Abstract
The adaptive resistance of Pseudomonas aeruginosa to aminoglycosides is known to occur during chronic lung infections in cystic fibrosis patients in response to nonlethal concentrations of aminoglycosides. Not only is it difficult to achieve high levels of drug throughout the dehydrated mucus in the lung, but also steep oxygen gradients exist across the mucus layer, further reducing the bactericidal activity of aminoglycosides. In this study, microarray analysis was utilized to examine the gene responses of P. aeruginosa to lethal, inhibitory, and subinhibitory concentrations of tobramycin under aerobic and anaerobic conditions. While prolonged exposure to subinhibitory concentrations of tobramycin caused increased levels of expression predominantly of the efflux pump genes mexXY, the greatest increases in gene expression levels in response to lethal concentrations of tobramycin involved a number of heat shock genes and the PA0779 gene (renamed here asrA), encoding an alternate Lon protease. Microarray analysis of an asrA::luxCDABE transposon mutant revealed that the induction of heat shock genes in response to tobramycin in this mutant was significantly decreased compared to that in the parent strain. The level of expression of asrA was induced from an arabinose-inducible promoter to 35-fold greater than wild-type expression levels in the absence of tobramycin, and this overexpression alone caused an increased expression of the heat shock genes, as determined by quantitative PCR (qPCR). This overexpression of asrA conferred short-term protection against lethal levels (4 μg/ml) of tobramycin but did not affect the tobramycin MIC. The RpoH heat shock sigma factor was found to be involved in the regulation of asrA in response to both heat shock and tobramycin at the posttranscriptional level. The results of this work suggest that the tobramycin concentration has a significant impact on the gene expression of P. aeruginosa, with lethal concentrations resulting in immediate adaptations conferring short-term protection, such as the induction of the heat shock response, and with subinhibitory concentrations leading to more sustainable long-term protection mechanisms, such as increased efflux.
INTRODUCTION
Adaptive resistance to antimicrobials can be defined as conditional and reversible resistance that occurs due to prevailing growth conditions (including the presence of subeffective antibiotic doses) and is increasingly being recognized as contributing to treatment outcomes (23). Adaptive resistance to aminoglycosides is a major concern for cystic fibrosis (CF) patients undergoing long-term therapy with the aerosolized aminoglycoside tobramycin. The exposure of localized microcolonies of Pseudomonas aeruginosa within the thick dehydrated mucus in the CF lung to subinhibitory and bacteriostatic concentrations of tobramycin provides the organism with the opportunity to adapt to the presence of this antibiotic. Indeed, it has been observed that genotypically related and identical bacteria with a variety of phenotypes can be isolated simultaneously from the lungs of CF patients chronically infected with P. aeruginosa (14, 16, 26, 33). This adaptation may reflect in part the increased frequency of hypermutator strains present in the CF lung; however, adaptive resistance of P. aeruginosa to aminoglycosides in the absence of mutations has been noted, and this type of resistance is more difficult to characterize in vivo. Nonetheless, a number of conditions occurring in the CF lung predispose P. aeruginosa to undergo adaptation. Importantly, although aminoglycosides such as tobramycin have potent bactericidal activity under aerobic conditions, this activity is significantly reduced under low-oxygen conditions (3, 13, 18). This reduction of bactericidal activity in hypoxic and anoxic environments is of relevance for the CF lung, where P. aeruginosa resides within the lower levels of the steep oxygen gradients existing across the thickened mucus layer (44).
The most commonly observed aminoglycoside adaptive resistance phenotype of P. aeruginosa is an impermeability phenotype. This phenotype has been attributed largely to the induction of the MexXY-OprM tripartite efflux pump, although the upregulation of MexXY-OprM does not account for all aminoglycoside adaptive resistance phenotypes observed (15, 41). In an attempt to identify factors that may contribute to these aminoglycoside resistance phenotypes, we recently identified a large number of factors that are capable of contributing to aminoglycoside resistance in vitro (39), and work reported previously by others demonstrated a cumulative effect on the MIC of several of these factors in double, triple, and quadruple mutants (12), suggesting that these factors are capable of contributing to resistance through independent mechanisms; however, contributions to adaptive resistance were not investigated in either study.
Moreover, the issue of adaptive resistance to aminoglycosides is complicated by the lack of an understanding of the mechanism of bactericidal activity of aminoglycosides. Early research ascribed the action of aminoglycosides to their interaction with ribosomes, either causing misreading and the incorporation of defective channel-forming proteins into the cytoplasmic membrane or inhibiting the association of the origin of initiation of DNA synthesis with the membrane, causing a defect in the partitioning of daughter chromosomes during cell division (11, 27, 42). Conversely, Kohanski et al. (19) recently proposed a mechanism of lethal action of aminoglycosides and other antibiotics in Escherichia coli involving hydroxyl radicals produced by the hyperactivation of the tricarboxylic acid (TCA) cycle and the electron transport chain. Components of the electron transport chain and the TCA cycle were found to be upregulated within 30 min after treatment with the aminoglycosides gentamicin and kanamycin (20). This response was mediated by the sensor kinase CpxA and the transcriptional regulator ArcA in response to envelope stress (20). In P. aeruginosa, the involvement of hydroxyl radicals has not been investigated, and no CpxA or ArcA homologues have been identified.
The purpose of this work was to characterize the effects of subinhibitory, inhibitory, and lethal concentrations of tobramycin on gene expression in P. aeruginosa in an attempt to understand the nature of the adaptations that occur in response to this drug. We utilized aerobic and anaerobic conditions in this study, as this allowed a direct comparison of responses to the inhibitory and lethal concentrations of tobramycin using the same concentrations to elicit both responses and also reflected a range of conditions more closely resembling those encountered within the CF lung.
MATERIALS AND METHODS
Bacterial strains and plasmids.
P. aeruginosa strains used included wild-type strain PAO1 and transposon mutants rpoH::IS;phoA/hah-Tcr, from the University of Washington mutant library (17), and PAO1_lux_15_F1, a PA0779 mutant from the PAO1 mini-Tn5-luxCDABE transposon mutant library (24); the latter gene locus has been renamed here “asrA,” for “aminoglycoside-induced stress response ATP-dependent protease,” and the mutant is termed the asrA::lux mutant. One Shot TOP10 chemically competent E. coli cells (Invitrogen) and plasmid pHERD20T (36) were used for cloning experiments.
