Protein delivery[1] has been considered as the most straightforward strategy for modulating cellular behavior without the safety concerns and expression performance issues associated with gene deliver approaches. Two major challenges remain to be overcome in order to enable practical applications in biology and medicine 1) how to foster cellular uptake of protein molecules and 2) how to retain their stabilities and functions[2] over the delivery process. Recently, attempts have been made to develop a variety of delivery vectors, including liposomes,[3] polymer micelles,[4] and nanoparticle,[5] to enhance the uptake of protein molecules in target cells, and at the same time, to stabilize the encapsulated proteins. Owing to the time-consuming procedures employed in optimization of delivery materials, significant endeavors have been made in search of better delivery systems, although there has been limited progress in the field to date. Alternatively, recombinant technology[6] can be utilized to conjugate cell-penetrating peptides[7] (CPPs) onto protein molecules, this is the most commonly used protein delivery system with improved delivery efficiency. In this case, the major bottlenecks associated with the complicated procedure of generating recombinant proteins and the lack of protection mechanism against protein denature need to be solved.
Transcription factor (TF) is a protein responsible for regulating gene transcription in cellular circuitry.[8] In general, TFs contain one or more DNA-binding domains (DBDs), which recognize matching DNA sequences adjacent to the genes they regulate. Apparently, highly efficient delivery of TFs can provide a powerful technology for modulating cellular behavior. One of the most important in-vitro applications that required highly efficient TF delivery is the generation of human induced pluripotent stem cells (hiPSCs) which has recently been demonstrated by introducing CPPs-fused reprogramming TFs (i.e., OCT4, SOX2, KLF4, and c-MYC)[9] into human somatic cells. The resulting hiPSCs have the potential to revolutionize regenerative medicine.[10] However, the high costs of the four reprogramming TFs in their recombinant forms, means it is unlikely that this approach can be used for large-scale hiPSCs generation without further improvement in the delivery performance of the reprogramming proteins. Therefore, it is crucial to develop a new type of vector capable of delivering intact (unmodified) TFs in a highly efficient manner.
Previously, we demonstrated a convenient, flexible, and modular self-assembly approach for the preparation of supramolecular nanoparticles (SNPs) from a small collection of molecular building blocks through a multivalent molecular recognition based on adamantane (Ad) and β-cyclodextrin (CD) motifs. Such a self-assembly synthetic strategy enables control upon the sizes, surfaces chemistry, zeta potentials, and payloads of the resulting SNPs, which open up many interesting opportunities for biomedical applications, for example, positron emission tomography (PET) imaging,[11] magnetic resonance imaging (MRI),[12] photothermal treatment of cancer cells,[13] and highly efficient gene delivery.[14]
Considering the unique role of TF, we attempted to explore the use of SNPs as a new type of nanoscale vector for delivering intact (unmodified) TFs with an efficiency superior to that of existing approaches. Our idea is to achieve the encapsulation of a TF into cationic SNP vectors by introducing anionic characteristics to the TF. A DNA plasmid with a matching recognition sequence specific to a TF can be employed to form an anionic TF·DNA complex, which can be subsequently encapsulated into SNPs, resulting in TF-encapsulated SNPs (TF·DNA⊂SNPs).
