Abstract
Cyclophilin inhibitors currently in clinical trials for hepatitis C virus (HCV) are all analogues of cyclosporine (CsA). Sanglifehrins are a group of naturally occurring cyclophilin binding polyketides that are structurally distinct from the cyclosporines and are produced by a microorganism amenable to biosynthetic engineering for lead optimization and large-scale production by fermentation. Preclinical characterization of the potential utility of this class of compounds for the treatment of HCV revealed that the natural sanglifehrins A to D are all more potent than CsA at disrupting formation of the NS5A-CypA, -CypB, and -CypD complexes and at inhibition of CypA, CypB, and CypD isomerase activity. In particular, sanglifehrin B (SfB) was 30- to 50-fold more potent at inhibiting the isomerase activity of all Cyps tested than CsA and was also shown to be a more potent inhibitor of the 1b subgenomic replicon (50% effective concentrations [EC50s] of 0.070 μM and 0.16 μM in Huh 5-2 and Huh 9-13 cells, respectively). Physicochemical and mouse pharmacokinetic analyses revealed low oral bioavailability (F < 4%) and low solubility (<25 μM), although the half-lives (t1/2) of SfA and SfB in mouse blood after intravenous (i.v.) dosing were long (t1/2 > 5 h). These data demonstrate that naturally occurring sanglifehrins are suitable lead compounds for the development of novel analogues that are less immunosuppressive and that have improved metabolism and pharmacokinetic properties.
INTRODUCTION
Hepatitis C virus (HCV) is a positive-strand RNA virus and is the major cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (39). The World Health Organization (WHO) estimates that there are more than 170 million chronic carriers of HCV, which is approximately 3% of the world population (28, 29). In developed countries, 50 to 76% of all cases of liver cancer and two-thirds of all liver transplants are due to chronic HCV infection (37).
The current standard of care (SOC) involves subcutaneous injections of pegylated alpha-interferon (peg-IFN-α) and oral dosing of the nonspecific antiviral drug ribavirin for a period of 24 to 48 weeks (9). Overall, response rates to the SOC depend upon the genotype and the pretreatment HCV RNA levels. Patients with genotypes 2 and 3 are more responsive to the SOC than patients infected with genotype 1 (particularly 1b) (25, 31). A significant number of HCV patients do not respond adequately to the SOC or cannot tolerate it due to side effects, leading to poor compliance and a sustained virological response (SVR) of about 50% (31).
Direct-acting antiviral (DAA) drugs are being developed to target viral proteins, such as HCV NS5B polymerase or NS3 protease (35). One concern with the increased use of virus-targeted agents is that, as was observed with HIV treatment, drug resistance emerges (47). However, compounds that target human proteins (i.e., cyclophilins), rather than viral targets, are also being developed, which should have higher hurdles for viral breakthrough and incidence of resistance during drug therapy (29, 38, 36).
Cyclophilins (Cyps) are a family of cellular proteins that display peptidyl-prolyl cis-trans isomerase activity, facilitating protein conformational changes and folding. Cyps are involved in cellular processes, such as transcriptional regulation, immune response, protein secretion, and mitochondrial function (48). During infection, HCV recruits Cyps for its replication. Although earlier resistance studies on cyclosporine (CsA) showed mutations in the NS5B polymerase and suggested that only cyclophilin B (CypB) was involved in the HCV replication process (42), recent studies have suggested an essential role for cyclophilin A (CypA) in HCV replication (2, 5, 24, 51) and also a potential role for a wider selection of cyclophilin pathways (17). The generation of knockouts in mice (7) and human T cells (1) indicates that CypA is not essential for cell growth and survival. Similar results have been observed with disruption of CypA homologues in bacteria (20), Neurospora sp. (45), and Saccharomyces cerevisiae (12). Therefore, inhibiting Cyps represents a novel and attractive host target for treating HCV infection. Most likely, these compounds will be combined with SOC or DAA drugs, which may result in an increase in the SVR and prevent the emergence of resistance.
Cyclosporine (23) and its structurally related nonimmunosuppressive clinical analogues alisporivir (DEBIO-025) (6, 16, 33), NIM811 (30), and SCY-635 (21) (Fig. 1B) are cyclophilin binding compounds. These compounds are active against multiple HCV genotypes and exhibit a significantly higher hurdle for the selection of resistance (6). However, adverse events, such as hyperbilirubinemia, were seen in a number of patients in early studies, particularly at higher dose levels (i.e., 1,000 mg), leading to treatment discontinuation with the most advanced candidate, alisporivir (15, 16). With lower doses (i.e., 400 mg orally [p.o.] once a day [q.d.]) of the compound, hyperbilirubinemia was less of an issue (32, 49). Interpatient variability, such as is seen with CsA itself (where careful drug level monitoring is needed to avoid adverse events), is also a concern (27).
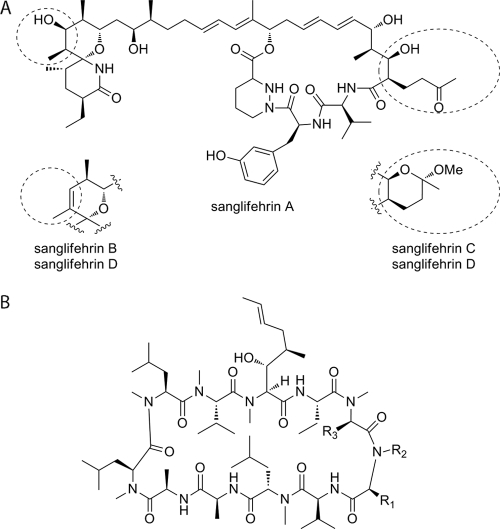
Fig. 1.