Tobramycin susceptibility assays.
MIC determinations were performed with BBL cation-adjusted Mueller-Hinton broth (CAMHB) for aerobic susceptibility testing by broth microdilution according to CLSI protocols (10). Under anaerobic conditions, MIC determinations were performed with CAMHB according to the broth macrodilution procedure described by the CLSI (10), with modifications including placing cultures into anaerobic jars containing a BBL GasPak Plus Disposable Hydrogen + Carbon Dioxide Generator Envelope (Becton Dickinson Biosciences) and supplementing the medium with 15 mM KNO3. Bactericidal concentrations were determined by spread plating 100 μl that was taken from each tobramycin dilution from the MIC assay plate after 20 h of incubation at 37°C. The bactericidal concentration was recorded as the lowest concentration at which no colonies formed.
Tobramycin kill curves were performed with cultures grown in CAMHB supplemented with 15 mM KNO3 to an optical density at 600 nm (OD600) of 0.5 to 0.6. Fifty-microliter samples were serially diluted in ice-cold CAMHB and plated at time zero to obtain the starting CFU/ml. Tobramycin was added at concentrations of either 1, 2, 4, or 20 μg/ml, and killing was assessed at specific time points by counting colonies and calculating percent survival relative to untreated cells at time zero.
To assess the growth of P. aeruginosa in the presence of tobramycin, cultures of P. aeruginosa PAO1 and the rpoH mutant grown overnight were diluted 1:100 in CAMHB with or without tobramycin at concentrations of 0.125, 0.25, 0.5, and 1 μg/ml. Two-hundred-milliliter aliquots were inoculated into 96-well microtiter plates and were then grown at 37°C with shaking in a Tecan Spectrofluor Plus instrument. The absorbance at 620 nm was monitored every 20 min for 18 h. This experiment was independently carried out three times with duplicate cultures of each strain.
Microarray conditions, slide preparation, and analysis.
Five separate microarray experiments were performed with 3 to 5 replicates assayed for each comparison. For each experiment described below, cultures grown overnight were diluted 1:50 into the appropriate medium and incubated with shaking at 37°C. Twelve-milliliter samples of a mid-log-phase culture at an OD600 of 0.5 to 0.6 were used for harvesting cells for the untreated controls. All cells were harvested by centrifugation at 5,000 × g at 4°C. RNA was isolated by using the RNeasy Midi kit (Qiagen) according to the manufacturer's instructions. Microarray hybridization and analysis were performed by using P. aeruginosa PAO1 DNA microarray epoxy-coated slides from The Institute for Genomic Research (TIGR) (now the J. Craig Venter Institute) Pathogenic Functional Genomics Resource Center and ArrayPipe (version 1.7), as previously described (32, 46). Validation of the microarray data was performed by using quantitative PCR (qPCR), as previously described (28), for select genes with altered expression from each microarray experiment, and a paired one-tailed Student's t test was used to assess significance.
A subinhibitory aerobic tobramycin microarray was performed to compare the gene expression of P. aeruginosa PAO1 grown to mid-log phase in CAMHB in the presence of 0.25 μg/ml tobramycin to expression occurring in the absence of tobramycin. A lethal/bactericidal aerobic tobramycin microarray compared the expression of P. aeruginosa PAO1 after a 30-min treatment with 2 μg/ml tobramycin to its expression prior to the addition of tobramycin. The CAMHB used for this experiment was supplemented with 15 mM KNO3 for the consistency of comparisons to subsequent anoxic microarrays. Anaerobic inhibitory and lethal microarray experiments each compared P. aeruginosa PAO1 gene expression occurring under anoxic conditions after a 30-min treatment with either 2 μg/ml or 20 μg/ml tobramycin, respectively, to PAO1 gene expression under anaerobic conditions in the absence of tobramycin. Anaerobic growth of cultures grown overnight was achieved by placing culture tubes into an anaerobic jar containing a BBL GasPak Plus Disposable Hydrogen + Carbon Dioxide Generator Envelope (Becton Dickinson Biosciences). Dilutions were performed with 125-ml flasks filled to within 2 cm of the brim with CAMHB supplemented with 15 mM KNO3 and plugged with rubber stoppers. To minimize the introduction of air, a syringe was used to remove control samples and to add tobramycin to the remaining cultures. A lethal tobramycin microarray of the asrA::lux mutant was performed to compare its expression in CAMHB supplemented with KNO3 after a 30-min treatment with 2 μg/ml tobramycin to its expression in the absence of tobramycin.
Plasmid construction and complementation.
Genomic DNA from P. aeruginosa PAO1 was isolated by using the PureLink genomic DNA minikit (Invitrogen). The entire PA0779 gene was PCR amplified from the isolated genomic DNA using Phusion high-fidelity DNA polymerase (New England BioLabs) and primers (AlphaDNA) containing flanking XbaI and HindIII restriction sites. The PCR amplicon and Escherichia-Pseudomonas shuttle vector pHERD20T were restricted with XbaI and HindIII, ligated, and transformed into E. coli. Blue-white selection was carried out with Luria-Bertani broth (LB) containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), isopropyl-β-d-1-thiogalactopyranoside (IPTG), and 100 μg/ml ampicillin. Plasmids were isolated by using a QIAprep Spin miniprep kit (Qiagen) and were verified by restriction analyses with EcoRV, XbaI, and HindIII (Invitrogen). The confirmed constructs were transformed by electroporation (9) into PAO1 and its asrA::lux mutant and selected with 300 μg/ml carbenicillin. Empty pHERD20T plasmids were also transformed via electroporation into PAO1 and the asrA::lux mutant for controls.
Overexpression of asrA.