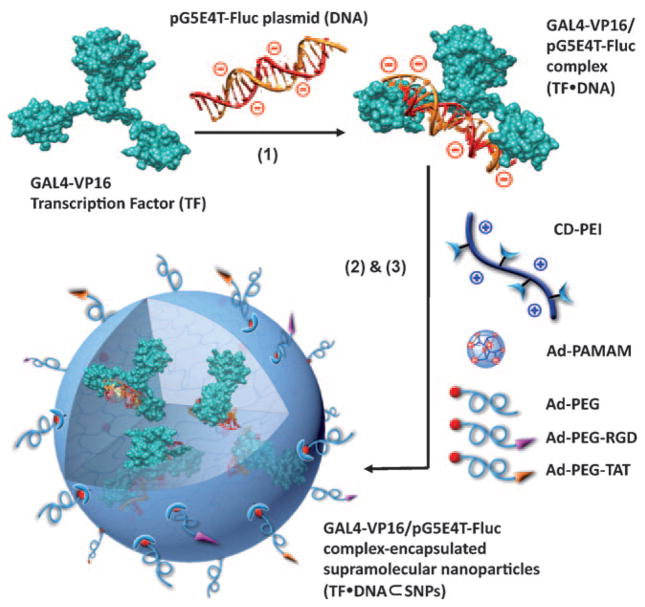
Herein, we introduce a new type of protein delivery system capable of highly efficient transduction of intact TFs. In this proof-of-concept study, a mammalian orthogonal fusion TF, GAL4-VP16 was chosen to serve as a model TF. Since GAL4-VP16 is an artificial transcription factor, there should be no background concentration in the mammalian cells employed in the delivery studies. To facilitate the encapsulation of the model TF into the SNP vectors, a DNA plasmid (i.e., pG5E4T-Fluc) that contains five tandem copies of GAL4-VP16 matching recognition sequences and a conjugated luciferase reporter was designed. The incorporation of multivalent recognition sequences enhances dynamic binding between GAL4-VP16 and pG5E4T-Fluc, allowing improved encapsulation and dynamic releasing of the intact TF. In addition, the conjugated luciferase reporter can be specifically activated by GAL4-VP16, providing a real-time readout reflecting the activities of the TF after its intracellular delivery. As shown in Figure 1, three types of molecular recognition mechanisms were employed to facilitate the preparation of TF-encapsulated SNP (TF·DNA⊂SNPs). First, the specific binding (the dissociate constant Kd ≈ 10 nM)[15] between GAL4-VP16 (TF) and pG5E4T-Fluc (DNA) led to the formation of an anionic TF·DNA complex. Second, the Ad/CD-based molecular recognition (K =1.1 × 105M−1)[16] was utilized to form the SNP vectors with cationic hydrogel cores. Third, electrostatic interactions assist the incorporation of TF·DNA into SNPs to give TF·DNA⊂SNPs. The preparation of TF·DNA⊂SNPs can be accomplished by simply mixing TF·DNA complex with other five functional building blocks (i.e., CD-PEI: CD-grafted branched poly-ethylenimine, Ad-PAMAM: Ad-grafted polyamidoamine dendrimer, Ad-PEG: Ad-grafted polyethylene glycol, Ad-PEG-RGD: Ad-grafted polyethylene glycol with RGD targeting ligand, and Ad-PEG-TAT: Ad-grafted polyethylene glycol with TAT-based CPP). Among the three ligand compounds, Ad-PEG plays a role of a capping/solvation reagent that can not only confine continuous propagation of the TF·DNA-encapsulated PEI/PAMAM hydrogel networks, but also impart desired water solubility, structural stability, and passivation performance to the resulting TF·DNA⊂SNPs. In addition, Ad-PEG-RGD and Ad-PEG-TAT, which were incorporated onto the surfaces of TF·DNA⊂SNPs during the one-pot mixing process,[14b] enable delivery specificity (to recognize a certain population of cells with αvβ3-integrin receptors) and cell transfusion capability (to foster internalization through membrane and releasing from endosome trapping), respectively, of TF·DNA⊂SNPs. The previous study revealed a set of optimal synthetic parameters[14a] that produce DNA-encapsulated SNPs which have good gene transfection performance. Additionally, the results suggested that the presence of both 5% RGD and 9% TAT ligands[17] is a crucial factor in the enhanced efficiency. In this study, we took the advantage of these optimal synthetic parameters for the preparation of TF·DNA⊂SNPs. We were able to demonstrated unprecedented performance for delivery intact TF when TF·DNA⊂SNPs is compared with the conventional CPPs-based protein delivery strategy. Moreover, the intracellular TF delivered by TF·DNA⊂SNPs retained its bioactivity, which was confirmed by monitoring the bioluminescence intensity of TF·DNA⊂SNPs-treated cells.
Figure 1.
Schematic representation of the self-assembly approach for the preparation of transcription factor-incorporated supramolecular nanoparticles (TF·DNA⊂SNPs). Three types of molecular recognition mechanisms, including 1) specific binding between GAL4-VP16 (a mammalian-orthogonal fusion TF) and pG5E4T-Fluc vector (with five tandem copies of GAL4-VP16 matching recognition sequences and a conjugated luciferase reporter) for formation of an anionic TF·DNA complex, 2) the Ad/CD-based molecular recognition for generation of SNP vectors with cationic PEI/PAMAM hydrogel cores, and 3) electrostatic interactions that facilitate incorporation of anionic TF·DNA into SNPs, were harnessed for the self-assembly of TF·DNA⊂SNPs by simply mixing TF·DNA with five functional molecular building blocks: CD-PEI, Ad-PAMAM, Ad-PEG, Ad-PEG-RGD, and Ad-PEG-TAT. See text for details. TAT provides the nanoparticle with the capacity to penetrate cell membranes, RGD with cell targeting, and PEG passivation.