Structural formulae of sanglifehrins and cyclosporines. (A) Structures of sanglifehrins A to D. (B) Structure of cyclosporines. CsA, R1 = CH2CH(CH3)2, R2 = CH3, R3 = H; alisporivir, R1 = CH(CH3)2, R2 = CH2CH3, R3 = CH3; SCY-635, R1 = CH2C(CH3)2OH, R2 = CH3, R3 = SCH2N(CH3)2; NIM-811, R1 = CH(CH3)CH2CH3, R2 = CH3, R3 = H.
Sanglifehrin A (SfA) and its natural analogues belong to a class of mixed nonribosomal peptides/polyketides, produced by the soil bacterium Streptomyces sp. strain A92-308110, which were originally discovered on the basis of their high affinity for CypA (Fig. 1A). SfA is the most abundant component in fermentation broths and exhibits approximately 20-fold-higher affinity for CypA than CsA (43). This led to the suggestion that sanglifehrins could be useful for the treatment of HCV (17).
Sanglifehrins exhibit immunosuppressive activity, the mechanism of action of which is not fully understood but has been shown not to involve calcineurin, the immunosuppressive target of CsA (52). The crystal structures of SfA and a semisynthetic derivative bound to CypA have been published (26) and reveal that they both bind to the same active site as CsA. SfA analogues devoid of immunosuppressive properties have been synthesized (44), indicating an opportunity for the design of nonimmunosuppressive Cyp inhibitors for potential use in HCV therapy.
Previous limited HCV replicon and Cyp binding studies, mostly directed toward SfA, sanglifehrin B (SfB), and a limited set of semisynthetic derivatives, showed moderate potency (17, 18, 50). A resistant genotype 1b replicon was also generated for a sanglifehrin derivative, SFA-1, and showed cross-resistance with NIM-811 and CsA (40). This study includes not only SfA, but its naturally produced analogues SfB, SfC, and SfD. SfB differs from SfA by the presence of a cis double bond from C-35–C-36 in place of the hydroxyl group at C-35, and SfC differs from SfA by the formation of a hemiketal involving the C-53 ketone, C-15 hydroxyl, and methanol. SfD incorporates both of these changes (Fig. 1A). This study characterizes SfA, SfB, SfC, and SfD and evaluates their potential as future treatments for HCV.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
SfA, SfB, SfC, and SfD were prepared at Biotica Technology Ltd. (Cambridge, United Kingdom) as described previously (13, 43) (see the supplemental material). CsA was purchased from LC Labotatories (Woburn, MA).
Enzyme-linked immunosorbent assay (ELISA) and Western blot analysis of the effects of sanglifehrins on Cyp-NS5A interactions.
The production and purification of recombinant glutathione S-transferase (GST), GST-CypA, GST-CypB, GST-CypD, and full-length NS5A Con1 (pET-Ub-NS5A Con1-His) proteins were carried out as described previously (4, 22). Nunc MaxiSorb 8-well strip plates were coated with GST, GST-CypA, GST-CypB, or GST-CypD for 16 h at 4°C and blocked. Recombinant NS5A-His (1 ng/ml) was added to wells in 50 μl of binding buffer (20 mM Tris, pH 7.9, 0.5 M NaCl, 10% glycerol, 10 mM dithiothreitol [DTT], and 1% NP-40) for 16 h at 4°C. Captured NS5A-His was subsequently detected using mouse anti-His antibodies (1 μg/ml) (anti-6×His; Clontech) and rabbit anti-mouse-horseradish peroxidase phosphatase (HRP) antibodies (1:1,000 dilution). All experiments were conducted twice using two different batches of recombinant CypA, CypB, CypD, and NS5A proteins, while CsA and bovine serum albumin (BSA) were included as positive and negative controls, respectively.
For Western blot analysis, glutathione beads were incubated for 2 h in dialysis buffer (50 mM Tris, pH 7.4, 100 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.5% NP-40, 1 mM DTT) with 5 mg/ml BSA and washed twice at 4°C in binding buffer (20 mM Tris, pH 7.9, 0.5 M NaCl, 10% glycerol, 10 mM DTT, and 1% NP-40). Meanwhile, 100 ng of GST-CypA, GST-CypB, GST-CypD, or GST was mixed with 10 ng of NS5A-His, along with 0 to 0.625 μM drug, in a total volume of 200 μl of binding buffer for 3 h at 4°C. Glutathione beads (25 μl) were added to the GST-Cyp–NS5A mixture for 30 min at 4°C and washed 3 times with 400 μl of binding buffer. The beads were pelleted for 30 s at 2,000 × g in a microcentrifuge, and bound material was eluted with 25 μl of 2× SDS sample buffer, heated for 5 min, and frozen at −20°C. The bound material was then analyzed by Western blotting using anti-GST to detect GST-Cyp and anti-His antibodies to detect NS5A-His.
Antiviral assays.
Cells carrying the HCV replicons I377NS3-3′/wt (Huh 9-13) and I389luc-ubi-neo/NS3-3′/5.1 (Huh 5-2) were kindly provided by R. Bartenschlager (University of Heidelberg, Heidelberg, Germany). Assays in replicon-containing Huh 5-2 and Huh 9-13 (genotype 1b) cells were performed as described previously (34, 46).
Anti-PPIase analysis of sanglifehrins.