The overexpression of the asrA gene from the araC-regulated PBAD promoter of pHERD20T was achieved by the addition of 1% arabinose to mid-log-phase cultures grown in CAMHB supplemented with 15 mM KNO3 and 300 μg/ml carbenicillin. qPCR was performed, as previously described (28), to compare the relative expression levels of asrA and the heat shock genes htpG, ibpA, groES, clpB, dnaJ, and hslV before and after a 30-min treatment with 1% arabinose. The effect of the overexpression of PA0779 on tobramycin susceptibility was assessed by comparisons of percent survivals of PAO1 complemented with pHERD20T::asrA and pHERD20T and treated with 1% arabinose for 30 min prior to the addition of 4 μg/ml tobramycin. Significant increases in percent survival and expression of heat shock genes in response to the overexpression of asrA were determined by using a one-tailed, paired Student's t test.
Induction of stress responses using heat shock and tobramycin.
For heat shock and antibiotic-induced stress studies, 4-ml cultures were prepared in CAMHB and grown to an OD600 of 0.5 to 0.6. Heat shock was induced by the incubation of the mid-log-phase cultures at 42°C for 10 min, and antibiotic stress was induced by using a 2-μg/ml tobramycin treatment for 30 min. RNA was isolated from three independent cultures using the RNeasy minikit (Qiagen) and utilized for reverse transcription-qPCR (RT-qPCR). RT-qPCR was performed for assessing the gene expressions of asrA, dnaK, and rpoH.
Microarray data accession numbers.
Array Express accession numbers for the microarrays presented in this study are E-FPMI-24 to E-FPMI-27.
RESULTS
P. aeruginosa PAO1 susceptibility to tobramycin under aerobic and anaerobic conditions.
Under aerobic conditions the MIC and bactericidal concentration of tobramycin for P. aeruginosa strain PAO1 were both 1 μg/ml. In an anaerobic environment, tobramycin activity was bacteriostatic, with an MIC for PAO1 of 1 μg/ml but a bactericidal concentration of 16 μg/ml when nitrate was provided as a terminal electron acceptor. In the absence of nitrate, killing was completely abolished, and tobramycin concentrations as high as 64 μg/ml were not bactericidal.
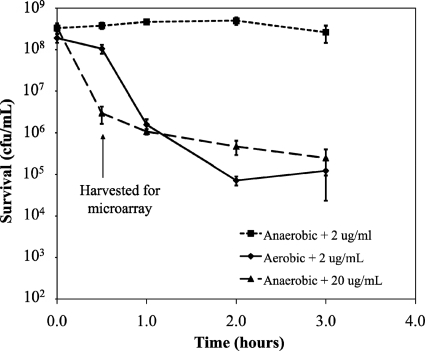
Under aerobic conditions, kill curves using 2 μg/ml of tobramycin (2× the MIC) demonstrated a 30-min lag phase prior to a phase of rapid cell death (Fig. 1). Under anaerobic conditions with 2 μg/ml of tobramycin, an inhibition of growth without apparent cell death was observed over a period of 3 h (Fig. 1). The treatment of PAO1 with a tobramycin concentration of 20 μg/ml under anaerobic conditions resulted in a level of killing similar to that seen with aerobically grown cells treated with 2 μg/ml tobramycin. Based on these data, cells were harvested at 30 min posttreatment for both the inhibitory and lethal microarrays.
Fig. 1.
Effect of anaerobiosis on tobramycin activity against P. aeruginosa PAO1. Log-phase cultures of PAO1 grown in CAMHB plus 15 mM KNO3 at 37°C were treated with tobramycin. Error bars represent standard deviations for five replicates.
Patterns of altered gene expression in P. aeruginosa strain PAO1 in response to lethal and inhibitory concentrations of tobramycin.
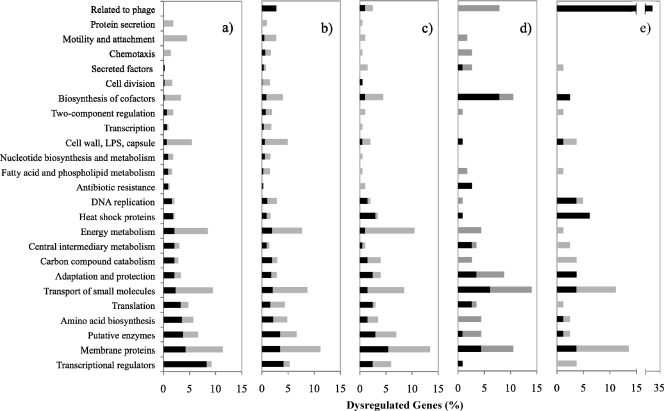
The gene expression of PAO1 was assessed by microarray analysis after 30 min of treatment with tobramycin at concentrations of 2 μg/ml under aerobic conditions and 20 μg/ml under anaerobic conditions. These concentrations were bactericidal, with a typical lag before the killing of PAO1 allowing the harvest of a sufficient number of bacteria for microarray analysis. Under aerobic conditions, the expression of 422 genes was affected, with 205 genes being upregulated and 217 genes being downregulated (see Table S1 in the supplemental material). Under the anaerobic conditions tested, the expression of 1,058 genes was affected, with 477 genes being upregulated and 581 genes being downregulated (see Table S1 in the supplemental material). While the total number of genes affected under anaerobic conditions was more than double the number observed under aerobic conditions, the general patterns of altered gene expression, as assessed by the distribution of functional classes (Fig. 2a and b), were similar between the two conditions. Under lethal conditions, the functional classes representing the majority of downregulated genes were related to energy metabolism, the transport of small molecules, and membrane proteins, while those most upregulated were transcriptional regulators. Genes classified as being involved in the heat shock response were very highly upregulated in response to lethal concentrations of tobramycin but were either not upregulated or only moderately upregulated in response to bacteriostatic concentrations. The asrA gene (PA0799), encoding a protein with 60% similarity (41% identity) to the Lon protease (PA1803), was also found to be upregulated in response to both bacteriostatic and lethal concentrations of tobramycin and was the most highly upregulated regulatory gene in the lethal microarrays.
Fig. 2.
PseudoCAP functional class distribution of nonhypothetical genes with altered expression in response to tobramycin. (a) PAO1 lethal aerobic. (b) PAO1 lethal anaerobic. (c) asrA::lux mutant lethal aerobic. (d) PAO1 subinhibitory aerobic. (e) PAO1 inhibitory anaerobic. Black bars indicate upregulation, and gray bars indicate downregulation. LPS, lipopolysaccharide.
