Abstract
Lacticin Q, a lactococcal pore-forming bacteriocin, shows activity toward Gram-positive bacteria but not Gram-negative bacteria. Lacticin Q did not induce permeability of the outer membrane of Gram-negative bacteria. Experiments using model membranes containing outer membrane components suggested that lacticin Q binds to the outer membrane of Gram-negative bacteria but is unable to penetrate it. The lack of activity of lacticin Q was attributed to physicochemical features of the outer membrane components.
INTRODUCTION
Cationic and membrane-permeabilizing antimicrobial peptides have been well characterized (30). While the antimicrobial spectra of individual peptides are varied, many peptides from multicellular eukaryotes uniformly kill Gram-positive and -negative bacteria and fungi in the micromolar concentration range (6). Cationic antimicrobial peptides recognize the negatively charged membrane of the target cells. Conversely, a higher peptide concentration is required for the inhibition of nontarget cells with electroneutral membranes. The selectivity of the killing activity is determined by the physicochemical features of the peptides and cell membranes (30).
Bacteria produce ribosomally synthesized antimicrobial peptides or proteins called bacteriocins (9), and the mode of action of small-peptide bacteriocins produced by lactic acid bacteria (LAB) has been studied (15). Nisin and pediocin PA-1 are the best characterized cationic and membrane-permeabilizing peptides (8, 11). Many LAB bacteriocins, including nisin and pediocin PA-1, exert their activity against Gram-positive bacteria but not against Gram-negative bacteria. Nisin and some bacteriocins require a bacterial peptidoglycan precursor, lipid II, for their pore-forming activity (2–5, 18, 25, 26); however, only Gram-positive bacteria display lipid II on the cell surface (24). In the case of Gram-negative bacteria, the lipid II-containing cytoplasmic membrane is covered by the outer membrane. Since increasing the outer membrane permeability leads the antimicrobial activity of nisin against Gram-negative bacteria (7), the selective toxicity of nisin is easily explained by the presence of receptor lipid II. The selective toxicity of nisin and other lipid II-targeting bacteriocins is probably determined by biochemical interactions between lipid II and the peptides (2). The selective toxicity of pediocin PA-1 and its homologs (pediocin-like bacteriocins) is also considered to occur through a similar mechanism. Some pediocin-like bacteriocins utilize a bacterial cytoplasmic membrane protein as a receptor (10, 13–15).
Recently, we discovered a new LAB bacteriocin, lacticin Q, produced by Lactococcus lactis QU 5 (12). Lacticin Q, a 53-amino-acid peptide containing abundant cationic residues (Fig. 1A), has strong antimicrobial activity in the nanomolar concentration range and high stability in various environments. We previously proposed a new model, named the huge toroidal pore (HTP), to account for the antimicrobial action of lacticin Q (Fig. 1B) (29). Lacticin Q-mediated HTP occurs in the absence of a specific receptor (28); meanwhile, lacticin Q does not show activity against Gram-negative bacteria (12). This study was designed to identify the factors necessary for the selective bactericidal activity of lacticin Q. Previous studies indicated that toroidal pore formation by some antimicrobial peptides, such as magainin 2, was inhibited by phosphatidylethanolamine (PE), a major component of the outer membrane, because the small, hydrophilic head of PE was not adaptive to form the positive curvature (19). We also focused on the outer membrane components of Gram-negative bacteria that affect the pore-forming activity of lacticin Q.
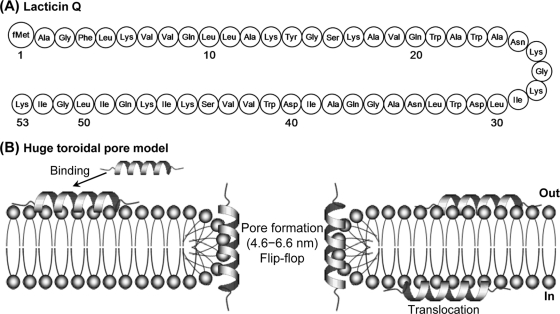
Fig. 1.
(A) Structure of lacticin Q. fMet, formylmethionine. (B) The action mechanism of lacticin Q was determined previously and termed the huge toroidal pore model. Lacticin Q rapidly binds to the outer leaflet of the cell membrane and forms huge toroidal pores (pore diameter, 4.6 to 6.6 nm) accompanied by lipid flip-flop. Some lacticin Q molecules migrate from the outer to the inner leaflet of the membrane.
