Abstract
We report the emergence of a novel VIM variant (VIM-24) in a Klebsiella pneumoniae isolate in Colombia. The isolate displays MICs for carbapenems below the resistance breakpoints, posing a real challenge for its detection. The blaVIM-24 gene was located within a class 1 integron carried on a large plasmid. Further studies are needed to clarify its epidemiological and clinical impact.
INTRODUCTION
The emergence of acquired metallo-β-lactamases (MBLs) among Gram-negative strains is a matter of significant concern worldwide, due to their successful dissemination capacity, increasing diversity, and variability of resistance phenotypes (5, 19, 24). Detection of MBL-producing strains is challenging (5), especially in Enterobacteriaceae, as some strains display MICs below the resistance breakpoint established by the Clinical and Laboratory Standards Institute (CLSI) (5, 22). In addition, discrepancies in susceptibility testing of carbapenems with MBL producers have been reported (9).
To the best of our knowledge, in Latin America, VIM-producing Enterobacteriaceae have been limited to Mexico (18) and Venezuela (15), and no IMP enzymes have been reported within this family. In Colombia, the first report of a VIM was in a carbapenem-resistant Pseudomonas aeruginosa strain harboring VIM-8 found in a single city (6), followed by several disseminated carbapenem-resistant P. aeruginosa strains producing VIM-2 (23). We now describe a novel VIM in a Klebsiella pneumoniae isolate, which confers a minimal increase in the MICs to selected β-lactams and exhibits variable MICs to imipenem when tested by different methods, posing a real challenge for its detection.
(Part of this work was presented at the 50th Annual Interscience Conference on Antimicrobial Agents and Chemotherapy, Boston, MA, 12 to 15 September 2010.)
K. pneumoniae 5639 was isolated from a blood culture of a 4-month-old Colombian female hospitalized in the intensive care unit (ICU) of a hospital located in Barranquilla, Colombia. The patient had an Escherichia coli intra-abdominal infection secondary to intestinal obstruction, requiring treatment with piperacillin-tazobactam followed by cefepime plus clindamycin. After a complicated clinical course, the abscess was surgically drained, and she was successfully treated with meropenem, which was given when blood cultures reported a K. pneumoniae (isolate 5639) susceptible (MIC of ≤1 μg/ml) to this antibiotic.
Isolate 5639 was sent to the International Center for Medical Research and Training (CIDEIM) as part of a carbapenemase surveillance study, where identification was confirmed by using the Vitek 2 test (bioMérieux, Marcy l'Etoile, France). Antibiotic susceptibility testing was performed by the broth microdilution (BMD) method (Sensititre panels; TREK Diagnostic Systems, Westlake, OH) and Vitek 2, and the results were interpreted according to the CLSI guidelines (3, 4). Tigecycline breakpoints for Enterobacteriaceae were defined based on the U.S. Food and Drug Administration guidelines (susceptible, MIC of ≤2 μg/ml), and polymyxin B breakpoints were based on the CLSI cutoff for Acinetobacter spp. (resistant, MIC of ≥4 μg/ml), as previously recommended (7).
The strain was tested in triplicate with a standardized inoculum, as discrepancies in cefepime MICs between the referral hospital and CIDEIM, as well as decreased susceptibility to imipenem (MIC of 2 μg/ml), were observed. Results from susceptibility testing for selected antibiotics are summarized in Table 1. Imipenem was additionally tested by Etest, obtaining variable MICs of 1 to 2 μg/ml. Further testing was performed, including characterization of β-lactamases by isoelectric focusing (IEF) as described by Matthew et al. (16), and three-dimensional (3D) testing as previously reported (21). IEF revealed the presence of a single β-lactamase with a pI of 7.9, and a positive 3D test indicated the presence of a carbapenemase. Screening by PCR of β-lactamases blaCTX-M, blaTEM, blaSHV, blaKPC, blaIMP, and blaVIM revealed the presence of a blaVIM gene, while the other resistance determinants were not detected, consistent with the IEF results. Even though there is a debate about the possible universality of SHV β-lactamase in K. pneumoniae (1, 10), our results suggested the absence of the SHV β-lactamase gene in the K. pneumoniae 5639 isolate.
Table 1.
