Abstract
GS-9191, a bis-amidate prodrug of the nucleotide analog 9-(2-phosphonylmethoxyethyl)-N6-cyclopropyl-2,6-diaminopurine (cPrPMEDAP), was designed as a topical agent for the treatment of papillomavirus-associated proliferative disorders, such as genital warts. In this study, we investigated the mechanism of conversion of GS-9191 to cPrPMEDAP. We observed that GS-9191 is hydrolyzed in the presence of the lysosomal carboxypeptidase cathepsin A (CatA) in vitro and is less efficiently metabolized in CatA-deficient fibroblasts than in control cells. In addition, knockdown of CatA by small interfering RNA (siRNA) reduced the intracellular accumulation of GS-9191 metabolites. However, intracellular CatA levels did not correlate with the susceptibility of tested cell lines to GS-9191, indicating that the CatA step is unlikely to be rate limiting for the activation of GS-9191. Further analysis showed that upon the hydrolysis of the carboxylester bond in one of the GS-9191 amidate moieties, the unmasked carboxyl group displaces l-phenylalanine 2-methylpropyl ester from the other amidate moiety. The cPrPMEDAP–l-phenylalanine conjugate (cPrPMEDAP-Phe) formed is not metabolized by Hint1 (histidine triad nucleotide binding protein 1) phosphoramidase but undergoes spontaneous degradation to cPrPMEDAP in acidic pH that can be significantly enhanced by the addition of SiHa cell extract. Pretreatment of SiHa cells with bafilomycin A or chloroquine resulted in an 8-fold increase in the intracellular concentration of cPrPMEDAP-Phe metabolite and the accumulation of GS-9191 metabolites in the lysosomal/endosomal fraction. Together, these observations indicate that the conversion of GS-9191 to cPrPMEDAP occurs in lysosomes via CatA-mediated ester cleavage, followed by the release of cPrPMEDAP, most likely through the combination of enzyme-driven and spontaneous pH-driven hydrolysis of a cPrPMEDAP-Phe intermediate.
INTRODUCTION
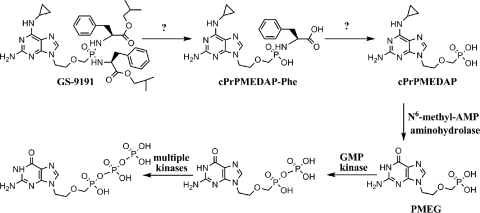
GS-9191 (Fig. 1) is a lipophilic bis-phosphonoamidate prodrug of 9-(2-phosphonylmethoxyethyl)-N6-cyclopropyl-2,6-diaminopurine (cPrPMEDAP) and a double prodrug of a potent cytotoxic nucleotide analog, 9-(2-phosphonylmethoxyethyl)guanine (PMEG). GS-9191 exhibits potent in vitro antiproliferative effect against a broad spectrum of cells transformed by human papillomavirus (HPV) and was specifically designed to increase the permeability and accumulation of PMEG and its active diphosphate metabolite (PMEG-DP) in skin cells following topical application (24). In the rabbit papillomavirus infection model, topical treatment with GS-9191 decreased the size of papilloma lesions in a dose-dependent manner, with cure achieved at 0.1% dose (24). Topical GS-9191 is currently being evaluated in clinics for the treatment of external genital warts. In a recently completed phase 2 study, GS-9191 was well tolerated and showed statistically superior effect compared to placebo treatment (7). These results suggest that GS-9191 could potentially have a clinical use for the treatment of skin conditions characterized by excessive cellular proliferation.
Fig. 1.
GS-9191 activation pathway.
In HPV-transformed cells, GS-9191 is metabolized to cPrPMEDAP, which is further converted to PMEG and the pharmacologically active metabolite PMEG-DP (24) (Fig. 1). PMEG-DP is a substrate and a potent inhibitor of host replicative DNA polymerases α, δ, and ε (8, 9), which are also responsible for the replication of the HPV genome (11). Following the incorporation of PMEG-DP into synthesized DNA, the DNA chain can no longer elongate because PMEG-DP lacks the 3′-like hydroxyl moiety (8, 9). Thus, PMEG-DP works as an obligate chain terminator blocking DNA synthesis in HPV-transformed cells. Inhibition of host DNA replication through this mechanism leads to the induction of apoptosis in treated cells (24). It has been shown that the conversion of cPrPMEDAP to PMEG is mediated by N6-methyl-AMP aminohydrolase (18, 19), but the mechanism leading to the formation of cPrPMEDAP from GS-9191 is poorly understood. The bis-amidate prodrug moiety of GS-9191 shares some structural similarity with GS-7340, a previously explored aryl-monoamidate prodrug of the antiretroviral nucleotide analog tenofovir (TFV) (12). After penetration into cells, the activation of GS-7340 is initiated by a lysosomal carboxypeptidase, cathepsin A (CatA), that cleaves the carboxyester bond in the prodrug moiety (3). In addition to CatA, various proteases were shown to hydrolyze the ester bond in GS-7340 and other TFV monoamidate prodrugs (2). Notably, the chymotrypsin-like proteases cleaved only prodrugs with phenylalanine and leucine, whereas elastase-like endopetidases activated only those with alanine, valine, and isoleucine, which is consistent with the substrate specificity of these enzymes (2). The hydrolysis of the ester bond in GS-7340 releases a metastable metabolite, from which the phenol group is believed to be spontaneously eliminated via intramolecular cyclization and hydrolysis (1, 22). The resulting metabolite is a conjugate of the nucleotide analog and amino acid with a P-N linkage between the phosphonate and the NH2 group of the amino acid. Phosphoramidate hydrolases belonging to the class of histidine triad nucleotide binding proteins (Hints) have been shown to catalyze the conversion of the amino acid conjugate to the corresponding parent nucleotide (4, 5). Alternatively, spontaneous pH-driven hydrolysis of the P-N bond in the acidic endosomal/lysosomal compartments of cells may play a role in the release of the parent nucleotide from the amino acid conjugate (2).
In this study, we investigated the mechanism of the conversion of GS-9191 to cPrPMEDAP. We assessed whether CatA and/or Hint1 hydrolase participates in this process and evaluated the role of the endosomal/lysosomal compartment in the activation of GS-9191.