Using a turbidimetric assay as previously described (27), purified lacticin Q showed antimicrobial activities in the range of 75 to 1,000 nM against Gram-positive bacteria (Table 1). Conversely, we did not identify any inhibitory activity of lacticin Q against Gram-negative bacteria under this experimental condition, as observed for many LAB bacteriocins (9).
Table 1.
MICs of lacticin Q against Gram-positive and -negative bacteria
| Indicator straina | MIC (nM) |
|---|---|
| Bacillus coagulans JCM 2257T | 75 |
| Lactococcus lactis IL1403 | 100 |
| Pediococcus pentosaceus JCM 5890T | 1,000 |
| Escherichia coli JM109 | >10,000 |
| Pseudomonas putida ATCC 12633 | >10,000 |
| Pseudomonas oleovorans ATCC 29347 | >10,000 |
| Ralstonia eutropha ATCC 17687T | >10,000 |
Abbreviations: JCM, Japan Collection of Microorganisms, Wako, Japan; ATCC, American Type Culture Collection, Rockville, MD. Bacillus coagulans JCM 2257T and all the Gram-negative indicator strains were grown in tryptic soy broth (Difco Laboratories, Detroit, MI) supplemented with 0.6% yeast extract (Difco Laboratories). Lactococcus lactis IL1403 and Pediococcus pentosaceus JCM 5890T were grown in MRS broth (Oxoid, Basingstoke, United Kingdom). The indicator strains were grown under the recommended conditions.
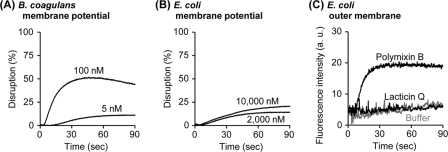
Peptide-inducing disruption of the membrane potential was measured using previously reported methods (5, 28), and a fluorescent probe, DiSC3(5) (Invitrogen, Carlsbad, CA), and an F-7000 spectrofluorometer (Hitachi High-Technologies, Tokyo, Japan) were used. Against Gram-positive Bacillus coagulans cells, 100 nM lacticin Q disrupted the membrane potential (Fig. 2A). A lower concentration of lacticin Q (5 nM) slightly disrupted the membrane potential. Conversely, 2,000 nM lacticin Q disrupted the membrane potential of Gram-negative Escherichia coli cells (Fig. 2B), but the disruption level was similar to that observed for 5 nM lacticin Q against B. coagulans. In addition, there was no significant difference in the membrane disruption of E. coli cells by treatment with 2,000 or 10,000 nM lacticin Q. We hypothesized that the outer membrane of Gram-negative bacteria prevented the membrane-permeabilizing activity of lacticin Q. To confirm this, the N-phenyl-1-naphthylamine (NPN; Sigma, St. Louis, MO) uptake assay was performed as previously described (17, 31). As shown in Fig. 2C, lacticin Q did not induce fluorescence in the indicator E. coli cells that would indicate outer membrane permeabilization. Conversely, polymyxin B, a peptide antibiotic used as a positive control, induced fluorescence. Although we previously reported that lacticin Q-mediated huge pores caused the leakage of macromolecules from the membrane of Gram-positive cells (29), even small molecules, such as NPN, could not pass through the outer membrane of Gram-negative bacteria.
Fig. 2.
Disruption of membrane potential and outer membrane permeabilization by lacticin Q. (A and B) Washed cells of B. coagulans (A) or E. coli (B) and DiSC3(5) were diluted using buffer M (250 mM sucrose, 5 mM MgSO4, 10 mM potassium phosphate, pH 7.0) in a 2-ml quartz cuvette incubated at 30°C [optical density of cells at 600 nm (OD600) = 0.05, 1 μM DiSC3(5)]. The excitation (EX) and emission (EM) wavelengths were set at 622 and 675 nm, respectively. At the beginning of the measurement, lacticin Q was added at the indicated concentrations. As controls, 0% leakage and 100% leakage were obtained by the addition of buffer and 0.1% Triton X-100, respectively. (C) The peptide-induced E. coli outer membrane permeabilization was determined with the NPN uptake assay. Washed cells of E. coli and NPN were diluted using buffer N (5 mM HEPES-NaOH, 5 mM glucose, 100 mM NaCl, 5 μM carbonyl cyanide m-chlorophenylhydrazone, pH 7.4) in a 2-ml quartz cuvette (OD600 = 0.5, 10 μM NPN). The EX and EM wavelengths were set at 350 and 420 nm, respectively. At the beginning of the measurement, 10 μM lacticin Q was added. The outer membrane-permeabilizing antibiotic polymyxin B (1 mg/ml) and the buffer N were applied for the positive and negative controls, respectively. a.u., arbitrary units.