Among the downregulated energy metabolism genes in the lethal microarrays were a number of genes that were shown previously to increase tobramycin resistance when insertionally inactivated (39). These genes included the genes of the nuo operon, a number of cytochrome genes, and a variety of dehydrogenases, oxidoreductases, and denitrification-related genes (Table 1). Genes involved in motility and attachment were also predominantly downregulated.
Table 1.
Comparison of the extents of altered expression of selected genes and operons observed for tobramycin microarrays and resistant mutant screense
| Gene or operon | Extent of altered regulationa |
TOB resistance geneb | ||||
|---|---|---|---|---|---|---|
| Aerobic WT (TOB at 2 μg/ml) | Anaerobic WT (TOB at 20 μg/ml) | Aerobic WT (TOB at 0.25 μg/ml) | Anaerobic WT (TOB at 2 μg/ml) | Aerobic PA0779 (asrA::lux) (TOB at TOB 2 μg/ml) | ||
| Heat shock | ||||||
| htpG-PA1597 | +++ | +++ | + | + | ||
| ibpA | ++++ | ++++ | + | + | ++ | |
| groES groEL | ++ | ++ | + | |||
| dnaJ dapB | ++ | +++ | + | + | ||
| grpE dnaK | +++ | +++ | + | + | ||
| hslVU-PA5055 | ++ | +++ | + | |||
| Energy metabolism | ||||||
| PA1547-PA1551 | − | − | Yes | |||
| ccoN1O1Q1P1 | − | − | ||||
| ccoN2O2Q2P2 | −− | + | Yes | |||
| sdhCDAB | − | |||||
| sucAB lpdG | − | |||||
| sucCD | − | Yes | ||||
| PA1600-PA1602 | −− | −−− | − | |||
| nuoABDEFGHIJKLMN | − | −−− | Yes | |||
| nqrABCDEF | − | − | Yes | |||
| Denitrification/nitrogen metabolism | ||||||
| nirSMCFDLH-PA0512-nirJEN | −− | −− | − | − | ||
| nirQ-PA0521-PA0522 | + | |||||
| norCB | + | − | − | |||
| norD | − | Yes | ||||
| nosRZDFYL | − | −− | −− | − | Yes | |
| narK1K2GHJI-PA3871 | − | −−− | −−− | −− | ||
| narXL | − | + | ||||
| Other | ||||||
| asrA | ++ | ++ | + | |||
| pqsABCDEphnA | + | |||||
| mexXY | +++ | |||||
| hcnABC | − | − | + | |||
| Pilus operonsc | − | −− | ||||
| Flagellin operonsd | − | − | ||||
−, downregulated 2- to 4-fold; −−, downregulated 5- to 10-fold; −−−, downregulated ≥10-fold; +, upregulated 2- to 9-fold; ++, upregulated 10- to 20-fold; +++, upregulated 20- to 50-fold; ++++, upregulated ≥50-fold. TOB, tobramycin; WT, wild type.
Determined to be resistant in a screen of inactivation mutants reported previously by Schurek et al. (39).
pilJK chpABCD pilB pilCD coaE-PA4530 fimU pilVWXY1Y2E PA4958-fimX.
flgFGHIJK PA1442-fliMNOPQR flhB.
The full list of dysregulated genes can be found in Table S1 in the supplemental material.
In contrast to these results, in the bacteriostatic anaerobic microarray, the expression of 81 genes was altered, with 50 genes being upregulated and 31 being downregulated (see Table S1 in the supplemental material). The distribution of the functional classes of affected genes in the inhibitory microarray was distinctly different from those in the lethal microarrays (Fig. 2e), although membrane protein genes and genes related to the transport of small molecules were largely downregulated, as was seen under lethal conditions.
Changes in gene expression of P. aeruginosa strain PAO1 in response to subinhibitory concentrations of tobramycin.
A total of 114 genes had at least a 2-fold change in expression in response to prolonged aerobic exposure to 0.25 μg/ml tobramycin, with 66 genes being downregulated and 48 genes being upregulated (see Table S1 in the supplemental material). Similarly to the lethal microarrays performed for cultures treated for only 30 min with tobramycin, the most downregulated genes were involved in energy metabolism, particularly those involved in nitrogen metabolism, and the aldehyde dehydrogenase and oxidoreductase genes in the PA1600-PA1602 operon. In contrast, several upregulated operons were associated with pathogenesis and demonstrated multiple genes being upregulated. These included pqsABCDE (Pseudomonas quinolone signal) and hcnABC (hydrogen cyanide [HCN]). The most highly upregulated genes (9- to 47-fold) encoded the known aminoglycoside efflux pump MexXY.
Gene regulation by asrA.
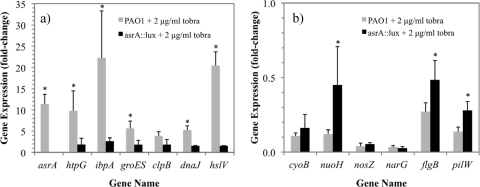
The asrA gene product displays homology to the Lon protease (PA1803) and was recently shown to be involved in the regulation of protection from nitric oxide (21, 22). As this gene was among the most highly upregulated under lethal conditions and was also moderately upregulated in the inhibitory arrays, its role in the tobramycin-induced regulon was investigated. A comparison of microarrays of asrA::lux mutant cells treated aerobically with 2 μg/ml tobramycin to those of PAO1 cells treated with tobramycin (see Table S1 in the supplemental material) revealed that the overall trends of gene expression by functional class were similar for the mutant and the wild type (Fig. 2a and c); however, notable differences were observed in the magnitudes of altered expression for a few specific genes. The most significant difference in gene expression in the asrA::lux mutant compared to the wild type was the reduced induction of the heat shock proteins in the asrA::lux mutant compared to the wild type. For this reason, we renamed the PA0779 gene asrA, for aminoglycoside-induced stress response ATP-dependent protease. Also of note, the downregulation of flagellum and pilus genes that was observed for the wild-type PAO1 lethal microarrays was not observed for the asrA::lux mutant microarray. These differences were confirmed for selected genes by using qPCR analysis (Fig. 3).