MATERIALS AND METHODS
Compounds.
PMEG, cPrPMEDAP, GS-7340, GS-8361 (2-amino-6-cyclopropylamino-9-[2-((bis-alanine n-butylester)phophonamidatemethoxy)ethyl]purine, GS-9191, GS-327421 (2-amino-6-cyclopropylamino-9-[2-(((phenylalanine i-butylester)phenoxy)phophonamidate-methoxy)ethyl]purine), and GS-340402 (9-[(R)-2((phenylalanine cyclobutylester)-phenoxy)phophonamidatemethoxy)propyl]adenine) were synthesized by the Department of Medicinal Chemistry, Gilead Sciences, Inc. All compounds were dissolved in dimethyl sulfoxide (DMSO). PMEG monophosphate (PMEGp) and PMEG diphosphate (PMEGpp) were synthesized by TriLink BioTechnologies (San Diego, CA) and dissolved in water.
TFV-Ala, TFV-Phe, cPrPMEDAP-Ala, and cPrPMEDAP-Phe conjugates were prepared by incubation of 100 μM GS-7340, GS-340402, GS-8361, and GS-9191 with purified CatA (5 μg/ml) in 20 mM HEPES, pH 7.4, for 2 h. The reactions were quenched by adding a 2-fold volume excess of ice-cold methanol, the samples were evaporated, and the pellets were reconstituted in water. Because of quantitative conversion by CatA, this process yielded highly pure nucleotide-amino acid conjugates.
Purine [8-14C]GS-9191 (55 mCi/mmol) was synthesized by Moravek Biochemicals (Brea, CA), dissolved in DMSO, and stored at −20°C.
Cell cultures.
A variety of cervical carcinoma cell lines (SiHa, CaSki, HeLa, MS-751, C-4I, ME-180, and HT-3), a tongue squamous carcinoma cell line (SCC-4), and embryonal lung (HEL) fibroblasts were obtained from the American Type Culture Collection (Manassas, VA) and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and antibiotics. Primary cells from healthy human donors, adult foreskin keratinocytes (PHK), and cervical keratinocytes (CK) were purchased from Cambrex (Walkersville, MD) and cultured in serum-free keratinocyte growth medium supplied by Cambrex. The fibroblast cell lines GM05076 and GM02438 from two CatA-deficient individuals with galactosialidosis and the control fibroblast cell line GM05400 with normal CatA expression were obtained from the NIGMS Human Genetic Cell Repository (Camden, NJ) and cultured in DMEM supplemented with 20% FBS, nonessential amino acids, and antibiotics (17).
Intracellular metabolism of GS-9191.
CatA-negative (GM05076 and GM02348) and CatA-positive (GM05757 and SiHa) cells were seeded into T75 flasks. The medium was changed the next day, and the cells were incubated for 1.5 h at 37°C in a humidified CO2 incubator before 14C-labeled GS-9191 was added to a final concentration of 10 μM (0.5 μCi/ml). SiHa cells were additionally preincubated in the presence or absence of 100 nM bafilomycin A or 100 μM chloroquine for 1 h, followed by incubation with 1 μM [14C]GS-9191 (0.05 μCi/ml). At selected time points (0, 0.5, 1, 2, 4, and 8.6 h), medium was removed, and the cells were washed with the cultivation medium and phosphate-buffered saline (PBS) and detached by incubation with trypsin-EDTA in PBS. Trypsin was neutralized with medium, and the cells were harvested by centrifugation for 5 min at 1,500 rpm (Beckman GPR; 4°C). The cell pellets were washed with 8 ml of PBS, and the metabolites were extracted in 1 ml of 80% methanol. The cell extracts were centrifuged at 13,000 × g for 30 min, and the supernatants were evaporated under vacuum and resuspended in 80 μl water. Aliquots (10 to 20 μl) were analyzed to determine the amount of total radioactivity present in the cell extracts. The total amount of metabolites in cells was calculated from the calibration curve for mass versus radioactivity.
The metabolites were separated on a Phenomenex Prodigy column (5 μm; octadecyl silica 3 [ODS(3)], 150 by 4 mm) using the Waters high-performance liquid chromatography (HPLC) system connected to a Radiomatic Flo-One Beta liquid scintillation detector (Packard series A-500). A gradient elution from buffer A (25 mM phosphate buffer, pH 6, and 5 mM tetrabutylammonium bromide) to 20% buffer A-80% buffer B (80% acetonitrile in 25 mM phosphate buffer, pH 6, and 5 mM tetrabutylammonium bromide) for 30 min and a flow rate of 1.2 ml/min at 40°C were used to separate the metabolites.
CatA knockdown in HeLa cells.
HeLa cells (5 × 105 per well) were transfected with 75 ng Flexitube gene solution small interfering RNA (siRNA) specific for CatA or with All Stars negative-control siRNA (Qiagen, Valencia, CA) according to the manufacturer's protocol or left untreated. At day 2 posttransfection, the level of CatA knockdown was analyzed by quantitative immunoblot analysis. At day 3 posttransfection, the cells were incubated with 10 μM [14C]GS-9191 (0.5 μCi/ml) at various time points (0, 0.5, 1, 2, and 4 h). The cells were washed 3 times with cold PBS before being harvested. To determine the total level of GS-9191 metabolites, cells were extracted with methanol and analyzed as described above.
Cell volume measurement.
Cells were washed with PBS, detached with trypsin (for 5 min at 37°C), and incubated for 30 min in the culture medium to regain their spherical shape. Cell diameters were measured from photographs using a hematocytometer grid. The cell volume was calculated using the mean diameter obtained from 150 cells.
Immunoblot analysis.