The peptidyl-prolyl cis-trans isomerase (PPIase) activity of recombinant Cyp, produced by thrombin cleavage of GST-Cyp, was determined by following the rate of hydrolysis of N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide by chymotrypsin. Cyp was equilibrated for 1 h at 5°C using a drug concentration range from 0.1 to 20 nM. The reaction was started by addition of the peptide, and the change in absorbance was monitored spectrophotometrically at 10 data points per second. The blank rates of hydrolysis (in the absence of Cyp) were subtracted from the rates in the presence of Cyp. The initial rates of the enzymatic reaction were analyzed by first-order regression analysis of the time course of the change in absorbance.
Cytotoxicity assays.
Huh7 and HepG2 cells (ATCC) were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, and 1% glutamine, whereas CEM cells (ATCC) were grown in RPMI 1640 medium with 10% FBS, 1% penicillin-streptomycin, and 1% glutamine. Fresh human peripheral blood mononuclear cells (PBMCs) were isolated from whole blood (see the supplemental material). Compound cytotoxicity was evaluated by testing half-log concentrations of each compound in triplicate against the cells described above. Untreated cells served as the controls. Huh7 and HepG2 cells were seeded at a concentration of 5 × 103 cells/well. After 24 h, the media containing 5% FBS and serial dilutions of compounds were added. After 72 h, the cells were processed for cytotoxicity assessment. PBMCs were diluted in fresh medium and plated at 5 × 104 cells/well. Similarly, CEM cells were plated at a density of 1 × 104 cells/well. Following compound addition, the cultures were maintained for 6 to 7 days and then processed for cytotoxicity determination. Cytotoxicity was determined using a CytoTox-One homogeneous membrane integrity assay kit (Promega).
Mouse intravenous and oral pharmacokinetic analysis.
Compounds were formulated in 5% ethanol, 5% cremophor EL, 90% saline for both p.o. and intravenous (i.v.) administration. Groups of 3 male CD1 mice were dosed with either 1 mg/kg of body weight i.v. or 10 mg/kg p.o. Whole-blood samples (40 μl) were taken via tail or saphenous vein predose and at 0.25, 0.5, 2, 8, and 24 h; diluted with an equal amount of distilled water (dH2O); and placed on dry ice immediately. The samples were stored at −70°C until analysis was performed. The concentration of the test article in the sample was then determined by liquid chromatography-tandem mass spectrometry (LC–MS-MS). The time course of blood concentrations was plotted and used to derive the area under the whole-blood concentration-time curve (AUC), which is directly proportional to the total amount of unchanged drug that reaches the systemic circulation. These values were used to generate pharmacokinetic (PK) parameters (with a noncompartmental model) using WinNonlin (version 5.2; Pharsight Corp., CA).
Solubility analysis.
Solubility was measured by diluting test compounds in dimethyl sulfoxide (DMSO) (10 mM) in phosphate-buffered saline (PBS) at pH 7.4 to a target concentration of 100 μM with a final DMSO concentration of 1%. The sample tubes were gently shaken for 4 h at room temperature and centrifuged, and the supernatants were diluted in PBS. The diluted samples were mixed with the same volume (1:1) of methanol and then the same volume (1:1) of acetonitrile with internal standard for LC–MS-MS analysis.
Log D analysis.
The partition coefficient (log D) was measured at pH 7.4 using a miniaturized shake flask method. Partition of test compounds was measured in a mixture of n-octanol and potassium phosphate buffer (pH 7.4) after shaking it for 1 h at 25°C. After centrifugation, water phase samples were diluted with additional water and then 50% ethanol with internal standard to achieve a 1:60 dilution. Samples of the n-octanol phase were diluted in 50% ethanol with internal standard to achieve a 1:2,400 dilution. Samples were then analyzed by LC-MS.
Hepatocyte stability analysis.
Cryopreserved hepatocytes (2 × 106 cells/ml) were seeded in Krebs-Henseleit bicarbonate (KHB) buffer, and test compound in 1% DMSO in KHB buffer was added to a level of 1 μM. The plates were incubated at 37°C, and samples were taken after 0, 15, 30, 60, and 120 min. The samples were added to an equal volume of acetonitrile with internal standard and centrifuged, and compound levels were analyzed by LC–MS-MS.
Microsome stability analysis.
Mouse or human liver microsomes (2.5 mg/ml) were prepared in 0.1 M potassium phosphate buffer, 1.0 mM EDTA, pH 7.4. Test compound was added to 1 μM. Following preincubation, the reaction was initiated by adding NADPH solution. Aliquots were removed at 0, 15, 30, 45, and 60 min and quenched with acetonitrile with internal standard. Protein was removed by centrifugation, and the sample plate was analyzed for the compound concentration by LC–MS-MS.
Anti-CD3/anti-CD28 stimulated PBMC proliferation.
Cryopreserved PBMCs (Seracare) in complete culture medium (CM) were used at 1 × 105 cells/well. The test article was added to 5, 50, 500, and 5,000 nM, and the plates were incubated at 37°C with 5% CO2 for 1 h. Human T-activator CD3/CD28 Dynabeads (Invitrogen) at 1 × 105 beads/well were added to half of the plates. The plates were incubated for 72 h at 37°C with 5% CO2. Cell proliferation was assayed using a luminescent ATP assay (CellTiter-Glo; Promega) according to the manufacturer's instructions. Values from triplicate culture wells were averaged, and values were analyzed by one-way analysis of variance (ANOVA) with Tukey's posttest comparing sample values to the vehicle value (Prism V 4.0; GraphPad Software). Fifty percent inhibitory concentrations (IC50s) were determined using sigmoidal dose-response curves with curve constraints (Prism V 4.0; GraphPad Software).
RESULTS
Sanglifehrins potently block the interaction between CypA, CypB, CypD, and HCV NS5A.