The model plasmid pG5E4T-Fluc and all other molecular building blocks (i.e. CD-PEI, Ad-PAMAM, Ad-PEG, Ad-PEG-RGD, and Ad-PEG-TAT), were synthesized and characterized as described in the Supporting Information. The model transcription factor, GAL4-VP16 was obtained from commercial sources. pG5E4T-Fluc is orthogonal to mammalian genome, thus cannot be activated to express luciferase in the absence of GAL4-VP16.[18] Prior to the preparation of TF·DNA⊂SNP, GAL4-VP16 was incubated with a slight excess amount of pG5E4T-Fluc (GAL4-VP16/pG5E4T-Fluc =1: 0.35 n/n, each pG5E4T-Fluc contains five tandem copies of GAL4-VP16 recognition sequences thus might accommodate more than one TF) for 30 min at 4°C to generate TF·DNA. Subsequently, TF·DNA⊂SNPs were prepared by slowly adding CD-PEI (4.32 μg) in 1 μL phosphate-buffered saline (PBS, pH 7.2) into a 19 μL of PBS solution containing TF·DNA complex (200 ng GAL4-VP16 and 2 μg pG5E4T-Fluc), Ad-PEG (5.94 μg), Ad-PEG-RGD (0.297 μg), Ad-PEG-TAT (0.535 μg), and Ad-PAMAM (0.528 μg). After a brief stirring, the mixture was incubated at 4°C for another 30 min.
To determine hydrodynamic size of the resulting TF·DNA⊂SNPs, we performed dynamic light scattering (DLS) measurements (Figure 2b), indicating a uniform size of (50 ± 3) nm. In parallel, the morphology of TF·DNA⊂SNPs was characterized by transmission electron microscopy (TEM), suggesting homogeneous, narrow size-distributed spherical nanoparticles with size of (40 ± 3) nm (Figure 2a). Finally, the encapsulation rate of TF in TF·DNA⊂SNPs was characterized by quantifying the SNP-encapsulated TF. For the convenience of using a florescence spectroscopy, Cy5-labeled GAL4-VP16 was prepared and employed (see detail procedure in Supporting Information). The result indicated that more than (81 ± 12)% of the TFs was successfully encapsulated into SNPs to give a TF·DNA⊂SNP under the synthetic parameters described above.
Figure 2.
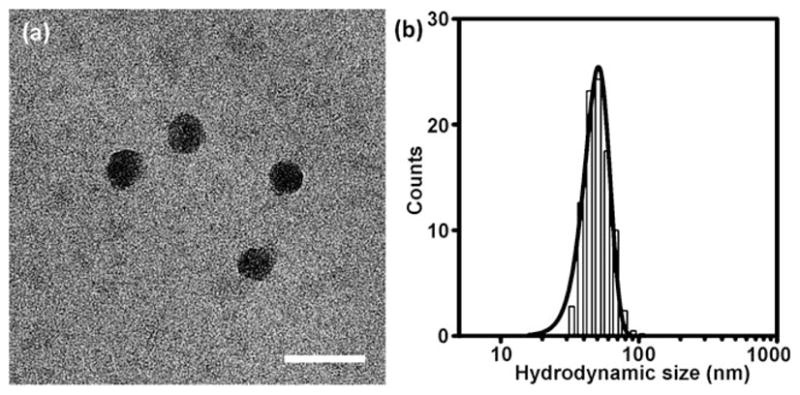
a) Transmission electron microscopy (TEM) micrographs of TF·DNA⊂SNPs. Scale bar: 80 nm. b) Histograms summarize the hydrodynamic size distribution obtained from DLS measurement of (50 ± 3) nm TF·DNA⊂SNPs.