Cells were grown in tissue culture flasks up to 90% confluence, washed with PBS, scraped into centrifuge tubes, pelleted, and lysed by incubation for 20 min on ice in a lysis buffer containing 10 mM Tris, pH 7.4, 20 mM MgCl2, 1 mM dithiothreitol (DTT), 150 mM NaCl, and 1% Nonidet P-40 (NP-40) detergent. Lysates were centrifuged at 14,000 rpm for 30 min at 4°C, and the supernatants were aliquoted in small volumes and stored at −80°C for single uses. Protein concentrations in the extracts were measured by the Lowry procedure, using a Protein Assay Kit (Pierce). Cell extracts (20 μg), in parallel with a dose range of recombinant CatA (100, 50, 25, 12.5, and 6.25 ng), were separated by SDS-PAGE (4 to 12% Bis-Tris; NuPAGE; Invitrogen, Carlsbad, CA) and transferred to a polyvinylidene difluoride (PVDF) membrane, following the protocol recommended by the manufacturer (Invitrogen, Carlsbad, CA). The membranes were blocked with 3% bovine serum albumin dissolved in 40 mM Tris-Cl, pH 7.4, 150 mM NaCl, and 0.1% Tween (TBS-Tween) overnight at 4°C. Goat polyclonal anti-CatA antibody (250 ng/ml; R&D Systems, Minneapolis, MN) was used, in combination with fluorescently labeled secondary antibody (Alexa Fluor 647 chicken anti-goat immunoglobulin G; 20 μg/ml; Invitrogen, Carlsbad, CA). The signal in cell extract samples was quantified using a Storm860 (Molecular Dynamics, Piscataway, NJ) and correlated with the signal from the titration of a CatA standard.
Hydrolysis of prodrugs with purified CatA or cellular extracts.
Purified CatA (1 μg/ml) or cellular extracts (2 mg/ml) were incubated with 30 μM GS-9191 in a reaction buffer containing 25 mM 2-[N-morpholino]ethanesulfonic acid (MES), pH 6.5, 100 mM NaCl, 1 mM DTT, 0.1% NP-40 at 37°C. At different time points, 65-μl aliquots were collected from the reaction mixture, and 180 μl ice-cold methanol was added to stop the reaction. Samples were incubated at −20°C for 30 min, followed by centrifugation at 13,000 rpm for 30 min at 4°C to remove denatured proteins. The supernatants were evaporated, resuspended in 100 μl buffer A (25 mM potassium phosphate, pH 6.0, 5 mM tetrabutylammonium bromide [TBAB]) and injected onto a C18 reverse-phase column [5 μm; 2.1 by 100 mm; ODS(2); Phenomenex, Torrance, CA] equilibrated with buffer A. Reaction substrates and products were eluted using a linear gradient of acetonitrile (0% to 70%; 15 min; 0.25 ml/min) in buffer A, followed by a 3-min column wash with 80% acetonitrile in buffer A (0.4 ml/min). The retention times for standards were as follows: GS-9191, 16.2 min; cPrPMEDAP-Phe, 10.2 min; l-phenylalanine 2-methylpropylester, 13.8 min; l-phenylalanine, 4.6 min. The rates of prodrug hydrolysis were quantified according to the corresponding peak areas.
Hydrolysis of nucleotide-amino acid conjugates.
To determine the effect of pH on the hydrolysis of nucleotide-amino acid conjugates, the metabolites (30 μM) were incubated in HEPES, pH 7.4, MES, pH 6.5, and acetate buffer, pH 5.5 or 4.5 (final concentration, 100 mM). At different time points, 100-μl aliquots were collected, and the pH of the samples was raised to 12 by adding 2 to 6 μl of 2 M NaOH. The reactions involving Hint1 were performed using 200 nM enzyme and 30 μM metabolites in 20 mM HEPES (pH 7.2), 1 mM DTT, 1 mM MgCl2, 200 ng/ml BSA, and 0.1% NP-40. The incubation of [14C]cPrPMEDAP-Phe (30 μM; 1.5 μCi/ml) with SiHa extracts (4 mg/ml) was carried out in HEPES, pH 7.4, MES, pH 6.5, and acetate buffer, pH 5.5 or 4.5 (final concentration, 100 mM). The reactions were quenched at different time points by adding methanol (60% final concentration) buffered with 50 mM Tris-HCl, pH 8.0. Sample processing and HPLC analysis were performed as described above.
Subcellular fractionation.
SiHa and HEL cells were incubated with 10 μM [14C]GS-9191 (0.52 μCi/ml) for 4 h. Where indicated, SiHa cells were preincubated in the presence or absence of 100 nM bafilomycin A for 1 h, followed by incubation with 10 μM [14C]GS-9191 for 30 min. Cells (1 × 107 to 2 × 107) were scraped from flasks and washed in a buffer containing 250 mM sucrose, 1 mM EDTA, 3 mM imidazole at pH 7.4 (sucrose buffer). The cells were suspended in 1 ml of cold sucrose buffer, placed on ice, and passed 20 times through a 26-gauge syringe needle. The disrupted-cell suspensions were diluted with the sucrose buffer to a total volume of 3 ml and centrifuged for 10 min at 1,000 × g to remove nuclei and unbroken cells. The volume of the supernatant was adjusted to 4.33 ml, and 1.67 ml of Percoll (Sigma) was added (final 25%). Samples were placed in Ultra Clear centrifuge tubes (0.5 by 2 in.) (Beckman-Coulter, Fullerton, CA) and centrifuged in a 50TI rotor at 32,000 rpm for 35 min. After centrifugation, 0.25-ml fractions were collected from the top of the tube into 96-well plates. The distribution of GS-9191 metabolites among organelles was evaluated by scintillation counting (23). Control fractions (no drug) were assayed for organelle-specific marker enzyme activities. The lysosomal enzyme marker β-hexosaminidase was assayed using 5 μM of the fluorescent substrate 4-methylumbelliferyl-2-acetamide-2-deoxy-β-O-glucopyranoside in 100 mM sodium acetate and 0.1% Triton X-100, pH 4.5 (13). Mitochondrial succinate dehydrogenase was assayed using iodonitrotetrazolium chloride according to a previously published protocol (15). NADPH cytochrome c reductase was used as a marker for the endoplasmic reticulum according to previously published methods (14).
RESULTS
Metabolism of GS-9191 in CatA-deficient cells.