Fig. 3.
Comparison of responses to tobramycin of selected genes in P. aeruginosa PAO1 and the asrA::lux mutant determined by qPCR. (a) Upregulated heat shock genes. (b) Energy metabolism and motility genes. *, P ≤ 0.05.
The total number of genes affected was only 202 for the asrA::lux mutant, compared to the 422 genes observed for the wild type (see Table S1 in the supplemental material); this may be attributable to the lack of an induction of the heat shock response, which would likely cause significant changes in gene regulation.
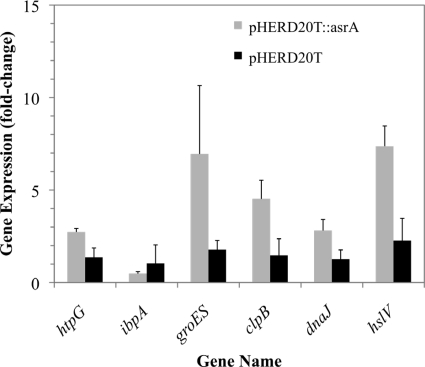
Overexpression of asrA led to induction of the heat shock response in the absence of tobramycin.
The asrA gene was conditionally expressed in trans on the pHERD20T vector from the araC-PBAD promoter by induction for 30 min with 1% arabinose. The level of expression of asrA as assessed by qPCR was 35-fold greater than the level of expression from its native promoter in PAO1 (P = 0.006). The heat shock genes assayed (with the exception of ibpA) were found to be significantly (P < 0.05) upregulated in response to 1% arabinose when asrA was overexpressed in trans but were not affected in the controls harboring the empty pHERD20T vector (Fig. 4).
Fig. 4.
Effect of overexpression of asrA on expression of heat shock genes in the P. aeruginosa asrA::lux mutant. Expression from the pHERD20T PBAD promoter was induced in mid-log-phase cultures of P. aeruginosa for 30 min with 1% arabinose.
Effect of asrA overexpression on tobramycin susceptibility.
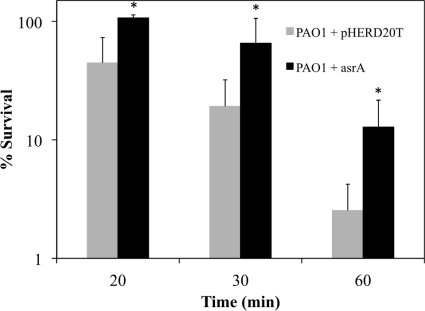
Killing by tobramycin was assessed by using 4 μg/ml tobramcyin to prevent an extended period of adaptation, since at this concentration the delay in killing was reduced to less than 10 min. A significant but quite modest difference (P ≤ 0.03) in percent survival was observed over the first 60 min in PAO1 cells with an overexpression of asrA due to arabinose induction (Fig. 5); however, for an MIC assay result read at 20 h, no difference was observed.
Fig. 5.
Effect of overexpression of asrA on tobramycin susceptibility. Expression from the PBAD promoter of pHERD20T was induced with 1% arabinose for 30 min in mid-log-phase cultures of P. aeruginosa PAO1 before treatment with 4 μg/ml tobramycin. *, P ≤ 0.03.
rpoH-mediated induction of asrA by tobramycin and heat shock.
Heat shock treatment of strain PAO1 induced the expression of asrA to an extent similar to that observed for treatment with tobramycin (Table 2). To determine if the known heat shock sigma factor rpoH was involved in mediating the stress response to tobramycin and heat shock through asrA, the expressions of asrA and dnaK in response to tobramycin and heat shock were assessed. Under the conditions tested, the induction of both genes was completely ablated in the rpoH mutant (Table 2). Analysis of the promoter region of asrA revealed the presence of a putative binding site for the transcription factor RpoH, 5′-GGATTGAAACCCGCCCCCGGGGCCCCAT-3′, which is highly similar to the E. coli RpoH consensus sequence 5′-GGCTTGA(12 to 20 bp)CCCCAT-3′ (47).
Table 2.
Involvement of RpoH in the induction of the heat shock response under heat shock and lethal tobramycin conditions
| Conditions compared | Fold change in gene expression (SD) |
||
|---|---|---|---|
| asrA | dnaK | rpoH | |
| WT with TOB vs WT | 9.0 (±0.2) | 6.0 (±0.5) | 1.5 (±0.4) |
| rpoH with TOB vs rpoH | 0.9 (±0.3) | 0.9 (±0.3) | |
| WT with TOB vs rpoH with Tb | 11.7 (±0.5) | 9 (±5) | |
| WT heat shock vs WT | 10 (±2) | 9.0 (±0.9) | 2.6 (±0.4) |
| rpoH heat shock vs rpoH | 1.3 (±0.2) | 1.0 (±0.2) | |
| WT heat shock vs rpoH heat shock | 11 (±4) | 12 (±3) | |
RpoH-mediated tobramycin susceptibility.
The susceptibility of the rpoH mutant to tobramycin was increased 2-fold compared to that of the wild type, as determined by MICs at 24 h. This difference was not due to differences in the growth rates of the mutant and wild type in the absence of tobramycin; however, at sublethal concentrations, the growth rate of the rpoH mutant in the presence of tobramycin was reduced compared to that of the wild type.
DISCUSSION
In this study we have provided data regarding global gene expression responses to tobramycin under a variety of conditions ranging from subinhibitory to lethal conditions. Recent studies based originally on analyses of gene expression responses led to the suggestion that aminoglycosides induce the production of lethal hydroxyl radicals in E. coli through a hyperactivation of the electron transport chain and of the TCA cycle (19, 20). The results of the present study are in stark contrast to those found for E. coli. Here, the components of the TCA cycle and electron transport chain regulated by ArcA in E. coli (the nuo and suc genes), specifically shown to be upregulated in work described previously by Kohanski et al., were found to be actually downregulated in response to tobramycin (Table 1). The results observed here do not generally conform to the model by Kohanski et al. for cell death in response to tobramycin. We hypothesized that a substantial proportion of the gene response to tobramycin represented the activation of defense mechanisms by Pseudomonas in an attempt to resist the lethal action of tobramycin. Indeed, 53 of the genes affected by tobramycin treatment (see Table S1 in the supplemental material) were previously found to be associated with tobramycin resistance when insertionally inactivated (39). Of these 53 genes, the majority were involved in energy metabolism, and all but 5 genes (none of these five were involved in energy metabolism) were downregulated in the presence of tobramycin. The major decrease in susceptibility to tobramycin observed between aerobically and anaerobically grown cells is indeed likely due to differences in uptake, as previously described (4, 5). Consistent with this suggestion, we saw a strong overlap between global gene expression changes at lethal concentrations of tobramycin under both aerobic and anaerobic conditions.