Four natural sanglifehrins, A to D, were evaluated for their capacities to block the interaction between CypA, CypB, CypD (also termed CypF or mitochondrial cyclophilin), and HCV NS5A by means of ELISA. To test the effect of sanglifehrins on Cyp-NS5A complex formation, increasing concentrations of each compound were added to GST-CypA, GST-CypB, or GST-CypD, together with recombinant NS5A. All of the sanglifehrins were more potent than CsA in disrupting complex formation (Cyp-NS5A), with SfB the most potent compound (Table 1).
Table 1.
Analysis of NS5A-CypA, CypB, and CypD complex prevention (by ELISA) and anti-PPIase activities of sanglifehrins and CsA
| Cyclophilin inhibitor | NS5A-CypA disruption (IC50) (μM) | CypA PPIase inhibition (Ki) (nM) | NS5A-CypB disruption (IC50) (μM) | CypB PPIase inhibition (Ki) (nM) | NS5A-CypD disruption (IC50) (μM) | CypD PPIase inhibition (Ki) (nM) |
|---|---|---|---|---|---|---|
| SfA | 0.35 ± 0.11 | 2.4 ± 0.11 | 0.42 ± 0.09 | 2.1 ± 0.1 | 0.37 ± 1.2 | 2.4 ± 0.2 |
| SfB | 0.11 ± 0.02 | 0.32 ± 0.08 | 0.15 ± 0.02 | 0.20 ± 0.02 | 0.09 ± 0.03 | 0.3 ± 0.1 |
| SfC | 0.56 ± 0.04 | 6.8 ± 1.1 | 0.61 ± 0.04 | 5.4 ± 0.5 | 0.53 ± 0.05 | 5.1 ± 0.03 |
| SfD | 0.53 ± 0.02 | 6.9 ± 1.4 | 0.49 ± 0.03 | 5.8 ± 0.4 | 0.55 ± 0.04 | 6.1 ± 0.5 |
| CsA | 0.98 ± 0.08 | 9.7 ± 1.3 | 0.83 ± 0.05 | 9.2 ± 0.8 | 0.91 ± 0.7 | 9.4 ± 1.1 |
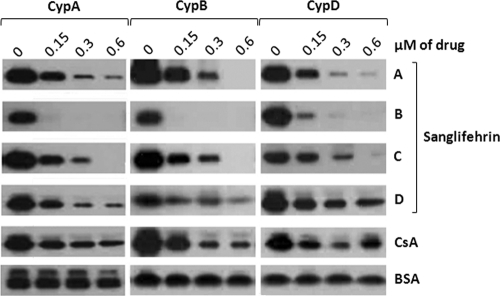
These data were further corroborated using GST-CypA, GST-CypB, or GST-CypD as bait to capture and pull down NS5A after the addition of glutathione beads. As previously reported (2, 14, 19), CsA (as a positive control) blocks the Cyp-NS5A interaction in a dose-dependent manner (Fig. 2, top), whereas BSA (as a negative control) does not (Fig. 2, bottom). All sanglifehrins tested were again more potent than CsA at blocking Cyp-NS5A complex formation for all cyclophilins tested (Fig. 2), with SfB the most potent (Fig. 2). In general, we observed good correlation between the ELISA and pulldown data (Table 1 and Fig. 2).
Fig. 2.
Sanglifehrins block recombinant Cyp-NS5A interactions in a dose-dependent manner. GST-CypA, GST-CypB, or GST-CypD (100 ng) was mixed with NS5A Con1-His (10 ng) for 3 h at 4°C in the presence of increasing concentrations of sanglifehrins, CsA, or BSA. Glutathione beads were added to the GST-Cyp/NS5A mixture for 30 min at 4°C and washed. Captured NS5A-His proteins were eluted and analyzed by Western blotting using anti-His antibodies.
Sanglifehrins potently inhibit the PPIase activity of CypA, CypB, and CypD.
Inhibition of the PPIase activity of CypA, CypB, and CypD was used to further analyze the inhibitory potential of sanglifehrins. CypA PPIase activity has previously been shown to be critical for HCV replication (2, 3). Sanglifehrins A to D inhibited the PPIase activity of CypA, CypB, and CypD more efficiently than CsA (Table 1), suggesting their affinities to the immunophilin are superior to that of CsA. SfB exhibited the most potent anti-PPIase activities (Ki = 0.32, 0.20, and 0.30 nM against CypA, CypB, and CypD, respectively), which was over 30-fold more potent than CsA in all cases (Ki = 9.7, 9.2, and 9.4 nM). Overall, a positive correlation was shown between the anti-PPIase activity of the sanglifehrins tested and their capacities to prevent Cyp-NS5A interactions analyzed by both ELISA and pulldown assays.
Sanglifehrins suppress HCV replicon 1b activity in a dose-dependent manner.
Sanglifehrins A to D were further tested in the Huh 5-2 and Huh 9-13 subgenomic replicon systems (genotype 1b) to measure their activities in a cell-based system. SfA exerted potency and cytotoxicity similar to those of CsA (SfA 50% effective concentration [EC50], 320 nM versus 306 nM for CsA in Huh 5-2 cells, and SfA EC50, 19 nM versus 415 nM for CsA in Huh 9-13 cells). Meanwhile, SfB and SfD were more potent, but also more cytotoxic, in Huh 5-2 (SfB EC50, 71 nM, and CC50 [the concentration of compound required to inhibit the exponential proliferation of the cells by 50%], 4.6 μM; SfD EC50, 306 nM, and CC50, 3.8 μM) (Table 2), suggesting that differences in rank order, compared with the NS5A-CypA and PPIase data, are most likely due to differing cell permeabilities. Further cytotoxicity analysis was carried out after incubation of SfA and SfB with two hepatocyte cell lines (Huh7 and HepG2), PBMCs, and the cancer cell line CEM and revealed that CC50 values were always above 10 μM (see the supplemental material).