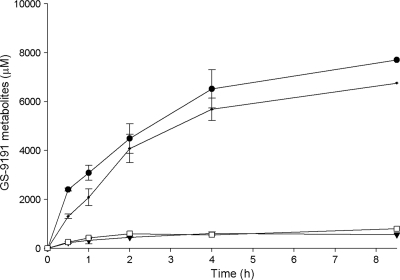
Human primary fibroblasts obtained from patients with galactosialidosis are deficient in CatA activity due to mutations present in the protein, preventing its proper folding (20, 21). When incubated in the presence of 14C-labeled GS-9191, both control and CatA-deficient cells showed time-dependent accumulation of the prodrug metabolites for up to 4 h (Fig. 2). However, the total concentration of G-9191 metabolites observed for the control cells (SiHa and GM05400) were 7 to 15 times greater than those for CatA-deficient fibroblasts (GM05076 and GM02346) (Fig. 2). cPrPMEDAP and cPrPMEDAP-Phe were the major metabolites detected in all cell lines (data not shown).
Fig. 2.
Metabolism of GS-9191 in CatA-deficient and control cells. CatA-deficient GM05076 (□) and GM02438 (▾) fibroblasts and GM05400 (•) and SiHa (•) control cells were incubated with 14C-labeled GS-9191 (10 μM; 0.521 μCi/ml) for 0 to 8.7 h. At selected time points, the medium was removed and the intracellular concentration of the metabolites was determined as described in Materials and Methods. The data are expressed as means ± standard deviations of two independent experiments.
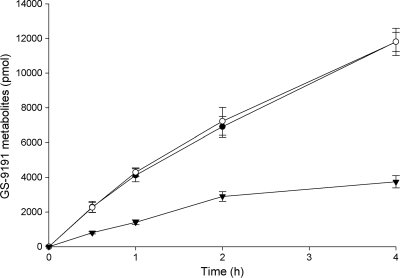
Furthermore, the knockdown of CatA expression in HeLa cells using RNA interference (RNAi) resulted in 3-fold-reduced cell loading with the GS-9191 metabolites compared to cells transfected with control siRNA (Fig. 3). The quantification of the CatA protein level in cells by immunoblot analysis showed that the siRNA knockdown reduced CatA expession approximately 4-fold compared to control cells (see Fig. S1 in the supplemental material).
Fig. 3.
Metabolism of GS-9191 in HeLa cells following CatA knockdown using siRNA. HeLa cells were mock transfected (○) or transfected with CatA-specific (▾) or control (●) siRNAs. The cells were incubated with 14C-labeled GS-9191 (10 μM; 0.521 μCi/ml) for 0 to 4 h on day 3. At selected time points, medium was removed, and the intracellular concentration of the metabolites was determined as described in Materials and Methods. The data are expressed as means ± standard deviations of two independent experiments.
Hydrolysis of GS-9191 by CatA.
To further understand the activation of GS-9191 by CatA, the compound was incubated in the presence of purified enzyme, and the reaction products were analyzed using HPLC. The hydrolysis of GS-9191 resulted in the appearance of a metabolite with the same retention time (13.8 min) as the cPrPMEDAP-Phe standard. In addition to cPrPMEDAP-Phe, l-phenylalanine 2-methylpropylester was detected as the only other product of the CatA-mediated hydrolysis of GS-9191, indicating that CatA can only hydrolyze one carboxylester bond in the GS-9191 molecule.
Intracellular levels of CatA and antiproliferative activity of GS-9191.
A potent antiproliferative effect of GS-9191 against a range of HPV-transformed cell lines has been recently reported (24). To understand the effect of CatA on the activity of GS-9191, we assessed the correlation between intracellular levels of CatA and the potency of GS-9191 in various cell lines. CatA protein levels in prepared cell extracts were quantified using immunoblot analysis with a fluorescently labeled secondary antibody (Table 1). The analytical conditions were optimized to achieve linear proportionality between the amount of CatA and the fluorescence signal on the immunoblot. In addition, the levels of GS-9191 hydrolysis in cellular extracts were also determined using HPLC-based quantification of GS-9191 and the product of its hydrolysis (cPrPMEDAP-Phe). While CatA protein levels determined by immunoblot analysis correlated well with the rate of GS-9191 hydrolysis in cell extracts, there was no correlation observed between GS-9191 potency (EC50) in tested cell lines and either CatA levels or the GS-9191 rate of hydrolysis in the corresponding cell extracts (R2 = 0.04). This suggests that the initial CatA-mediated hydrolysis of GS-9191 is unlikely to be the only rate-limiting step in the compound activation pathway.
Table 1.
Cathepsin A expression and antiproliferative activity of GS-9191 in different cell lines.
| Cell line | EC50a (nM) | CatA (ng/μg cellular protein)b,c | GS-9191 hydrolysis (pmol/min · μg)b,d |
|---|---|---|---|
| SiHa | 0.03 ± 0.01 | 4.5 ± 1.1 | 38 ± 8.2 |
| MS-751 | 0.04 ± 0.02 | 2.7 ± 1.1 | ND |
| HEL | 7.44 ± 8.62 | 2.4 ± 0.6 | 27.3 ± 6 |
| CaSki | 2.03 ± 0.97 | 1.12 ± 0.6 | 6.6 ± 1.2 |
| HeLa | 0.71 ± 0.46 | 0.8 ± 0.4 | ND |
| HT-3 | 5.42 ± 4.75 | 0.7 ± 0.3 | ND |
| SCC-4 | 5.03 ± 6.76 | 0.5 ± 0.3 | ND |
| ME-180 | 1.83 ± 1.52 | 0.5 ± 0.2 | ND |
| PHK | 2.91 ± 3.33 | <0.3 | 3.9 ± 3.7 |
| C-4I | 0.44 ± 0.07 | <0.3 | 2.6 ± 0.5 |
EC50s were published previously (24).
The results represent the means ± standard deviations of two independent experiments.
Cell lysates were analyzed by quantitative immunoblotting using anti-cathepsin A antibody.
GS-9191 was incubated with cell lysates, and the formation of cPrPMEDAP-Phe was monitored by HPLC.
Subcellular localization of GS-9191 metabolites.
Since CatA is a lysosomal carboxylpeptidase, the hydrolysis of GS-9191 should occur primarily in lysosomes. In contrast, the downstream activation steps of GS-9191, including the deamination of cPrPMEDAP to PMEG and the phosphorylation of PMEG, occur in cytosol (18, 24), indicating that cPrPMEDAP or the upstream metabolite cPrPMEDAP-Phe should undergo translocation from lysosomes to the cytosol. Previous findings indicating that all major metabolites of the tenofovir amidate prodrug GS-7340 localize to cytosol support this hypothesis (2).