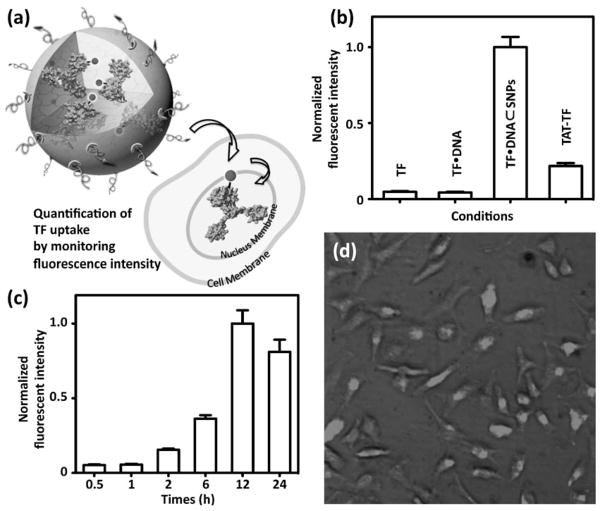
To examine the delivery performance of TF·DNA⊂SNPs, we perform their cell uptake studies using by incubating TF·DNA⊂SNPs (10 ng TF per well) with HeLa cells in a 96-well plate (104 cells per well). Again, GAL4-VP16 was labeled by Cy5 dye to allow quantitative monitoring of the delivery performance of TF·DNA⊂SNPs. Control experiments based on Cy5-labeled-TF alone (TF), Cy5-labeled-TF·DNA complex and Cy5-labeled-TF with TAT-conjugation (TAT-TF) were carried out in parallel under the same experimental conditions. After incubation for various periods (i.e., 0.5, 1, 2, 6, 12, and 24 h) and removal of non-uptaken reagents in the media, the delivery performances of individual studies were quantified by measuring their fluorescence intensities in a plate reader (Fujifilm BAS-5000). As shown in Figure 3b, Cy5-labeled TF·DNA⊂SNPs exhibited dramatically enhanced delivery performance in contrast to those observed in the control studies. It is noteworthy that the delivery efficiency of TF·DNA⊂SNPs was approximately five-times greater than that of TAT-TF, which was commonly used as a standard method for TF delivery. The time-dependent uptake studies (Figure 3c) of TF·DNA⊂SNPs revealed that accumulation of the fluorescence signals increased with the incubation time and reached saturation at 12 h. Fluorescence micrographs (Figure 3d) indicated that localization of Cy5-labeled TF in the cell nuclei, suggesting that the TF molecules were delivered to cell nuclei, where TF functioned as a regulator by controlling the translation of specific gene(s). This result was also confirmed by the co-localization of Cy5-labeled TF and 43,6-diamidino-2-phenylindole (DAPI) stained cell nuclei using fluorescence microscopy (Supporting Information).
Figure 3.
a) Quantification studies on the delivery performance of TF·DNA⊂SNPs. b) Delivery efficiency of Cy5-labeled TF·DNA⊂SNPs, Cy5-labeled-TF alone (TF), Cy5-labeled-TF·DNA complex, and Cy5-labeled-TF with TAT-conjugation (TAT-TF). c) Time-dependent uptake studies of TF·DNA⊂SNPs. d) Fluorescence micrographs of HeLa cells after incubating with TF·DNA⊂SNPs for 12 h. Cy5-labeled TF was localized in the cell nuclei, where TF functioned as a regulator to control the translation of a specific gene.
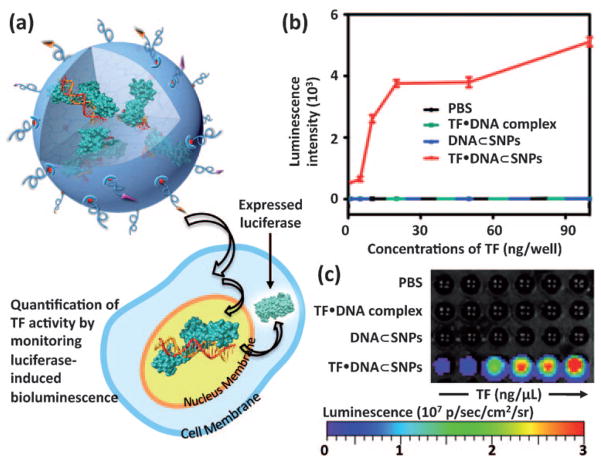
To confirm that the GAL4-VP16 (TF) retained its activity after delivery, we quantified the luciferase expression by measuring the bioluminescence intensity of TF·DNA⊂SNPs-treated cells (Figure 4). Again, the pG5E4T-Fluc (DNA) used in our study contains a luciferase reporter that can be specifically activated by GAL4-VP16. Therefore, the activity of GAL4-VP16 is reflected in the bioluminescence intensity of TF·DNA⊂SNPs-treated cells as a result of luciferase expression. After the incubation of HeLa cells with TF·DNA⊂SNPs and the control reagents (including SNP vector, TF·DNA, and DNA⊂SNPs), the cells were lysed for quantification of bioluminescence. After incubation with luciferin for 2 min, the bioluminescence intensities were recorded by both a plate reader (Figure 4b) and a cooled charge-coupled device (CCD) camera (IVIS, Xenogen; Figure 4c). Compared to the background-level bioluminescence intensities observed from the control experiments, that observed for TF·DNA⊂SNPs-treated cells is significantly higher, suggesting that the GAL4-VP16 retains its activity to trigger the luciferase expression after intracellular delivery. The dose-dependent studies (Figure 4b) indicated that bio-luminescence intensities of the TF·DNA⊂SNPs-treated cells increased with the TF dosages. In addition, we also conducted a set of control studies, where the functional gene (pG5E4T-Fluc) and TF are delivered separately using the respective SNP-based delivery systems at different ratios. We were able to observe very similar bioluminescent outcomes as shown in Figure 4, validating the release of TF from the SNP vector, as well as the dominance of TF amount to the expression level of luciferase (Supporting Information). Moreover, the cell viability assays carried out at different doses of TF·DNA⊂SNPs indicated that the TF·DNA⊂SNPs exhibit negligible toxicity. (Supporting Information)
Figure 4.