Nonetheless, similarities to data for E. coli reported previously by Kohanski et al. were observed with regard to the upregulation of heat shock response genes, particularly those involved in protein stabilization and protein folding. The heat shock response is typically induced by protein misfolding resulting from increased temperature stress. In keeping with this notion, we have found that aminoglycosides that cause decreased translation accuracy also resulted in the upregulation of this stress pathway. In contrast, as anticipated, antibiotics that do not target translation, including ciprofloxacin, ceftazidime, and polymyxin B, were not found to upregulate the heat shock genes (data not shown). Additionally, other protein synthesis inhibitors, including erythromycin and tetracycline, which do not cause a reduction of translation fidelity, also did not trigger the heat shock response (data not shown). The upregulation of heat shock genes by subinhibitory concentrations of gentamicin was previously observed for Bacillus subtilis (25). In Pseudomonas, however, this response appears to be linked to exposure to lethal concentrations.
In addition to the heat shock genes, asrA, encoding a putative Lon-type protease, was found to be highly upregulated in response to lethal concentrations of tobramycin under both aerobic and anaerobic conditions but was only moderately upregulated in response to inhibitory (bacteriostatic) concentrations. Lon proteases were previously identified as participating in the heat shock response in E. coli and other organisms to degrade misfolded proteins (2, 31, 35). The results observed here suggest that the AsrA protein is not merely part of the heat shock response but is in fact a key mediator of this response, particularly in response to stress induced by tobramycin. Consistent with this conclusion, the induction of the heat shock genes did not occur in the asrA::lux mutant in response to tobramycin, and the expression of most heat shock genes assayed (with the exception of ibpA) was induced in the absence of tobramycin when asrA was overexpressed from an inducible promoter (Fig. 4).
Lon proteases belong to the multifunctional family of AAA+ proteins (ATPases associated with a variety of cellular activities) that are involved in DNA replication, transcription, membrane fusion, and proteolysis (38). The P. aeruginosa PAO1 genome encodes a Lon protease, PA1803, that is 84% similar to the E. coli Lon protease and one (AsrA/PA0799) that shows 53% similarity to E. coli Lon. It was suggested previously that Lon proteases play a role in general stress responses (31); however, the presence of multiple Lon proteases in P. aeruginosa and the induction of only asrA and not other ATP-dependent proteases in response to tobramycin suggests a more specialized role for these proteases. While the gene product encoded by asrA has only 60% similarity (41% identity) to the PA1803 Lon protease, close homologues of asrA include Lon-like proteases in the denitrifying bacteria Azotobacter vinelandii (93% similarity) and Thiobacillus denitrificans (81% similarity), which would appear to be consistent with data from recent studies demonstrating the involvement of asrA in regulating the protective response to NO (21, 22). However, using the NO scavenger carboxy-PTIO [2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl 3-oxide], we observed that the level of NO production by P. aeruginosa was decreased in response to tobramycin and that this decrease was not affected by the loss of a functional asrA product (data not shown). Furthermore, the downregulation of denitrification genes in the asrA::lux mutant occurred in response to tobramycin, as it did in PAO1. This finding indicates that the proposed involvement of asrA in regulating NO detoxification does not offer substantial protection against tobramycin.
The induction of heat shock, in response to temperature increases and the presence of lethal concentrations of tobramycin, requires the presence of the heat shock sigma factor RpoH. The transcription of rpoH in P. aeruginosa is known to be under the control of the AlgU sigma factor (40). Of note, however, the expression of rpoH is not upregulated by tobramycin, and an algU mutant exhibited similar levels of induction of the heat shock genes in response to tobramycin (data not shown). Thus, regulation at the transcriptional level does not seem to be necessary for RpoH to mediate the tobramycin-induced heat shock response. This highlights the importance of posttranscriptional regulation by stabilization and increased translation of RpoH, as was previously reported for E. coli (47).
The increased susceptibility of the rpoH mutant to tobramycin demonstrated that the development of a heat shock response indeed mediates some degree of resistance to this aminoglycoside. This result agrees with the results observed for the overexpression of the asrA gene. A greater ability to survive and multiply at concentrations of tobramycin around the lethal range can be very important during infection by enabling the bacterium to upregulate long-term adaptive mechanisms or to acquire resistance genes by horizontal transfer from other bacteria.
Interestingly, we found that the uncharacterized Lon-like protease encoded by PA0779, AsrA, is also induced by RpoH as part of the heat shock response. Indeed, there is a conserved binding site for this sigma factor in the promoter of the asrA gene. Moreover, our results show that the presence of a functional AsrA protease is necessary to trigger a full response. Thus, once RpoH is activated, it would induce the transcription of a series of heat shock genes encoding principally chaperones and proteases but also including asrA. At that point, the protease AsrA would, by means of an as-yet-unknown mechanism, amplify the effect of the signal. The result of this would be an even higher level of expression than that achieved by RpoH alone in the absence of the participation of the protease. It is possible that this induction is due to the degradation by AsrA of a protein that somehow inhibits the activation or translation of RpoH or by the proteolytic cleavage of a negative regulator of the genes involved in the response. In any case, more research needs to be done in order to identify the substrates of this ATP-dependent protease.