Table 2.
Effects of sanglifehrins and CsA on replicons Huh 5-2 and Huh 9-13 (1b)
| Cyclophilin inhibitor | Huh 5-2 |
Wild-type Huh 9-13 |
CsAres Huh 9-13 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| EC50a (nM) | CC50b (μM) | SIc | EC50 (nM) | CC50 (μM) | SI | EC50 (nM) | CC50 (μM) | Wt/res ratiod | |
| SfA | 320 | 9.1 | 29 | 619 | 7.3 | 12 | 308 | 6.8 | 0.5 |
| SfB | 71 | 4.6 | 64 | 164 | 4.2 | 26 | 51 | 6.7 | 0.3 |
| SfC | 1,600 | >100 | >62 | 1,100 | 40 | 38 | 1,300 | >20 | 1.3 |
| SfD | 262 | 3.8 | 14 | 1,000 | 332 | 30 | 871 | 3.3 | 0.9 |
| CsA | 306 | 4.4 | 14 | 415 | 8.8 | 21 | 3,880 | 7.9 | 9.3 |
EC50 is the concentration of compound required to reduce the replicon content by 50% as measured by either the luciferase (Huh 5-2) or the RT-qPCR assay (Huh 9-13).
CC50 is the concentration of compound required to inhibit the exponential proliferation of the cells by 50% as determined by the MTS assay.
In vitro selectivity index (SI) = CC50/EC50.
Wt, wild type; res, resistant replicon.
Activity against CsA-resistant replicons.
To test whether sanglifehrins are active against subgenomic replicons containing mutations conferring resistance to CsA, SfA to -D were evaluated against a CsA-resistant Huh 9-13 subgenomic replicon. While CsA was shown to be 9 times less potent against the CsA-resistant replicon, sanglifehrins A to D were found to be as active against the resistant replicon as against the wild type (Table 2).
Physicochemical and DMPK analysis.
Optimal future HCV therapies will require oral dosing. Therefore, we analyzed the solubility in PBS and carried out initial in vitro drug metabolism and pharmacokinetic (DMPK) studies (Table 3). All sanglifehrins showed poor PBS solubility and log D values of >4. Human and mouse microsome stability was relatively low, with all sanglifehrins displaying human microsome half-lives of <8 min. In contrast, SfA and SfB were more stable in human hepatocytes, showing half-lives in excess of 100 min.
Table 3.
Physicochemical parametersa
| Cyclophilin inhibitor | Solubility (μM) | log D, pH 7.4 | HLM t1/2 (min) | MLM t1/2 (min) | Human hepatocyte t1/2 (min) |
|---|---|---|---|---|---|
| SfA | 45 | >4.6 | 7.9 | 4.0 | 145 |
| SfB | 16 | >4.5 | 7.0 | 2.4 | 156 |
| SfC | 10 | >4.2 | 3.4 | 11.0 | 40 |
| SfD | 25 | 4.1 | 4.4 | 9.1 | 54 |
Data are shown for solubility in PBS (pH 7.4), lipophilicity (log D, pH 7.4), and human (HLM) and mouse (MLM) microsome and hepatocyte stability (t1/2).
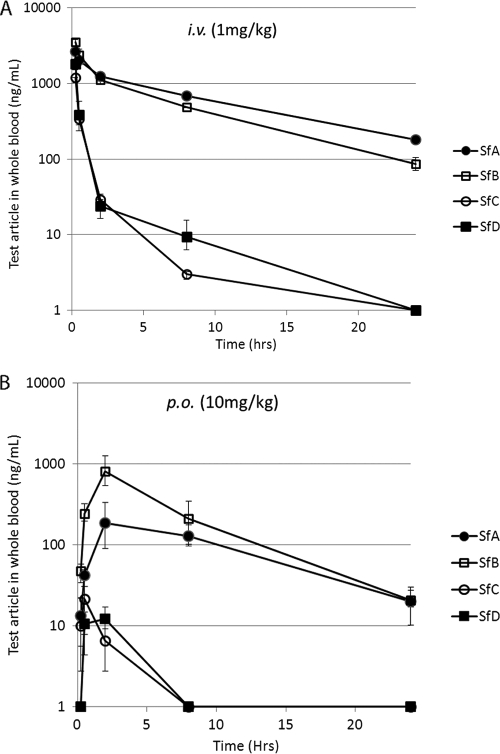
An initial examination of in vivo pharmacokinetics was performed, analyzing mouse whole blood after i.v. (1 mg/kg) and p.o. (10 mg/kg) dosing (Table 4 and Fig. 3). Although all sanglifehrins showed low oral bioavailability (F < 4%), both SfA and SfB had low clearances, at only 1 to 12% of hepatic blood flow for a mouse (4.5 liters/h/kg), with long half-lives (t1/2 > 4 h). The volume of distribution (Vss) was low (<0.8 liters/kg) for all sanglifehrins, suggesting that the long half-lives were indeed due to the low clearances and, if allometrically scaled up (41), would be expected to lead to human half-lives in excess of 24 h.
Table 4.