a) Bioluminescence study on TF·DNA⊂SNPs-treated cells. The activity of GAL4-VP16 can be reflected in the bioluminescence intensity as a result of luciferase expression. b) Dose-dependent profile and c) bioluminescence imaging of TF·DNA⊂SNPs-treated cells along with the controlled experiments based on TF·DNA complex and DNA⊂SNPs. Error bars in (b) were obtained from three independent experiments.
In conclusion, we have successfully demonstrated the feasibility of applying TF·DNA⊂SNPs for delivery of intact (unmodified) transcription factor (TF) in a highly efficient manner. The uniqueness of our self-assembly synthetic strategy for the preparation of TF·DNA⊂SNPs has to do with the combined use of three types of molecular recognition mechanisms, including 1) specific binding between TF and matching DNA plasmid for formation of an anionic TF·DNA complex, 2) the Ad/CD-based molecular recognition for generation of SNP vectors with cationic hydrogel cores, and 3) electrostatic interactions that facilitate encapsulation of anionic TF·DNA into SNPs. We believe such a TF delivery approach provides a powerful method for manipulating cellular behaviors. A potential application is for generating human induced pluripotent stem cells (hiPSCs), which required the delivery of four reprogramming TFs. We note that, in conjunction with the use of a miniaturized high-throughput screening platform[19] and biological assays,[20] to achieve hiPSCs generation in a highly efficient manner, it is feasible to optimize the ratios of the four reprogramming TFs, something that could be possible through the use of TF·DNA⊂SNPs.
Supplementary Material
Footnotes
This research was supported by National Institutes of Health (NIH), Defense Threat Reducing Agency (DTRA), and National Science Foundation of China, NSFC (50625310 and 50830103).
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201005740.
Contributor Information
Yang Liu, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), Institute for Molecular Medicine (IMED) University of California, Los Angeles 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770 (USA). Department of Chemical & Biomolecular Engineering University of California, Los Angeles 420 Westwood Plaza, BH5573G, Los Angeles, CA 90095 (USA). Laboratory of Functional Polymer Materials, Ministry of Education (China) and Institute of Polymer Chemistry and Physics, Nankai University Tianjin (China).
Dr. Hao Wang, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), Institute for Molecular Medicine (IMED) University of California, Los Angeles 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770 (USA).
Dr. Ken-ichiro Kamei, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), Institute for Molecular Medicine (IMED) University of California, Los Angeles 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770 (USA). Institute for Integrated Cell-Material Sciences, Kyoto University (Japan)
Dr. Ming Yan, Department of Chemical & Biomolecular Engineering University of California, Los Angeles 420 Westwood Plaza, BH5573G, Los Angeles, CA 90095 (USA)
Kuan-Ju Chen, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), Institute for Molecular Medicine (IMED) University of California, Los Angeles 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770 (USA).
Qinghua Yuan, Institute for Integrated Cell-Material Sciences, Kyoto University (Japan).
Prof. Linqi Shi, Laboratory of Functional Polymer Materials, Ministry of Education (China) and Institute of Polymer Chemistry and Physics, Nankai University Tianjin (China).
Prof. Yunfeng Lu, Department of Chemical & Biomolecular Engineering University of California, Los Angeles 420 Westwood Plaza, BH5573G, Los Angeles, CA 90095 (USA).
Prof. Hsian-Rong Tseng, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), Institute for Molecular Medicine (IMED) University of California, Los Angeles 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770 (USA).