While asrA was a major player in the lethal and inhibitory responses to tobramycin, it did not appear to have a significant role in adaptive responses occurring during prolonged exposure to subinhibitory concentrations of tobramycin. Indeed, mexX and mexY were the most upregulated genes during prolonged exposure to subinhibitory concentrations of tobramycin. It was demonstrated previously that the MexXY-OprM tripartite multidrug efflux pump can mediate aminoglycoside efflux, and derepressed mutants in this system explain impermeability-type resistance to tobramycin. The upregulation of the MexXY pump in response to ribosome-targeting agents, including the aminoglycosides, is mediated through the PA5471 gene product (29, 30, 45). Consistent with this, our microarray results with subinhibitory concentrations revealed a 2.2-fold increase in the level of PA5471 expression in response to tobramycin. No other genes were upregulated to as great an extent as mexXY, consistent with this being the predominant mechanism of adaptive resistance to tobramycin.
Other genes and operons observed to have moderate upregulation under conditions of treatment with subinhibitory concentrations were involved in hydrogen cyanide production and quorum sensing. While these systems are typically associated with pathogenesis, their potential roles in adaptive resistance to aminoglycosides have not been investigated. Hydrogen cyanide is a respiratory inhibitor, and its production by P. aeruginosa typically occurs under low-oxygen conditions (7, 8) and has been associated with increased iron levels (1) as well as quorum sensing (34, 37). While HCN has been found in CF sputum (6), its physiological role in P. aeruginosa is not well understood. It was suggested previously that the P. aeruginosa hydrogen cyanide synthase is merely a respiration-linked amino acid dehydrogenase (43), and consistent with this, its synthesis is accompanied by the production of a cyanide-resistant electron transport chain. Given the findings of this study, which show that a large number of anaerobic and aerobic respiratory elements are downregulated in response to lethal, inhibitory, and subinhibitory concentrations of tobramycin and are also related to tobramycin susceptibility, this may indicate a switch to an alternate pathway of energy generation that might impact the efficiency of aminoglycoside respiration-dependent uptake and thus provide further protection against the effects of tobramycin.
Concluding remarks.
Adaptive resistance is a complex phenomenon that remains poorly understood. While mechanisms imparting an impermeability phenotype, such as the upregulation of antibiotic efflux pumps, likely provide the greatest adaptive protection against aminoglycosides over the long term, P. aeruginosa also alters its gene expression in a more immediate fashion in response to a lethal hit by aminoglycosides. We have shown here that some of these immediate adaptations include the downregulation of energy metabolism genes (including those involved in nitrogen metabolism) as well as the upregulation of the heat shock response mediated by RpoH and the asrA gene product. The combined effects of a number of adaptations, each providing some modest degree of resistance independently, may provide sufficient short-term protection until more effective long-term adaptations are established.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Canadian Cystic Fibrosis Foundation and the Canadian Institutes of Health Research. K.N.K. was supported by a Natural Sciences and Engineering Council of Canada postgraduate scholarship and a Michael Smith Foundation for Health Research senior graduate studentship. L.F. received a postdoctoral fellowship from the Fundacion Alfonso Martin Escudero (Spain).
R.E.W.H. holds a Canada Research Chair.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 28 February 2011.
REFERENCES
- 1. Askeland R. A., Morrison S. M. 1983. Cyanide production by Pseudomonas fluorescens and Pseudomonas aeruginosa. Appl. Environ. Microbiol. 45:1802–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bissonnette S. A., Rivera-Rivera I., Sauer R. T., Baker T. A. 2010. The IbpA and IbpB small heat-shock proteins are substrates of the AAA+ Lon protease. Mol. Microbiol. 75:1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Borriello G., Richards L., Ehrlich G. D., Stewart P. S. 2006. Arginine or nitrate enhances antibiotic susceptibility of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 50:382–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bryan L. E., Kwan S. 1981. Aminoglycoside-resistant mutants of Pseudomonas aeruginosa deficient in cytochrome d, nitrite reductase, and aerobic transport. Antimicrob. Agents Chemother. 19:958–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bryan L. E., Kwan S. 1983. Roles of ribosomal binding, membrane potential, and electron transport in bacterial uptake of streptomycin and gentamicin. Antimicrob. Agents Chemother. 23:835–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carterson A. J., et al. 2004. The transcriptional regulator AlgR controls cyanide production in Pseudomonas aeruginosa. J. Bacteriol. 186:6837–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Castric P. A. 1983. Hydrogen cyanide production by Pseudomonas aeruginosa at reduced oxygen levels. Can. J. Microbiol. 29:1344–1349 [DOI] [PubMed] [Google Scholar]
- 8. Castric P. A. 1975. Hydrogen cyanide, a secondary metabolite of Pseudomonas aeruginosa. Can. J. Microbiol. 21:613–618 [DOI] [PubMed] [Google Scholar]
- 9. Choi K. H., Kumar A., Schweizer H. P. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391–397 [DOI] [PubMed] [Google Scholar]
- 10. Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 7th ed. (M7-A7) CLSI, Wayne, PA [Google Scholar]
- 11. Davis B. D., Chen L. L., Tai P. C. 1986. Misread protein creates membrane channels: an essential step in the bactericidal action of aminoglycosides. Proc. Natl. Acad. Sci. U. S. A. 83:6164–6168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. El'Garch F., Jeannot K., Hocquet D., Llanes-Barakat C., Plesiat P. 2007. Cumulative effects of several nonenzymatic mechanisms on the resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 51:1016–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Field T. R., White A., Elborn J. S., Tunney M. M. 2005. Effect of oxygen limitation on the in vitro antimicrobial susceptibility of clinical isolates of Pseudomonas aeruginosa grown planktonically and as biofilms. Eur. J. Clin. Microbiol. Infect. Dis. 24:677–687 [DOI] [PubMed] [Google Scholar]
- 14. Foweraker J. E., Laughton C. R., Brown D. F., Bilton D. 2005. Phenotypic variability of Pseudomonas aeruginosa in sputa from patients with acute infective exacerbation of cystic fibrosis and its impact on the validity of antimicrobial susceptibility testing. J. Antimicrob. Chemother. 55:921–927 [DOI] [PubMed] [Google Scholar]