Pharmacokinetic parameters after i.v. and p.o. dosing of CD1 micea
| Cyclophilin inhibitor | Clearance (liter/h/kg) | t1/2 (h) | Vss (liter/kg) | AUClastb (h · ng/ml) |
p.o. Cmaxc (ng/ml) | F (%) | |
|---|---|---|---|---|---|---|---|
| I.v. | P.o. | ||||||
| SfA | 0.054 | 8.02 | 0.522 | 16,473 | 2,332 | 192 | 1.42 |
| SfB | 0.068 | 5.89 | 0.439 | 14,067 | 5,693 | 809 | 3.94 |
| SfC | 0.824 | 1.11 | 0.383 | 1,257 | 26 | 22 | 0.21 |
| SfD | 0.545 | 1.64 | 0.244 | 2,157 | 20 | 12 | 0.09 |
Data were generated after analysis of whole-blood samples, following doses of 1 mg/kg i.v. and 10 mg/kg p.o. to CD1 mice.
AUClast, area under the concentration-time curve.
Cmax, maximum concentration of drug in serum.
Fig. 3.
Pharmacokinetics after i.v. and p.o. dosing of CD1 mice. (A) Concentration in whole blood after i.v. dosing (1 mg/kg) of SfA, SfB, SfC, and SfD. (B) Concentration in whole blood after p.o. dosing (10 mg/kg) of SfA, SfB, SfC, and SfD. The error bars indicate standard deviations.
To confirm liver penetration, blood and liver samples were taken after a single i.v. dose of 1 mg/kg SfA to CD1 mice. The measured concentrations of SfA in the liver were 8 to 21 times greater than the corresponding concentrations in whole blood (under the assumption that the density of liver tissue is approximately equal to that of the whole-blood fraction), with calculated liver/blood ratios of 14:1 (see the supplemental material).
Immunosuppressive activity.
SfA has been shown to have immunosuppressive activity via a currently unknown mechanism (52). This is an undesirable property for an HCV therapy, and we tested all compounds for activity in an anti-CD3/anti-CD28-stimulated human PBMC assay. SfB and SfD were the more potent sanglifehrins at inhibition of stimulated cell proliferation, with IC50s of 480 and 740 nM, respectively, compared to SfA (1,340 nM) and SfC (1,780 nM); however, these compounds were still less potent than CsA in this assay (IC50, 140 nM). None of the sanglifehrins were seen to inhibit the growth of unstimulated PBMCs, whereas CsA caused some inhibition (cell growth, 77% of control at 5 μM).
DISCUSSION
While the current SOC for chronic HCV infection (pegylated interferon and ribavirin) has shown encouraging results for many patients, around 50% do not show an SVR, especially genotype 1 patients. In early clinical studies of the newer DAA drugs, while there have been improvements in response rates, a potential reason for a lack of SVR in some cases may be the selection of resistant viral populations (28). This has led to a search for targeted antiviral drugs with reduced potential for resistance. As CypA is a human target that seems essential for viral replication but nonessential for human cellular growth (1), it may represent an ideal target for therapy. A number of Cyp inhibitors have now been found to be associated with very low levels of resistance both in vitro and in vivo and showed additive or synergistic effect in combination. More importantly, they lack cross-resistance with SOC and the most advanced DAA drugs. This has led to the suggestion that they could form part of future combination therapies for chronic HCV (8).
Sanglifehrins were first described in 1999 (13, 43). Since then, the majority of studies have focused on their potential use as immunosuppressants, although there have been some suggestions of their potential utility in HCV treatment. Even so, this work is the first, and most detailed, analysis focused entirely on the potential of sanglifehrins for the treatment of HCV.
Characterization of naturally occurring sanglifehrins confirms that they have potential as agents for the treatment of HCV, making them a distinct opportunity compared to CsA and analogues. While there are a number of reports showing the potential relevance of multiple cyclophilins to the replication of HCV (and indeed a number of other viruses), this report shows that sanglifehrins are equally potent against all three cyclophilins tested, both in terms of inhibiting the formation of HCV NS5A-Cyp complexes and direct Cyp isomerase inhibition. All of the sanglifehrins tested disrupt all of these complexes more potently than CsA, and the most potent, SfB, is also significantly more potent against 1b HCV replicons. Not only are sanglifehrins inherently more potent cyclophilin inhibitors, they provide an additional opportunity to avoid class issues associated with cyclosporine analogues. Indeed, this work suggests that SfB may be a more promising natural candidate than the typically analyzed compound from the class, SfA.
Data are also presented suggesting that natural sanglifehrins are active against CsA-resistant replicons, in contrast to the limited data previously presented (18). It is interesting that the mutations found in a replicon selected for resistance to the sanglifehrin analogue SFA-1 (40) did not include the D320E mutation, the most commonly generated mutation to CsA and analogues (6). Taken together, this suggests that sanglifehrins, while potent cyclophilin inhibitors, disrupt NS5A-Cyp complexes in a subtly different way than the cyclosporines.
However, while natural sanglifehrins have long in vivo mouse blood half-lives, they have a number of poor physicochemical and pharmacokinetic properties for clinical HCV treatment (low solubility, low oral bioavailability, and some immunosuppressive activity). Initial characterization of novel sanglifehrin analogues, developed as part of a lead optimization program, using semisynthetic and biosynthetic engineering methods, has shown that each of these properties can be improved (M. Gregory, unpublished data). Sanglifehrin analogues with improved potency against all HCV genotypes tested (1a, 1b, and 2a), reduced cytotoxicity, improved physicochemical properties for oral administration, and substantially reduced immunosuppression have now been generated. These compounds are being actively advanced toward clinical assessment for the treatment of HCV infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bruce Campbell for helpful discussions regarding physicochemical and pharmacokinetic parameters; Chen Weiqing, David Xie, Yan Liping, Chen Ping, Qian Mingxin, and Shi Junwei for in vitro ADME, physicochemical, and mouse PK analysis; Chen Yongsheng, Liu Jinlun, Chen Yufeng, Li Xin, and Xie Deijan for chemistry support at Shanghai Chempartner; Zhuhui Huang at Southern Research Institute for cytotoxicity analysis; and Eric Bensen at MDBiosciences for T-cell proliferation studies.
