Nucleic Acids Res., 30, 301–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarai A., Gromiha,M.M, An,J., Prabakaran,P., Selvaraj,S., Kono,H., Oobatake,M. and Uedaira,H. (2002) Thermodynamic databases for proteins and protein–nucleic acid interactions. Biopolymers, 61, 121–126. [DOI] [PubMed] [Google Scholar]
- 6.Gromiha M.M., Oobatake,M., Kono,H., Uedaira,H. and Sarai,A. (1999) Role of structural and sequence information in the prediction of protein stability changes: comparison between buried and partially buried mutations. Protein Eng., 12, 549–555. [DOI] [PubMed] [Google Scholar]
- 7.Gromiha M.M., Oobatake,M., Kono,H., Uedaira,H. and Sarai,A. (1999) Relationship between amino acid properties and protein stability: buried mutations. J. Protein Chem., 18, 565–578. [DOI] [PubMed] [Google Scholar]
- 8.Gromiha M.M., Oobatake,M., Kono,H., Uedaira,H. and Sarai,A. (2000) Importance of surrounding residues for protein stability of partially buried mutations. J. Biomol. Struct. Dyn., 18, 281–295. [DOI] [PubMed] [Google Scholar]
- 9.Gromiha M.M., Oobatake,M., Kono,H., Uedaira,H. and Sarai,A. (2002) Importance of mutant position in Ramachandran plot for predicting protein stability of surface mutations. Biopolymers, 64, 210–220. [DOI] [PubMed] [Google Scholar]
- 10.Ooi T. and Shimizu,S. (2002) Molecular life of proteins: folding and stability. In Gromiha,M.M. and Selvaraj,S. (eds), Recent Research Developments in Protein Folding, Stability and Design. Research Signpost, Trivandrum, India, pp. 243–264. [Google Scholar]
- 11.Guerois R., Nielsen,J.E. and Serrano,L. (2002) Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol., 320, 369–387. [DOI] [PubMed] [Google Scholar]
- 12.Kortemme T. and Baker,D. (2002) A simple physical model for binding energy hot spots in protein–protein complexes. Proc. Natl Acad. Sci. USA, 99, 14116–14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou H. and Zhou,Y. (2002) Stability scale and atomic solvation parameters extracted from 1023 mutation experiments. Proteins, 49, 483–492. [DOI] [PubMed] [Google Scholar]
- 14.An J., Nakama,T., Kubota,Y. and Sarai,A. (1998) 3DinSight: an integrated relational database and search tool for structure, function and property of biomolecules. Bioinformatics, 14, 188–195. [DOI] [PubMed] [Google Scholar]
- 15.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cathy H.W., Lai-Su,L., Huang,Y.H., Arminski,L., Castro-Alvear,J., Chen,Y., Hu,H., Ledley,R.S., Kourtesis,P., Suzek,B.E. et al. (2003) The Protein Information Resource (PIR). Nucleic Acids Res., 31, 345–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O’Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]