References
- 1.a) Cross BCS, Sinning I, Luirink J, High S. Nat Rev Mol Cell Biol. 2009;10:255–264. doi: 10.1038/nrm2657. [DOI] [PubMed] [Google Scholar]; b) Brasn-jevic I, Steinbusch HWM, Schmitz C, Martinez-Martinez P. Prog Neurobiol. 2009;87:212–251. doi: 10.1016/j.pneurobio.2008.12.002. [DOI] [PubMed] [Google Scholar]; c) Leader B, Baca QJ, Golan DE. Nat Rev Drug Discovery. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]; d) Tessmar JK, Goepferich AM. Adv Drug Delivery Rev. 2007;59:274–291. doi: 10.1016/j.addr.2007.03.020. [DOI] [PubMed] [Google Scholar]; e) Lee KY, Yuk SH. Prog Polym Sci. 2007;32:669–697. [Google Scholar]; f) Galan JE, Wolf-Watz H. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]; g) Kost T, Condreay J, Jarvis D. Nat Biotechnol. 2005;23:567–575. doi: 10.1038/nbt1095. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Muth T, Caplan M. Annu Rev Cell Dev Biol. 2003;19:333–366. doi: 10.1146/annurev.cellbio.19.110701.161425. [DOI] [PubMed] [Google Scholar]; i) Ford KG, Souberbielle BE, Darling D, Farzaneh F. Gene Ther. 2001;8:1–4. doi: 10.1038/sj.gt.3301383. [DOI] [PubMed] [Google Scholar]; j) Cao Z, Tong R, Mishra A, Xu W, Wong G, Cheng J, Lu Y. Angew Chem. 2009;121:6616–6620. doi: 10.1002/anie.200901452. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2009;48:6494–6498. doi: 10.1002/anie.200901452. [DOI] [PubMed] [Google Scholar]
- 2.a) Tokuriki N, Tawfik DS. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]; b) Haidar ZS, Hamdy RC, Tabrizian M. Biomaterials. 2008;29:1207–1215. doi: 10.1016/j.biomaterials.2007.11.012. [DOI] [PubMed] [Google Scholar]; c) Frokjaer S, Otzen DE. Nat Rev Drug Discovery. 2005;4:298–306. doi: 10.1038/nrd1695. [DOI] [PubMed] [Google Scholar]
- 3.a) Rezler EM, Khan DR, Lauer-Fields J, Cudic M, Baronas-Lowell D, Fields GB. J Am Chem Soc. 2007;129:4961–4972. doi: 10.1021/ja066929m. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cheong I, Huang X, Bettegowda C, Diaz LA, Kinzler KW, Zhou S, Vogelstein B. Science. 2006;314:1308–1311. doi: 10.1126/science.1130651. [DOI] [PubMed] [Google Scholar]; c) Torchilin VP. Nat Rev Drug Discovery. 2005;4:145–160. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- 4.a) Lee Y, Ishii T, Kim H, Nishiyama N, Hayakawa Y, Itaka K, Kataoka K. Angew Chem. 2010;122:2606–2609. doi: 10.1002/anie.200905264. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2010;49:2552–2555. doi: 10.1002/anie.200905264. [DOI] [PubMed] [Google Scholar]; b) De Cock LJ, De Koker S, De Geest BG, Grooten J, Vervaet C, Remon JP, Sukhorukov GB, Antipina MN. Angew Chem. 2010;122:7108–7127. doi: 10.1002/anie.200906266. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2010;49:6954–6973. doi: 10.1002/anie.200906266. [DOI] [PubMed] [Google Scholar]; c) van Dongen SFM, Verdurmen WPR, Peters RJRW, Nolte RJM, Brock R, van Hest JCM. Angew Chem. 2010;122:7371–7374. [Google Scholar]; Angew Chem Int Ed. 2010;49:7213–7216. doi: 10.1002/anie.201002655. [DOI] [PubMed] [Google Scholar]; d) Amidi M, Mastrobattista E, Jiskoot W, Hennink WE. Adv Drug Delivery Rev. 2010;62:59–82. doi: 10.1016/j.addr.2009.11.009. [DOI] [PubMed] [Google Scholar]; e) Lee Y, Ishii T, Cabral H, Kim H, Seo JH, Nishiyama N, Oshima H, Osada K, Kataoka K. Angew Chem. 2009;121:5413–5416. doi: 10.1002/anie.200900064. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2009;48:5309–5312. doi: 10.1002/anie.200900064. [DOI] [PubMed] [Google Scholar]; f) George M, Abraham TE. J Controlled Release. 2006;114:1–14. doi: 10.1016/j.jconrel.