Work in the laboratory of Johan Neyts is supported by grant G.0728.09N from the FWO.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 7 March 2011.
REFERENCES
- 1. Braaten D., Luban J. 2001. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J. 20:1300–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chatterji U., et al. 2009. The isomerase active site of cyclophilin A is critical for hepatitis C virus replication. J. Biol. Chem. 284:16998–17005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chatterji U., et al. 2005. Naturally occurring capsid substitutions render HIV-1 cyclophilin A independent in human cells and TRIM-cyclophilin-resistant in owl monkey cells. J. Biol. Chem. 280:40293–40300 [DOI] [PubMed] [Google Scholar]
- 4. Chatterji U., et al. 2010. HCV resistance to cyclosporin A does not correlate with a resistance of the NS5A-cyclophilin A interaction to cyclophilin inhibitors. J. Hepatol. 53:50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ciesek S., et al. 2009. Cyclosporine A inhibits hepatitis C virus nonstructural protein 2 through cyclophilin A. Hepatology 50:1638–1645 [DOI] [PubMed] [Google Scholar]
- 6. Coelmont L., et al. 2009. Debio 025, a cyclophilin binding molecule, is highly efficient in clearing hepatitis C virus (HCV) replicon-containing cells when used alone or in combination with specifically targeted antiviral therapy for HCV (STAT-C) inhibitors. Antimicrob. Agents Chemother. 53:967–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Colgan J., Asmal M., Luban J. 2000. Isolation, characterization and targeted disruption of mouse Ppia: cyclophilin A is not essential for mammalian cell viability. Genomics 68:167–178 [DOI] [PubMed] [Google Scholar]
- 8. Crabbé R., et al. 2009. An evaluation of the cyclophilin inhibitor Debio 025 and its potential as a treatment for chronic hepatitis C. Expert Opin. Investig. Drugs 18:211–220 [DOI] [PubMed] [Google Scholar]
- 9. Delang L., Coelmont L., Neyts J. 2010. Antiviral therapy for hepatitis C virus: beyond the standard of care. Viruses (Basel) 2:826–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reference deleted.
- 11. Reference deleted.
- 12. Dolinski K., Muir S., Cardenas M., Heitman J. 1997. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 94:13093–13098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fehr T., Kallen J., Oberer L., Sanglier J. J., Schilling W. 1999. Sanglifehrins A, B, C and D, novel cyclophilin-binding compounds isolated from Streptomyces sp. A92-308110. II. Structure elucidation, stereochemistry and physico-chemical properties. J. Antibiot. (Tokyo) 52:474–479 [DOI] [PubMed] [Google Scholar]
- 14. Fernandes F., Ansari I. U., Striker R. 2010. Cyclosporine inhibits a direct interaction between cyclophilins and hepatitis C NS5A. PLoS One 5:e9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Flisiak R., et al. 2009. The cyclophilin inhibitor Debio 025 combined with PEG IFNalpha2a significantly reduces viral load in treatment-naive hepatitis C patients. Hepatology 49:1460–1468 [DOI] [PubMed] [Google Scholar]
- 16. Flisiak R., et al. 2008. The cyclophilin inhibitor Debio-025 shows potent anti-hepatitis C effect in patients coinfected with hepatitis C and human immunodeficiency virus. Hepatology 47:817–826 [DOI] [PubMed] [Google Scholar]
- 17. Gaither L. A., et al. 2010. Multiple cyclophilins involved in different cellular pathways mediate HCV replication. Virology 397:43–55 [DOI] [PubMed] [Google Scholar]
- 18. Goto K., Watashi K., Inoue D., Hijikata M., Shimotohno K. 2009. Identification of cellular and viral factors related to anti-hepatitis C virus activity of cyclophilin inhibitor. Cancer Sci. 100:1943–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hanoulle X., et al. 2009. Hepatitis C virus NS5A protein is a substrate for the peptidyl-prolyl cis/trans isomerase activity of cyclophilins A and B. J. Biol. Chem. 284:13589–13601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Herrler M., Bang H., Marahiel M. A. 1994. Cloning and characterization of ppiB, a Bacillus subtilis gene which encodes a cyclosporin A-sensitive peptidyl-prolyl cis-trans isomerase. Mol. Microbiol. 11:1073–1083 [DOI] [PubMed] [Google Scholar]
- 21. Hopkins S., et al. 2010. SCY-635, a novel nonimmunosuppressive analog of cyclosporine that exhibits potent inhibition of hepatitis C virus RNA replication in vitro. Antimicrob. Agents Chemother. 54:660–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang L., et al. 2004. Purification and characterization of hepatitis C virus non-structural protein 5A expressed in Escherichia coli. Protein Expr. Purif. 37:144–153 [DOI] [PubMed] [Google Scholar]
- 23. Inoue K., et al. 2007. Evaluation of a cyclophilin inhibitor in hepatitis C virus-infected chimeric mice in vivo. Hepatology 45:921–928 [DOI] [PubMed] [Google Scholar]
- 24. Ishii N., et al. 2006. Diverse effects of cyclosporine on hepatitis C virus strain replication. J. Virol. 80:4510–4520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jacobson I. M., et al. 2007. Impact of weight-based ribavirin with peginterferon alfa-2b in African Americans with hepatitis C virus genotype 1. Hepatology 46:982–990 [DOI] [PubMed] [Google Scholar]