MICs of selected antibiotics for K. pneumoniae strain 5639 and E. coli strain J53 and its transconjugants
| Strain or TC | Method | MIC or MIC range (μg/ml) ofa: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETP | IPM | MEM | FEP | CAZ | CTX | TZP | SAM | AMK | TGC | PMB | CIP | ||
| K. pneumoniae 5639 | BMD | 0.25 | 0.25–2.0 | ≥0.12 | ≤1 | 4 | 4 | 16/4 | 32/16 | 32 | 0.5–1 | 0.5–1 | 2 |
| Vitek 2 | ≤0.5 | ≤1.0 | ≤0.25 | ≤1 | 4 | NDb | ND | ≥32 | ≥64 | 1 | ND | 1 | |
| E. coli J53 | BMD | ≤0.12 | ≤0.12 | ≤0.12 | ≤1 | ≤2 | ≤1 | ≤8/4 | ND | ≤8 | ≤0.12 | 2 | ≤0.5 |
| Vitek 2 | ≤0.5 | ≤1.0 | ≤0.25 | ≤1 | ≤1 | ND | ND | 4 | ≤1 | ≤0.5 | ND | ≤0.25 | |
| E. coli J53 VIM-24 TCc | BMD | 0.25–0.5 | 0.5 | ≤0.12 | 2–4 | 16–32 | 8 | 16/4–32/4 | ND | ≤8 | ≤0.12 | 1–2 | ≤0.5 |
| Vitek 2 | ≤0.5 | 2–4 | ≤0.25 | ≤1 | 8–16 | ND | ND | ≥32 | 8–16 | ≤0.5 | ND | 0.5 | |
ETP, ertapenem; IPM, imipenem; MEM, meropenem; FEP, cefepime; CAZ, ceftazidime; CTX, cefotaxime; TZP, piperacillin-tazobactam; SAM, ampicillin-sulbactam; AMK, amikacin; TGC, tigecycline; PMB, polymixin B; and CIP, ciprofloxaxin.
ND, not determined.
Shown are the MIC ranges for four transconjugants.
Sequencing revealed a novel blaVIM gene, designated blaVIM-24 (nucleotide sequence accession no. HM855205). This novel gene differed from that coding for VIM-2, the closest related enzyme, and the previously reported variants by a substitution at position 614 from G to T, changing the amino acid from Arg to Leu (Arg205→Leu). Detection and mapping of the class 1 integron were carried out by using specific primers for the 5′ and 3′ conserved segments (13, 17), as well as primers targeting the VIM gene cassette. The analysis revealed that blaVIM-24 was the first gene cassette of a class 1 integron, followed by aacA7, catB3, and arr-3 gene cassettes. The promoter region analysis indicated that these gene cassettes were preceded by a strong P1 promoter and the inactive form of P2.
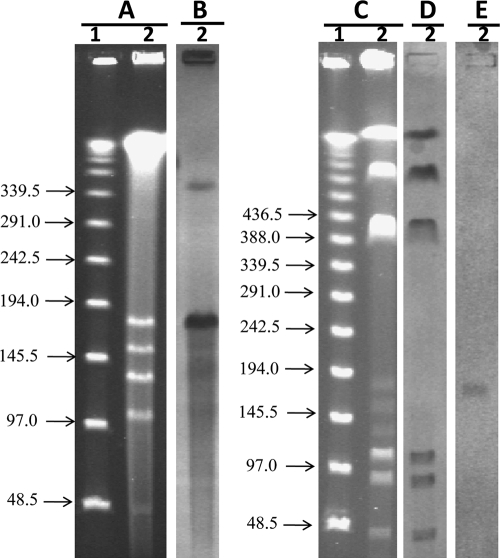
S1 nuclease coupled with pulsed-field gel electrophoresis (PFGE) was used for the detection and size estimation of plasmids (2). This methodology revealed the presence of five plasmids ranging in size between 46 and 172 kb (Fig. 1A). The blaVIM-specific probe hybridized with the plasmid band of approximately 172 kb (Fig. 1B), indicating that the gene was carried on a large plasmid, as previously reported (11, 20). The extrachromosomal location of blaVIM-24 was confirmed by digesting K. pneumoniae 5639 DNA with I-Ceu-I endonuclease (14), followed by PFGE (Fig. 1C) and hybridization with probes for 16S rRNA genes (Fig. 1D) and blaVIM (Fig. 1E). When comparing 16S rRNA gene and blaVIM hybridization results, the blaVIM gene was localized in a position different from that of the chromosomal fragments. Mating experiments using Escherichia coli J53 as the recipient strain and amikacin as the selection marker (8 μg/ml) confirmed that blaVIM-24 was encoded in a conjugative plasmid. Transconjugants (TC) were PCR positive for the blaVIM gene and display a MIC for imipenem of between 2 and 4 μg/ml when tested by Vitek 2 and 0.5 μg/ml when tested by BMD (Table 1).
Fig. 1.
Localization of the blaVIM-24 gene in K. pneumoniae isolate 5639. Shown are the results of S1 nuclease and I-Ceu-I digestions, pulsed-field gel electrophoresis, and hybridizations. (A and B) S1 restriction and PFGE of total DNA (A) and hybridization with a probe specific to blaVIM (B). (C to E) I-Ceu-I fragment restriction pattern of total DNA after PFGE (C) and hybridization with a probe specific to 16S rRNA genes (D) and blaVIM (E). Lane 1, lambda ladder (molecular sizes in kilobases are shown to the left); lane 2, K. pneumoniae isolate 5639.