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 5.a) Yan M, Du J, Gu Z, Liang M, Hu Y, Zhang W, Priceman S, Wu L, Zhou ZH, Liu Z, Segura T, Tang Y, Lu Y. Nat Nanotechnol. 2010;5:48–53. doi: 10.1038/nnano.2009.341. [DOI] [PubMed] [Google Scholar]; b) Ghosh P, Yang X, Arvizo R, Zhu ZJ, Agasti SS, Mo Z, Rotello VM. J Am Chem Soc. 2010;132:2642–2645. doi: 10.1021/ja907887z. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ghosh P, Yang XC, Arvizo R, Zhu ZJ, Agasti SS, Mo ZH, Rotello VM. J Am Chem Soc. 2010;132:2642–2645. doi: 10.1021/ja907887z. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Li H, Ma Y, Chen Y, Sang Y, Zhou T, Qiu M, Huang X, Zhou C, Su Z. Angew Chem. 2010;122:5050–5053. doi: 10.1002/anie.201000287. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2010;49:4930–4933. doi: 10.1002/anie.201000287. [DOI] [PubMed] [Google Scholar]; e) Skwarczynski M, Zaman M, Urbani C, Lin IC, Jia Z, Batzloff M, Good M, Monteiro M, Toth I. Angew Chem. 2010;122:5878–5881. doi: 10.1002/anie.201002221. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2010;49:5742–5745. doi: 10.1002/anie.201002221. [DOI] [PubMed] [Google Scholar]; f) Huang Y, Park Y, Moon C, David A, Chung H, Yang V. Angew Chem. 2010;122:2784–2787. doi: 10.1002/anie.200906153. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2010;49:2724–2727. doi: 10.1002/anie.200906153. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Giljohann D, Seferos D, Daniel W, Massich M, Patel P, Mirkin C. Angew Chem. 2010;122:3352–3366. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2010;49:3280–3294. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Liu J, Stace-Naughton A, Jiang X, Brinker CJ. J Am Chem Soc. 2009;131:1354–1355. doi: 10.1021/ja808018y. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Ghosh P, Han G, De M, Kim CK, Rotello VM. Adv Drug Delivery Rev. 2008;60:1307–1315. doi: 10.1016/j.addr.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 6.Fothergill-Gilmore LA. In: Protein Biotechnology. Franks F, editor. Humana; New York: 1993. pp. 467–487. [Google Scholar]
- 7.a) Asoh S, Ohta S. Adv Drug Delivery Rev. 2008;60:499–516. doi: 10.1016/j.addr.2007.09.011. [DOI] [PubMed] [Google Scholar]; b) Fittipaldi A, Giacca M. Adv Drug Delivery Rev. 2005;57:597–608. doi: 10.1016/j.addr.2004.10.011. [DOI] [PubMed] [Google Scholar]; c) Wadia J, Dowdy S. Adv Drug Delivery Rev. 2005;57:579–596. doi: 10.1016/j.addr.2004.10.005. [DOI] [PubMed] [Google Scholar]; d) Abbing A, Blaschke UK, Grein S, Kretschmar M, Stark CMB, Thies MJW, Walter J, Weigand M, Woith DC, Hess J, Reiser COA. J Biol Chem. 2004;279:27410–27421. doi: 10.1074/jbc.M313612200. [DOI] [PubMed] [Google Scholar]; e) Console S, Marty C, Garc%a-Echeverr%a C, Schwendener R, Ballmer-Hofer K. J Biol Chem. 2003;278:35109–35114. doi: 10.1074/jbc.M301726200. [DOI] [PubMed] [Google Scholar]
- 8.Kumagai Y, Sumi D. Annu Rev Pharmacol Toxicol. 2007;47:243–262. doi: 10.1146/annurev.pharmtox.47.120505.105144. [DOI] [PubMed] [Google Scholar]
- 9.Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Maherali N, Hochedlinger K. Cell Stem Cell. 2008;3:595–605. doi: 10.1016/j.stem.2008.11.008. [DOI] [PubMed] [Google Scholar]; b) Takahashi K, Yamanaka S. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Wang S, Su H, Chen KJ, Armijo AL, Lin WY, Wang Y, Sun J, Kamei KI, Czernin J, Radu CG, Tseng HR. Angew Chem. 2009;121:4408–4412. [Google Scholar]; Angew Chem Int Ed. 2009;48:4344–4348. doi: 10.1002/anie.200900063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen KJ, Wolahan SM, Wang H, Hsu CH, Chang HW, Durazo A, Hwang LP, Garcia MA, Jiang ZK, Wu L, Lin YY, Tseng HR. Biomaterials. 