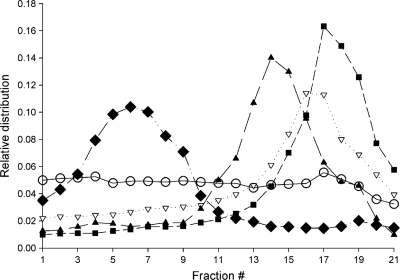
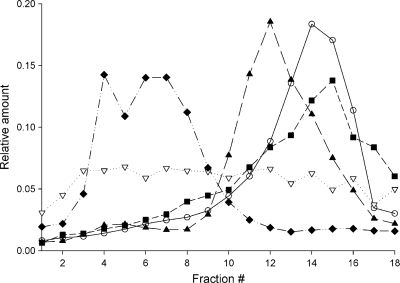
To test the intracellular distribution of GS-9191 metabolites, HEL and SiHa cells were incubated with [14C]GS-9191 for 4 h, disintegrated, and fractionated using Percoll density gradient ultracentrifugation (23). Each gradient fraction was examined for total radioactivity, representing GS-9191 metabolites and the activities of specific enzymatic markers for the endoplasmic reticulum, mitochondria, and lysosomes. As shown in Fig. 4, the peak radioactivity in fractions from HEL cells was copurified with the peak activity of the lysosomal marker β-hexosaminidase, indicating trapping of GS-9191 metabolites in the lysosomal/late endosomal compartment. In contrast, no significant lysosomal trapping of GS-9191 metabolites was observed in SiHa cells (Fig. 4).
Fig. 4.
Subcellular localization of GS-9191-derived metabolites in SiHa and HEL cells. SiHa (○) and HEL (▿) cells were incubated with 14C-labeled GS-9191 (10 μM; 0.521 μCi/ml) for 4 h. The cells were lysed, and the organelles were separated on a Percoll gradient as described in Materials and Methods. Shown are the lysosomal marker hexosaminidase (■), the mitochondrial marker succinate dehydrogenase (▴), and the endoplasmic reticulum marker NADPH cytochrome c reductase ( ).
).
To further investigate the role of lysosomes in the activation of GS-9191 in SiHa cells, the cell cultures were incubated with [14C]GS-9191 in the presence and absence of bafilomycin A, an agent that increases the lysosomal pH through the inhibition of the lysosomal H+ pump (6). The results of the Percoll gradient fractionation indicated that treatment with bafilomycin A induces substantial accumulation of GS-9191 metabolites in lysosomes/endosomes (Fig. 5). To establish the identity of these metabolites, the intracellular metabolism of GS-9191 was investigated in SiHa cells treated with 100 nM bafilomycin A. In parallel, the same experiment was also performed in the presence of 100 μM chloroquine, a compound that increases lysosomal pH via a mechanism different from that of bafilomycin A (16). Fluorescein-labeled dextran was used as a pH probe to optimize the concentration of both compounds so that they increase the lysosomal pH to 7.4 (16). As shown in Table 2, the pretreatment of cells with either of the two agents increased the total level of GS-9191 metabolites in SiHa cells approximately 3-fold, with cPrPMEDAP-Phe and cPrPMEDAP being two predominant metabolic forms. In addition, the neutralization of the lysosomal pH resulted in an approximately 8-fold increase in the cPrPMEDAP-Phe intracellular levels compared to control cells, indicating that the hydrolysis of cPrPMEDAP-Phe is affected by the lysosomal pH and thus likely occurs in this compartment following the cleavage of GS-9191 by CatA.
Fig. 5.
Subcellular localization of GS-9191-derived metabolites in SiHa cells pretreated with 100 nM bafilomycin. SiHa cells were pretreated with 100 nM bafilomycin A (○) or DMSO (▿) for 60 min and then incubated with 14C-labeled GS-9191 (10 μM; 0.521 μCi/ml) for another 30 min. The cells were lysed, and the organelles were separated on a Percoll gradient as described in Materials and Methods. Shown are the lysosomal marker hexosaminidase (■), the mitochondrial marker succinate dehydrogenase (▴), and the endoplasmic reticulum marker NADPH cytochrome c reductase ( ).
).
Table 2.
Effects of bafilomycin A and chloroquine on intracellular levels of cPrMEDAP-Phe and cPrMEDAP in SiHa cells
| Time (min) | Treatment of SiHa cells | Intracellular concn (μM)a (fold change relative to untreated control) |
|
|---|---|---|---|
| cPrPMEDAP | cPrPMEDAP-Phe | ||
| 30.0 | Control | 199.2 ± 21.1 | 38.0 ± 4.5 |
| 100 nM bafilomycin | 372.6 ± 42.4 (1.9) | 303.7 ± 21.0 (8.0) | |
| 100 μM chloroquine | 391.2 ± 29.6 (2.0) | 306.7 ± 18.9 (8.0) | |
| 60.0 | Control | 337.3 ± 32.0 | 54.3 ± 3.0 |
| 100 nM bafilomycin | 482.5 ± 51.5 (1.4) | 296.4 ± 23.1 (5.5) | |
| 100 μM chloroquine | 709.3 ± 59.1 (2.1) | 299.7 ± 27.6 (5.5) | |
| 120.0 | Control | 476.1 ± 56.8 | 59.0 ± 5.1 |
| 100 nM bafilomycin | 642.8 ± 78.4 (1.3) | 195.7 ± 26.0 (3.3) | |
| 100 μM chloroquine | 895.1 ± 91.1 (1.9) | 175.9 ± 23.4 (3.0) | |
The results represent the means ± standard deviations of two independent experiments.
Mechanism of conversion of cPrPMEDAP-Phe to cPrPMEDAP.