The production of metallo-β-lactamases (MBL) is a growing health problem worldwide (5, 22) that poses challenges for the treatment of infections due to Gram-negative bacteria. The increase in diversity, as evidenced by the growing number of new variants of the already known MBLs, as well as the recent detection of the New Delhi MBL (12), is particularly worrisome. We now report the emergence of a VIM-producing K. pneumoniae strain in Colombia, which caused the first documented case of an infection with MBL-producing Enterobacteriaceae in the country. Surveillance for similar strains is warranted considering the known association of blaVIM with mobile genetic elements and the fact that Klebsiella pneumoniae carbapenemase (KPC) is already disseminated in Colombia, which could lead to the emergence of K. pneumoniae isolates coproducing KPC and VIM, as has been reported in Greece (8).
Although VIM-producing Enterobacteriaceae isolates were initially recognized by their in vitro resistance to carbapenems (22), this strain displays MICs for carbapenems below the current resistant breakpoints (ertapenem, ≥1 μg/ml; imipenem, ≥4 μg/ml; and meropenem, ≥4 μg/ml) (4). This represents a real challenge for its detection and hinders the estimation of its real incidence. Because MICs may be unreliable in detecting MBLs, special tests (such as the 3D test) could be helpful in detecting the resistance mechanisms involved. It seems prudent that infection control procedures should apply to all proven MBL-producing isolates regardless of their actual level of susceptibility.
Acknowledgments
We are grateful to Gabriel Gutkind for providing the CTX-M-2 isolate and Cesar Arias for experimental advice. We thank the institutions that are part of the Colombian Nosocomial Resistance Study Group, whose members are as follows: Henry Mendoza, Flor Angela Cubides, Martha Patricia Melendez, Luz Angelica Quintero, and Ilba Liliana Galeano at Hospital Central de la Policía; Beatriz Porras, Guillermo Ortiz, and Luz Mila Lopez at Hospital Santa Clara; Henry Oliveros, Maria Nilse Gonzalez, Angela Pescador, Monica Ballesteros, Sandra Valderrama, Alirio Rodriguez, and Jairo Perez at Hospital Militar Central; Carlos Ignacio Gomez, Jaime Lopez, Jorge Donado, Monica Cuartas, Ana Lucia Correa, and Lina Marcela Castañeda at Hospital Pablo Tobon Uribe; Julian Betancourth, Juan David Villa, Jorge Nagles, Nancy Estella Gonzalez, Magda Orjuela, Ana Cristina Quiroga, Rodrigo Agudelo, and Carolina Rios at Clínica de las Américas; Martha Vallejo, Luz Marina Melguizo, Dora Rivas, and Sergio Velez at Hospital General de Medellín; Ernesto Martinez, Jose Millan Oñate, Christian Pallares, Luz Marina Gallardo, Alba Lucia Bohorques, and Nancy Villamarin at Hospital Universitario del Valle; Fernando Rosso, Juan Diego Velez, Jose Garcia, Monica Recalde, Alejandra Toala, and John Jairo Echeverry at Clínica Fundación Valle del Lili; Beatriz Lopez and Claudia Barcenas at La Foscal; Karol Monsalve, Luis Angel Villar, and Mayerly Anaya at Fundación Cardiovascular; Agustin Vega, Miriam Fanny Amaya, Matha Jacome, Rocio Abaunza, and Martin Mejía at Hospital Universitario de Santander; Angela Mendoza at Hospital Universitario San Jorge; Carmen Elisa Llano, Myriam Gomez, and Rodolfo Cabrales at Clínica General del Norte; and Amparo Ovalle, Claudia Echeverry, M. del Rosario Aldana, Pablo Lopez, and Luis Gonzalez at Hospital Federico Lleras Acosta.
The conformation of the network of institutions of the Colombian Nosocomial Resistance Study Group has been made possible thanks in part to the support of Merck Sharp & Dohme, Janssen-Cilag SA, Pfizer SA, AstraZeneca Colombia SA, Merck Colombia, Novartis, and Baxter SA.
No financial or commercial interests were involved in the development of this study, except that J. P. Quinn is an employee of Pfizer Global Research and Development (New London, CT) and M. V. Villegas has received consulting fees and research grants from Merck Sharp & Dohme, Pfizer SA, Janssen-Cilag SA, Novartis, Merck Colombia, and AstraZeneca Colombia SA.
Footnotes
Published ahead of print on 31 January 2011.