- 15. Hocquet D., et al. 2003. MexXY-OprM efflux pump is necessary for a adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 47:1371–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Irvin R. T., Govan J. W., Fyfe J. A., Costerton J. W. 1981. Heterogeneity of antibiotic resistance in mucoid isolates of Pseudomonas aeruginosa obtained from cystic fibrosis patients: role of outer membrane proteins. Antimicrob. Agents Chemother. 19:1056–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jacobs M. A., et al. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:14339–14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. King P., Citron D. M., Griffith D. C., Lomovskaya O., Dudley M. N. 2010. Effect of oxygen limitation on the in vitro activity of levofloxacin and other antibiotics administered by the aerosol route against Pseudomonas aeruginosa from cystic fibrosis patients. Diagn. Microbiol. Infect. Dis. 66:181–186 [DOI] [PubMed] [Google Scholar]
- 19. Kohanski M. A., Dwyer D. J., Hayete B., Lawrence C. A., Collins J. J. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810 [DOI] [PubMed] [Google Scholar]
- 20. Kohanski M. A., Dwyer D. J., Wierzbowski J., Cottarel G., Collins J. J. 2008. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135:679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koskenkorva T., et al. 2008. Transcriptional activity of Pseudomonas aeruginosa fhp promoter is dependent on two regulators in addition to FhpR. Arch. Microbiol. 189:385–396 [DOI] [PubMed] [Google Scholar]
- 22. Koskenkorva-Frank T. S., Kallio P. T. 2009. Induction of Pseudomonas aeruginosa fhp and fhpR by reactive oxygen species. Can. J. Microbiol. 55:657–663 [DOI] [PubMed] [Google Scholar]
- 23. Levin B. R., Rozen D. E. 2006. Non-inherited antibiotic resistance. Nat. Rev. Microbiol. 4:556–562 [DOI] [PubMed] [Google Scholar]
- 24. Lewenza S., et al. 2005. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res. 15:583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin J. T., Connelly M. B., Amolo C., Otani S., Yaver D. S. 2005. Global transcriptional response of Bacillus subtilis to treatment with subinhibitory concentrations of antibiotics that inhibit protein synthesis. Antimicrob. Agents Chemother. 49:1915–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. MacLeod D. L., et al. 2000. Aminoglycoside-resistance mechanisms for cystic fibrosis Pseudomonas aeruginosa isolates are unchanged by long-term, intermittent, inhaled tobramycin treatment. J. Infect. Dis. 181:1180–1184 [DOI] [PubMed] [Google Scholar]
- 27. Matsunaga K., Yamaki H., Nishimura T., Tanaka N. 1986. Inhibition of DNA replication initiation by aminoglycoside antibiotics. Antimicrob. Agents Chemother. 30:468–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McPhee J. B., et al. 2006. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J. Bacteriol. 188:3995–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morita Y., Gilmour C., Metcalf D., Poole K. 2009. Translational control of the antibiotic inducibility of the PA5471 gene required for mexXY multidrug efflux gene expression in Pseudomonas aeruginosa. J. Bacteriol. 191:4966–4975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morita Y., Sobel M. L., Poole K. 2006. Antibiotic inducibility of the MexXY multidrug efflux system of Pseudomonas aeruginosa: involvement of the antibiotic-inducible PA5471 gene product. J. Bacteriol. 188:1847–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ngo J. K., Davies K. J. 2009. Mitochondrial Lon protease is a human stress protein. Free Radic. Biol. Med. 46:1042–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Overhage J., Bains M., Brazas M. D., Hancock R. E. W. 2008. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J. Bacteriol. 190:2671–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pai V. B., Nahata M. C. 2001. Efficacy and safety of aerosolized tobramycin in cystic fibrosis. Pediatr. Pulmonol. 32:314–327 [DOI] [PubMed] [Google Scholar]
- 34. Pessi G., Haas D. 2001. Dual control of hydrogen cyanide biosynthesis by the global activator GacA in Pseudomonas aeruginosa PAO1. FEMS Microbiol. Lett. 200:73–78 [DOI] [PubMed] [Google Scholar]
- 35. Phillips T. A., VanBogelen R. A., Neidhardt F. C. 1984. lon gene product of Escherichia coli is a heat-shock protein. J. Bacteriol. 159:283–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qiu D., Damron F. H., Mima T., Schweizer H. P., Yu H. D. 2008. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl. Environ. Microbiol. 74:7422–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reimmann C., et al. 1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24:309–319 [DOI] [PubMed] [Google Scholar]
- 38. Rotanova T. V., et al. 2006. Slicing a protease: structural features of the ATP-dependent Lon proteases gleaned from investigations of isolated domains. Protein Sci. 15:1815–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schurek K. N., et al. 2008. Novel genetic determinants of low-level aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52:4213–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schurr M. J., Deretic V. 1997. Microbial pathogenesis in cystic fibrosis: co-ordinate regulation of heat-shock response and conversion to mucoidy in Pseudomonas aeruginosa. Mol. Microbiol. 24:411–420 [DOI] [PubMed] [Google Scholar]
- 41. Sobel M. L., McKay G. A., Poole K. 2003. Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 47:3202–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tanaka N., Matsunaga K., Yamaki H., Nishimura T. 1984. Inhibition of initiation of DNA synthesis by aminoglycoside antibiotics. Biochem. Biophys. Res. Commun. 122:460–465 [DOI] [PubMed] [Google Scholar]
- 43. Williams H. D., Zlosnik J. E., Ryall B. 2007. Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv. Microb. Physiol. 52:1–71 [DOI] [PubMed] [Google Scholar]
- 44. Worlitzsch D., et al. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109:317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yamamoto M., et al. 2009. Role of MexZ and PA5471 in transcriptional regulation of mexXY in Pseudomonas aeruginosa. Microbiology 155:3312–3321 [DOI] [PubMed] [Google Scholar]
- 46. Yeung A. T., et al. 2009. Swarming of Pseudomonas aeruginosa is controlled by a broad spectrum of transcriptional regulators, including MetR. J. Bacteriol. 191:5592–5602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhao K., Liu M., Burgess R. R. 2005. The global transcriptional response of Escherichia coli to induced sigma 32 protein involves sigma 32 regulon activation followed by inactivation and degradation of sigma 32 in vivo. J. Biol. Chem. 280:17758–17768 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.