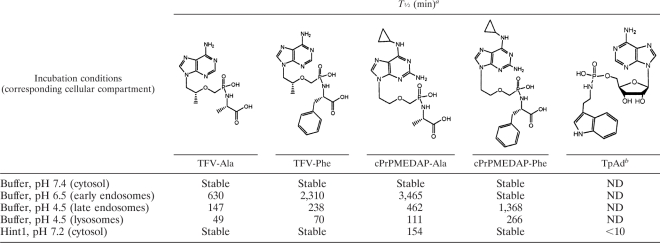
It has been reported previously that a conjugate of alanine with TFV has limited stability in acidic pH (2) but is stable at pHs above 7.0. To investigate the stability of the conjugates of nucleotides with amino acids in greater detail, the spontaneous hydrolysis of cPrPMEDAP-Ala, cPrPMEDAP-Phe, TFV-Ala, and TFV-Phe was measured in buffers at different pHs ranging from 4.5 (lysosomes) to 7.4 (cytosol). Tenofovir-amino acid conjugates proved to be less stable in acidic pH than the corresponding amino acid conjugates of cPrPMEDAP (Table 3). Furthermore, conjugates derived from prodrugs with alanine exhibited lower stability in acidic pH than those with phenylalanine.
Table 3.
Half-lives (min) of nucleotide-amino acid conjugates at different pHs and in the presence of recombinant Hint1
Conjugates were incubated with recombinant Hint1 or in buffers of different pHs, and the formation of parent nucleotides was monitored by HPLC.
TpAd, tryptamine adenosine phosphoramidate monoester.
We also measured the hydrolysis of metabolites by recombinant human Hint1 (hHint1), an enzyme with phosphoramidase activity that has been shown to hydrolyze the P-N bond in conjugates of amines with natural nucleotides (4, 5). However, human Hint1 was able to hydrolyze only cPrPMEDAP-Ala and a synthetic control substrate, tryptamine adenosine phosphoramidate monoester (TpAd) (Table 3), but not the GS-9191-derived metabolite cPrPMEDAP-Phe. These results are consistent with the significantly reduced hHint1 catalytic turnover observed for substrates containing substitutions α to the amine, as well as loss of the ribosyl 2′-hydroxyl (5).
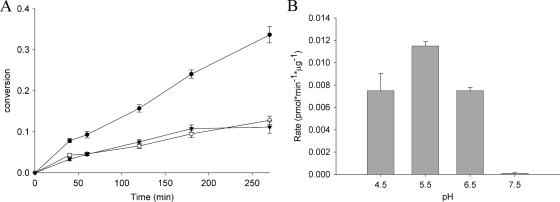
We therefore incubated cPrPMEDAP-Phe in the presence of extract prepared from SiHa cells at different pHs. In acidic pH, cPrPMEDAP-Phe was hydrolyzed significantly faster in the presence of SiHa cell extract than with buffer alone (Fig. 6A), and this enzymatically driven hydrolysis showed a pH optimum of approximately 5.5 (Fig. 6B). These data clearly indicate the presence of an acidic phosphoamidase activity in SiHa cell extract capable of hydrolyzing cPrPMEDAP-Phe.
Fig. 6.
Hydrolysis of cPrPMEDAP-Phe in the presence of SiHa cell extracts. (A) [14C]cPrPMEDAP-Phe (30 μM; 1.5 μCi/ml) was incubated in a buffer (pH 5.5) containing extract from SiHa cells (●), denatured extract (○), or buffer only (▾). The conversion to cPrPMEDAP was monitored by HLPC. (B) Effect of pH on the cPrPMEDAP-Phe-hydrolyzing activity present in SiHa extracts. The data are expressed as means ± standard deviations of two independent experiments.
DISCUSSION
Results from our initial experiments indicated that CatA is capable of hydrolyzing the isobutyl-ester bond in GS-9191 in vitro, which constitutes the initial step in the prodrug activation pathway (24). In addition, previous studies established the pathways leading to the formation of the active metabolite PMEGpp from cPrPMEDAP (10, 18, 19). In the present study, we further investigated the role of CatA in the metabolism of GS-9191 and mapped the mechanism of the formation of cPrPMEDAP.
To assess the role of CatA in the activation of GS-9191 in intact cells, we compared the rates of GS-9191 hydrolysis in fibroblasts isolated from individuals with wild-type CatA and those with CatA deficiency. In addition, levels of GS-9191 were assessed in HeLa cells with decreased CatA expression following the siRNA knockdown. Significantly reduced levels of GS-9191 metabolites both in cells naturally deficient in CatA and in cells in which the enzyme expression was artificially reduced indicate the major role of CatA in the hydrolysis of GS-9191. Thus, similarly to monoamidate prodrugs of the nucleotide HIV reverse transcriptase inhibitors tenofovir and GS-9148 (3), CatA also plays an important role in the intracellular activation of the bis-amidate GS-9191. It should be noted that cPrPMEDAP and cPrPMEDAP-Phe can still be formed in cells with CatA deficiency, albeit with much lower efficiency. This indicates that, in addition to CatA, some other enzyme(s) may contribute to the hydrolysis of GS-9191. The identity of this secondary enzyme(s) has yet to be determined. Similar to what has been observed with amidate prodrugs of tenofovir (2), additional cellular proteases may contribute to the hydrolysis of GS-9191.
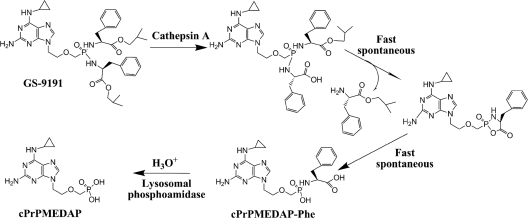
The steps immediately following the carboxylester bond hydrolysis in monoamidate prodrugs are well understood. The freed carboxylate mediates a nucleophilic attack on the phosphorus, leading to the formation of a metastable cyclic intermediate, from which the phenol group is spontaneously eliminated (1, 22). Thus, the product of the hydrolysis of monoamidate prodrugs is a conjugate of amino acid and nucleotide. In the case of symmetrical bisamidate prodrugs, like GS-9191, CatA might cleave one or both carboxylester bonds. Moreover, the freed carboxylate might not be able to efficiently eliminate the whole amino acid ester moiety from the intermediate formed. In order to address these questions, we incubated GS-9191 with purified CatA and analyzed the reaction products. cPrPMEDAP-Phe and l-phenylalanine-2-methylpropylester were the only products detected, proving that the initial activation of bisamidate prodrugs follows the same pathway established with monoamidates (1, 22).