REFERENCES
- 1. Babini G. S., Livermore D. M. 2000. Are SHV beta-lactamases universal in Klebsiella pneumoniae? Antimicrob. Agents Chemother. 44:2230 (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barton B. M., Harding G. P., Zuccarelli A. J. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235–240 [DOI] [PubMed] [Google Scholar]
- 3. Clinical and Laboratory Standards Institute. 2010. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed. CLSI document M07-A8. CLSI, Wayne, PA [Google Scholar]
- 4. Clinical and Laboratory Standards Institute. 2011. Performance standards for antimicrobial susceptibility testing: 20th informational supplement. CLSI document M100-S21. CLSI, Wayne, PA [Google Scholar]
- 5. Cornaglia G., et al. 2007. Metallo-beta-lactamases as emerging resistance determinants in Gram-negative pathogens: open issues. Int. J. Antimicrob. Agents 29:380–388 [DOI] [PubMed] [Google Scholar]
- 6. Crespo M. P., et al. 2004. Outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-8, a novel metallo-beta-lactamase, in a tertiary care center in Cali, Colombia. J. Clin. Microbiol. 42:5094–5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gales A. C., Reis A. O., Jones R. N. 2001. Contemporary assessment of antimicrobial susceptibility testing methods for polymyxin B and colistin: review of available interpretative criteria and quality control guidelines. J. Clin. Microbiol. 39:183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Giakkoupi P., et al. 2009. Emerging Klebsiella pneumoniae isolates coproducing KPC-2 and VIM-1 carbapenemases. Antimicrob. Agents Chemother. 53:4048–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Giakkoupi P., et al. 2005. Discrepancies and interpretation problems in susceptibility testing of VIM-1-producing Klebsiella pneumoniae isolates. J. Clin. Microbiol. 43:494–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haeggman S., Lofdahl S., Paauw A., Verhoef J., Brisse S. 2004. Diversity and evolution of the class A chromosomal beta-lactamase gene in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:2400–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ktari S., et al. 2006. Emergence of multidrug-resistant Klebsiella pneumoniae isolates producing VIM-4 metallo-beta-lactamase, CTX-M-15 extended-spectrum beta-lactamase, and CMY-4 AmpC beta-lactamase in a Tunisian university hospital. Antimicrob. Agents Chemother. 50:4198–4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kumarasamy K. K., et al. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Levesque C., Piche L., Larose C., Roy P. H. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu S. L., Hessel A., Sanderson K. E. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci.U. S. A. 90:6874–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marcano D., et al. 2008. First isolation of a VIM-producing Klebsiella pneumoniae from a seven-year-old child in Venezuela. J. Infect. Dev. Ctries. 2:241–244 [DOI] [PubMed] [Google Scholar]
- 16. Matthew M., Harris A. M. 1976. Identification of beta-lactamases by analytical isoelectric focusing: correlation with bacterial taxonomy. J. Gen. Microbiol. 94:55–67 [DOI] [PubMed] [Google Scholar]
- 17. Miriagou V., Tzelepi E., Gianneli D., Tzouvelekis L. S. 2003. Escherichia coli with a self-transferable, multiresistant plasmid coding for metallo-beta-lactamase VIM-1. Antimicrob. Agents Chemother. 47:395–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morfin-Otero R., Rodriguez-Noriega E., Deshpande L. M., Sader H. S., Castanheira M. 2009. Dissemination of a bla(VIM-2)-carrying integron among Enterobacteriaceae species in Mexico: report from the SENTRY Antimicrobial Surveillance Program. Microb. Drug Resist. 15:33–35 [DOI] [PubMed] [Google Scholar]
- 19. Queenan A. M., Bush K. 2007. Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20:440–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Robin F., Aggoune-Khinache N., Delmas J., Naim M., Bonnet R. 2010. Novel VIM metallo-beta-lactamase variant from clinical isolates of Enterobacteriaceae from Algeria. Antimicrob. Agents Chemother. 54:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thomson K. S., Sanders C. C. 1992. Detection of extended-spectrum beta-lactamases in members of the family Enterobacteriaceae: comparison of the double-disk and three-dimensional tests. Antimicrob. Agents Chemother. 36:1877–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vatopoulos A. 2008. High rates of metallo-beta-lactamase-producing Klebsiella pneumoniae in Greece—a review of the current evidence. Euro. Surveill. 13:8023. [PubMed] [Google Scholar]
- 23. Villegas M. V., et al. 2006. First detection of metallo-beta-lactamase VIM-2 in Pseudomonas aeruginosa isolates from Colombia. Antimicrob. Agents Chemother. 50:226–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Walsh T. R., Toleman M. A., Poirel L., Nordmann P. 2005. Metallo-beta-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306–325 [DOI] [PMC free article] [PubMed] [Google Scholar]