2010;32:2160–2165. doi: 10.1016/j.biomaterials.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang S, Chen KJ, Wu TH, Wang H, Lin WY, Ohashi M, Chiou PY, Tseng HR. Angew Chem. 2010;122:3865–3869. doi: 10.1002/anie.201000062. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2010;49:3777–3781. doi: 10.1002/anie.201000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Wang H, Liu K, Chen KJ, Lu Y, Wang S, Lin WY, Guo F, Kamei K, Chen YC, Ohashi M, Wang M, Zhao XZ, Shen CKF, Tseng HR. ACS Nano. 2010;4:6235–6243. doi: 10.1021/nn101908e. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang H, Chen KJ, Wang S, Ohashi M, Kamei KI, Sun J, Ha JH, Liu K, Tseng HR. Chem Commun. 2010;46:1851–1853. doi: 10.1039/b923711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodgers KK, Coleman JE. Protein Sci. 1994;3:608–619. doi: 10.1002/pro.5560030409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a) Ludden MJW, Sinha JK, Wittstock G, Reinhoudt DN, Huskens J. Org Biomol Chem. 2008;6:1553–1557. doi: 10.1039/b718940k. [DOI] [PubMed] [Google Scholar]; b) Rekharsky MV, Inoue Y. Chem Rev. 1998;98:1875–1918. doi: 10.1021/cr970015o. [DOI] [PubMed] [Google Scholar]
- 17.According to the previous study, the optimal delivery efficiency was achieved when the DNA⊂SNP surface was covered with 5% RGD targeting ligand and 9% TAT CPPs. For details see Ref. [14a].
- 18.Sadowski I, Ma J, Triezenberg S, Ptashne M. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 19.a) Lee CC, Sui G, Elizarov A, Shu CJ, Shin YS, Dooley AN, Huang J, Daridon A, Wyatt P, Stout D, Kolb HC, Witte ON, Satyamurthy N, Heath JR, Phelps ME, Quake SR, Tseng HR. Science. 2005;310:1793–1796. doi: 10.1126/science.1118919. [DOI] [PubMed] [Google Scholar]; b) Wang J, Sui G, Mocharla VP, Lin RJ, Phelps ME, Kolb HC, Tseng HR. Angew Chem. 2006;118:5402–5407. doi: 10.1002/anie.200601677. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2006;45:5276–5281. doi: 10.1002/anie.200601677. [DOI] [PubMed] [Google Scholar]; c) Lin WY, Wang Y, Wang S, Tseng HR. Nano Today. 2009;4:470–481. doi: 10.1016/j.nantod.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Wang Y, Lin WY, Liu K, Lin RJ, Selke M, Kolb HC, Zhang N, Zhao XZ, Phelps ME, Shen CKF, Faull KF, Tseng HR. Lab Chip. 2009;9:2281–2285. doi: 10.1039/b907430a. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Liu K, Wang H, Chen KJ, Guo F, Lin WY, Chen YC, Phung DL, Tseng HR, Shen CK. Nanotechnology. 2010;21:445603–445608. doi: 10.1088/0957-4484/21/44/445603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.a) Kamei K-i, Guo S, Yu ZTF, Takahashi H, Gschweng E, Suh C, Wang X, Tang J, McLaughlin J, Witte ON, Lee K-B, Tseng H-R. Lab Chip. 2009;9:555–563. doi: 10.1039/b809105f. [DOI] [PubMed] [Google Scholar]; b) Kamei K-i, Ohashi M, Gschweng E, Ho Q, Suh J, Tang J, For Yu ZT, Clark AT, Pyle AD, Teitell MA, Lee K-B, Witte ON, Tseng H-R. Lab Chip. 2010;10:1113–1119. doi: 10.1039/b922884e. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Sun J, Masterman-Smith MD, Graham NA, Jiao J, Mottahedeh J, Laks DR, Ohashi M, DeJesus J, Kamei K-i, Lee K-B, Wang H, Yu ZTF, Lu Y-T, Hou S, Li K, Liu M, Zhang N, Wang S, Angenieux B, Panosyan E, Samuels ER, Park J, Williams D, Konkankit V, Nathanson D, van Dam RM, Phelps ME, Wu H, Liau LM, Mischel PS, Lazareff JA, Kornblum HI, Yong WH, Graeber TG, Tseng H-R. Cancer Res. 2010;70:6128–6138. doi: 10.1158/0008-5472.CAN-10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.