The important role of CatA in GS-9191 metabolism raised the question of whether CatA could serve as a marker for sensitivity to GS-9191. We therefore tested this hypothesis by developing a quantitative immunoblot analysis of CatA expression in the cellular extracts. Somewhat surprisingly, we did not observe any correlation between CatA levels and 50% effective concentrations (EC50s) for GS-9191 in the tested cell lines. On the other hand, the GS-9191 hydrolytic activity in the cell extracts correlated well with CatA expression levels, supporting the conclusion that Cat A is the main hydrolase involved in the intracellular metabolism of GS-9191 in these cell lines. There are various reasons that can explain the lack of correlation between sensitivity to GS-9191 and CatA levels in the tested set of cells. In this and previous studies, we observed that the main metabolite in cells exposed to GS-9191 is cPrPMEDAP (24), indicating that the rate-limiting step in the activation of GS-9191 is the conversion of cPrPMEDAP to PMEG, which is affected by the cellular levels of N6-methyl-AMP aminohydrolase (C. Frey and G. Birkus, unpublished data). Therefore, the high rate of hydrolysis of GS-9191 does not necessarily translate into high levels of the active metabolite PMEGpp. In addition, the slow conversion of cPrPMEDAP to PMEG increases the likelihood of cPrPMEDAP elimination from cells by membrane efflux transporters, such as MRP4 or MRP5, whose expression may also vary in the tested cell lines. Last but not least, our present data suggest that the transport of cPrPMEDAP and/or cPrPMEDAP-Phe from lysosomes to the cytosol might represent another possible bottleneck in the activation of GS-9191 in some cell types, such as HEL fibroblasts (see below). Thus, the sensitivity of cells to GS-9191 can be affected by factors other than the level of CatA expression.
As CatA is a lysosomal enzyme, we further characterized the role of lysosomes in the metabolism of GS-9191. We assessed the intracellular distribution of GS-9191 metabolites in organelles of HEL and SiHa cells. While metabolites in HEL cells were detected almost exclusively in dense fractions that copurified with a marker for lysosomes, the metabolites in SiHa cells were distributed in the cytosol. The trapping of GS-9191 metabolites in lysosomes of HEL cells is the likely explanation for the 100-fold-higher potency of the compound in SiHa cells than in HEL cells, even though the CatA levels in the two cell lines are comparable (Table 1). Additional evidence for the role of lysosomes in the activation of GS-9191 was generated using bafilomycin A, an agent that increases the lysosomal pH through the inhibition of the lysosomal H+ pump (6). Treatment of SiHa cells with bafilomycin A resulted in almost complete retention of GS-9191 metabolites in lysosomes/endosomes, further confirming that the generation of cPrPMEDAP-Phe from GS-9191 occurs in this cellular compartment.
Several mechanisms were considered for the subsequent hydrolysis of cPrPMEDAP-Phe to cPrPMEDAP. It is known that the P-N bond in nucleotide-amino acid conjugates is labile in acidic pH, suggesting that this step might be a spontaneous process driven by low lysosomal/endosomal pH. Alternatively, the conjugate might be enzymatically hydrolyzed, either directly in lysosomes or following its translocation into the cytosol, where it might be a substrate for phosphoramidases, such as Hint1 (4, 5). To test these different alternatives, we prepared cPrPMEDAP-Phe, together with some other conjugates, and measured their stability under various conditions. At lysosomal pH, the half-life of cPrPMEDAP-Phe was 270 min, which is a rate substantially lower than that at which cPrPMEDAP emerges in intact cells treated with GS-9191 (Table 2), indicating that an enzymatic process is involved in the hydrolysis of cPrPMEDAP-Phe. While Hint1 was unable to release cPrPMEDAP from its conjugate with phenylalanine, a phosphoamidase activity capable of hydrolyzing cPrPMEDAP-Phe was detected in extract from SiHa cells. Its acidic pH optimum indicates that the enzyme, which remains to be identified, is likely localized in the endosomal/lysosomal compartment. Furthermore, the increase of lysosomal pH using the treatment with bafilomycin A or chloroquine resulted in up to 8-fold-higher intracellular levels of cPrPMEDAP-Phe, with the majority trapped in lysosomes.
Taken together, the present data indicate that both the initial hydrolysis of GS-9191 by CatA and the subsequent release of cPrPMEDAP from cPrPMEDAP-Phe occur in the lysosomal/endosomal compartment. It is likely that the latter step is driven primarily by enzymatic hydrolysis, but the spontaneous cleavage of P-N at acidic pH may also contribute to the generation of cPrPMEDAP (Fig. 7). cPrPMEDAP is subsequently translocated to the cytosol, where it undergoes deamination and phosphorylation, yielding the active metabolite PMEGpp. The fact that cPrPMEDAP can be trapped in lysosomes by artificially increasing the lysosomal pH indicates the presence of an active lysosomal efflux. However, all of the observations presented were obtained with highly passaged cell lines, and their in vivo relevance has to be established.
Fig. 7.
Proposed pathway for GS-9191 conversion to cPrPMEDAP occurring in lysosomes.
Phosphonoamidates recently emerged as novel prodrugs of nucleoside phosphonates with significant pharmacological and therapeutic advantages compared to ester-based nucleotide prodrugs (12, 18). GS-7340, a monoamidate of the nucleotide HIV reverse transcriptase inhibitor tenofovir, has shown potent antiretroviral efficacy in phase I clinical studies. In addition, two bis-amidates of cPrPMEDAP are currently in clinical development as potential treatments for anti-proliferative disorders. While the highly lipophilic prodrug GS-9191 is being explored as a topical treatment for genital warts, another, more hydrophilic prodrug, GS-9219, is being developed for the systemic treatment of hematological malignancies (18). Detailed understanding of intracellular metabolic pathways involved in the activation of phosphonoamidates, including the identification of specific activation bottlenecks, is essential for successful exploration of the full therapeutic potential offered by this class of nucleoside phosphonate prodrugs.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 7 March 2011.