- 26. Kallen J., Sedrani R., Zenke G., Wagner J. 2005. Structure of human cyclophilin A in complex with the novel immunosuppressant sanglifehrin A at 1.6 A resolution. J. Biol. Chem. 280:21965–21971 [DOI] [PubMed] [Google Scholar]
- 27. Kapturczak M. H., Meier-Kriesche H. U., Kaplan B. 2004. Pharmacology of calcineurin antagonists. Transplant Proc. 36:25S–32S [DOI] [PubMed] [Google Scholar]
- 28. Kronenberger B., Zeuzem S. 2009. Current and future treatment options for HCV. Ann. Hepatol. 8:103–112 [PubMed] [Google Scholar]
- 29. Manns M. P., et al. 2007. The way forward in HCV treatment—finding the right path. Nat. Rev. Drug Discov. 6:991–1000 [DOI] [PubMed] [Google Scholar]
- 30. Mathy J. E., Ma S., Compton T., Lin K. 2008. Combinations of cyclophilin inhibitor NIM811 with hepatitis C virus NS3-4A protease or NS5B polymerase inhibitors enhance antiviral activity and suppress the emergence of resistance. Antimicrob. Agents Chemother. 52:3267–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Melnikova I. 2008. Hepatitis C therapies. Nat. Rev. Drug Discov. 7:799–800 [Google Scholar]
- 32. Nelson D. R., et al. 2009. Efficacy and safety of the cyclophilin inhibitor DEBIO-025 in combination with pegylated interferon alpha-2A and ribavirin in previously null-responder genotype 1 HCV patients. J. Hepatol. 50:S40 [Google Scholar]
- 33. Paeshuyse J., et al. 2006. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology 43:761–770 [DOI] [PubMed] [Google Scholar]
- 34. Paeshuyse J., et al. 2008. Comparative in vitro anti-hepatitis C virus activities of a selected series of polymerase, protease, and helicase inhibitors. Antimicrob. Agents Chemother. 52:3433–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Parfieniuk A., Jaroszewicz J., Flisiak R. 2007. Specifically targeted antiviral therapy for hepatitis C virus. World J. Gastroenterol. 13:5673–5681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pawlotsky J. M. 2005. Current and future concepts in hepatitis C therapy. Semin. Liver Dis. 25:72–83 [DOI] [PubMed] [Google Scholar]
- 37. Pawlotsky J. M., Chevaliez S., McHutchison J. G. 2007. The hepatitis C virus life cycle as a target for new antiviral therapies. Gastroenterology 132:1979–1998 [DOI] [PubMed] [Google Scholar]
- 38. Pockros P. 2008. Emerging therapies for chronic hepatitis C virus. Gastroenterol. Hepatol. 4:729–734 [PMC free article] [PubMed] [Google Scholar]
- 39. Poynard T., Yuen M. F., Ratziu V., Lai C. L. 2003. Viral hepatitis C. Lancet 362:2095–2100 [DOI] [PubMed] [Google Scholar]
- 40. Puyang X., et al. 2010. Mechanism of resistance of hepatitis C virus replicons to structurally distinct cyclophilin inhibitors. Antimicrob. Agents Chemother. 54:1981–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ritschel W. A., Vachharajani N. N., Johnson R. D., Hussain A. S. 1992. The allometric approach for interspecies scaling of pharmacokinetic parameters. Comp. Biochem. Physiol. C 103:249–253 [DOI] [PubMed] [Google Scholar]
- 42. Robida J. M., Nelson H. B., Liu Z., Tang H. 2007. Characterization of hepatitis C virus subgenomic replicon resistance to cyclosporine in vitro. J. Virol. 81:5829–5840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sanglier J. J., et al. 1999. Sanglifehrins A, B, C and D, novel cyclophilin-binding compounds isolated from Streptomyces sp. A92-308110. I. Taxonomy, fermentation, isolation and biological activity. J. Antibiot. (Tokyo) 52:466–473 [DOI] [PubMed] [Google Scholar]
- 44. Sedrani R., et al. 2003. Sanglifehrin-cyclophilin interaction: degradation work, synthetic macrocyclic analogues, X-ray crystal structure, and binding data. J. Am. Chem. Soc. 125:3849–3859 [DOI] [PubMed] [Google Scholar]
- 45. Tropschug M., Barthelmess I. B., Neupert W. 1989. Sensitivity to cyclosporin A is mediated by cyclophilin in Neurospora crassa and Saccharomyces cerevisiae. Nature 342:953–955 [DOI] [PubMed] [Google Scholar]
- 46. Vrolijk J. M., et al. 2003. A replicon-based bioassay for the measurement of interferons in patients with chronic hepatitis C. J. Virol. Methods 110:201–209 [DOI] [PubMed] [Google Scholar]
- 47. Wainberg M. A., Martinez-Cajas J. L. 2009. Sequencing of therapy to avoid resistance and the need for new antiretroviral drugs in the treatment of HIV disease. Infect. Disord. Drug Targets 9:172–190 [DOI] [PubMed] [Google Scholar]
- 48. Wang P., Heitman J. 2005. The cyclophilins. Genome Biol. 6:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Watashi K. 2010. Alisporivir, a cyclosporin derivative that selectively inhibits cyclophilin, for the treatment of HCV infection. Curr. Opin. Investig. Drugs 11:213–224 [PubMed] [Google Scholar]
- 50. Watashi K., et al. 2005. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell 19:111–122 [DOI] [PubMed] [Google Scholar]
- 51. Yang F., et al. 2008. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J. Virol. 82:5269–5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zenke G., et al. 2001. Sanglifehrin A, a novel cyclophilin-binding compound showing immunosuppressive activity with a new mechanism of action. J. Immunol. 166:7165–7171 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.