REFERENCES
- 1. Balzarini J., et al. 1996. Mechanism of anti-HIV action of masked alaninyl d4T-MP derivatives. Proc. Natl. Acad. Sci. U. S. A. 93:7295–7299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Birkus G., et al. 2008. Activation of 9-[(R)-2-[[(S)-[[(S)-1-(Isopropoxycarbonyl)ethyl]amino] phenoxyphosphinyl]-methoxy]propyl]adenine (GS-7340) and other tenofovir phosphonoamidate prodrugs by human proteases. Mol. Pharmacol. 74:92–100 [DOI] [PubMed] [Google Scholar]
- 3. Birkus G., et al. 2007. Cathepsin A is the major hydrolase catalyzing the intracellular hydrolysis of the antiretroviral nucleotide phosphonoamidate prodrugs GS-7340 and GS-9131. Antimicrob. Agents Chemother. 51:543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chou T. F., Cheng J., Bieganowski P., Brenner C., Wagner C. R. 2005. 31P NMR and genetic analysis establish hinT as the only E. coli purine nucleoside phosphoramidase and as essential for growth under high salt conditions. J. Biol. Chem. 280:15356–15361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chou T. F., et al. 2007. Phosphoramidate pronucleotides: a comparison of the phosphoramidase substrate specificity of human and Escherichia coli histidine triad nucleotide binding proteins (Hint1). Mol. Pharm. 4:208–217 [DOI] [PubMed] [Google Scholar]
- 6. Dröse S., Altendorf K. 1997. Bafilomycins and concanamycins as inhibitors of V-ATPase and P-ATPase. J. Exp. Biol. 200:1–8 [DOI] [PubMed] [Google Scholar]
- 7. Graceway Pharmaceuticals 6 November 2009. posting date Graceway Pharmaceuticals acquires worldwide rights to GS 9191 from Gilead Sciences. Graceway Pharmaceuticals, Bristol, TN: http://www.gracewaypharma.com/news/0035-graceway-pharmaceuticals-acquires-worldwide-rights-gs-9191-gilead-sciences [Google Scholar]
- 8. Kramata P., Votruba I., Otova B., Holy A. 1996. Different inhibitory potencies of acyclic phosphonomethoxyalkyl nucleotide analogs toward DNA polymerases α, δ and ε. Mol. Pharmacol. 49:1005–1011 [PubMed] [Google Scholar]
- 9. Kramata P., Downey K. M., Paborsky L. R. 1998. Incorporation and excision of 9-(2-phosphonylmethoxyethyl)guanine (PMEG) by DNA polymerase delta and epsilon in vitro. J. Biol. Chem. 273:21966–21971 [DOI] [PubMed] [Google Scholar]
- 10. Krejčová R., Horská K., Votruba I., Holý A. 1999. Isoenzymes of GMP kinase from L1210 cells: isolation and characterization. Collect. Czech. Chem. Commun. 64:559–570 [Google Scholar]
- 11. Kuo S. R., Liu J. S., Broker T. R., Chow L. T. 1994. Cell-free replication of the human papillomavirus DNA with homologous viral E1 and E2 proteins and human cell extracts. J. Biol. Chem. 269:24058–24065 [PubMed] [Google Scholar]
- 12. Lee W. A., et al. 2005. Selective intracellular activation of a novel prodrug of the human immunodeficiency virus reverse transcriptase inhibitor tenofovir leads to preferential distribution and accumulation in lymphatic tissue. Antimicrob. Agents Chemother. 49:1898–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Merion M., Sly W. S. 1983. The role of intermediate vesicles in the adsorptive endocytosis and transport of ligand to lysosomes by human fibroblasts. J. Cell Biol. 96:644–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ouar Z., et al. 2003. Inhibitors of vacuolar H+-ATPase impair the preferential accumulation of daunomycin in lysosomes and reverse the resistance to anthracyclines in drug-resistant renal epithelial cells. Biochem. J. 370:185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pennington R. J. 1961. Biochemistry of dystrophic muscle mitochondrial succinatetetrazolium reductase and adenosine triphosphatase. Biochem. J. 80:649–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Poole B., Ohkuma S. 1981. Effect of weak bases on the intralysosomal pH in mouse peritoneal macrophages. J. Cell Biol. 90:665–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pshezhetsky A. V., Potier M. 1996. Association of N-acetylgalactosamine-6-sulfate sulfatase with the multienzyme lysosomal complex of beta-galactosidase, cathepsin A, and neuraminidase. Possible implication for intralysosomal catabolism of keratan sulfate. J. Biol. Chem. 271:28359–28365 [DOI] [PubMed] [Google Scholar]
- 18. Reiser H., et al. 2008. GS-9219—a novel prodrug of the acyclic nucleotide PMEG with potent anti-neoplastic activity in dogs with spontaneous non-Hodgkin's lymphoma. Clin. Cancer Res. 14:2824–2832 [DOI] [PubMed] [Google Scholar]
- 19. Schinkmanová M., et al. 2008. Human N6-methyl-AMP/DAMP aminohydrolase (abacavir 5′-monophosphate deaminase) is capable of metabolizing N6-substituted purine acyclic nucleoside phosphonates. Collect. Czech. Chem. Commun. 73:275–291 [Google Scholar]
- 20. Shimmoto M., et al. 1993. Protective protein gene mutations in galactosialidosis. J. Clin. Invest. 91:2393–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tranchemontagne J., Michaud L., Potier M. 1990. Deficient lysosomal carboxypeptidase activity in galactosialidosis. Biochem. Biophys. Res. Commun. 168:22–29 [DOI] [PubMed] [Google Scholar]
- 22. Valette G., et al. 1996. Decomposition pathways and in vitro HIV inhibitory effects of isoddA pronucleotides: toward a rational approach for intracellular delivery of nucleoside 5′-monophosphates. J. Med. Chem. 39:1981–1990 [DOI] [PubMed] [Google Scholar]
- 23. Wex T., et al. 2001. Human cathepsin W, a cysteine protease predominantly expressed in NK cells, is mainly localized in the endoplasmic reticulum. J. Immunol. 167:2172–2178 [DOI] [PubMed] [Google Scholar]
- 24. Wolfgang G. H., et al. 2009. GS-9191 is a novel topical prodrug of the nucleotide analog 9-(2-phosphonylmethoxyethyl)guanine with antiproliferative activity and possible utility in the treatment of human papillomavirus lesions. Antimicrob. Agents Chemother. 53:2777–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.