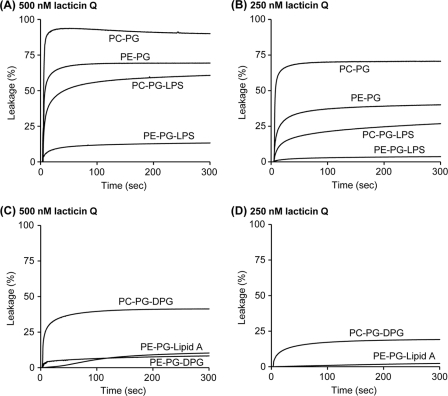
Next, we examined the outer membrane components responsible for calcein leakage induced by lacticin Q. Liposomes, large unilamellar vesicles (LUVs), entrapping calcein (Dojindo, Kumamoto, Japan) were prepared as previously described (28, 29). The lipids (Sigma) and the composition of liposomes used in this study are listed in Table 2. The lipid concentration of LUVs was determined using a phosphorous assay (1). The main components of the outer membrane, PE and lipopolysaccharide (LPS), were added to the basal l-α-phosphatidylcholine–l-α-phosphatidyl-dl-glycerol (PC-PG) LUVs, and lacticin Q-mediated calcein leakage was monitored (Fig. 3A and B). Following treatment with 500 nM peptide for 5 min (Fig. 3A), lacticin Q induced high (90.0%) and rapid calcein leakage from PC-PG LUVs, as we previously observed (28, 29). Against PE-PG and PC-PG-LPS LUVs, lacticin Q induced 69.4% and 60.8% leakage, respectively. PE-PG-LPS LUVs, containing PE and LPS, clearly inhibited the lacticin Q-mediated calcein leakage, and the leakage reached only 13.3%. Following treatment with 250 nM peptide for 5 min (Fig. 3B), the leakage from LUVs was at a level similar to that observed for 500 nM peptide (Fig. 3A). Although 250 nM lacticin Q induced 70.6% calcein leakage from PC-PG LUVs, only 3.6% leakage was observed from PE-PG-LPS LUVs. The effect of PE on toroidal pore formation has been previously investigated (19). We examined the inhibitory effect of lipid A, LPS without the hydrophilic sugar composition and the O-antigen (Fig. 3C and D). Lacticin Q (500 nM) induced 10.3% leakage from PE-PG-lipid A LUVs (Fig. 3C), and this was very similar to the levels from PE-PG-LPS LUVs (Fig. 3A). This result indicated that the hydrophilic sugar composition and O-antigen of LPS were not essential for the inhibition. We then assessed LUVs containing diphosphatidylglycerol (DPG), consisting of two PG molecules connected by a condensation reaction, which contains a high density of acyl chains, such as LPS and lipid A. Lacticin Q (500 nM) induced 8.3% leakage from PE-PG-DPG LUVs, similar to the levels observed from PE-PG-LPS LUVs; furthermore, lacticin Q (500 nM) induced a higher rate of leakage (41.3%) from PC-PG-DPG LUVs. These results indicated that a combination including PE was necessary for the DPG-mediated strong inhibition, as was observed for LPS (Fig. 3A and B). Although lacticin Q (250 nM) induced 19.1% calcein leakage from PC-PG-DPG LUVs, no leakage was observed from PE-PG-DPG LUVs (Fig. 3D). Conversely, it was reported that LPS enhanced the membrane-binding and pore formation of several toroidal pore-type antimicrobial peptides from multicellular eukaryotes, such as tachyplesin I (16, 30). Unlike lacticin Q, the strong interaction between peptides and LPS is considered to be important for animals to recognize bacterial invasion because LPS is a well-known endotoxin (23, 30).
Table 2.
Lipids and LUVs used in this study
| Membrane and LUV component(s)a | Charge | Remarks |
|---|---|---|
| Lipids (abbreviation/source) | ||
| Egg yolk l-α-phosphatidylcholine (PC) | ± | A basal lipid |
| Egg yolk l-α-phosphatidyl-dl-glycerol (PG) | − | A main component of the bacterial membrane |
| Egg yolk l-α-phosphatidylethanolamine (PE) | ± | A main component of the inner leaflet of the outer membrane |
| Lipopolysaccharide (LPS; from Salmonella enterica serotype Minnesota Re 595) | − | A main component of the outer leaflet of the outer membrane |
| Lipid A (from Salmonella enterica serotype Minnesota Re 595) | − | A hydrophobic part of LPS |
| Diphosphatidylglycerol (DPG; from bovine heart) | − | A component of the bacterial membrane |
| LUVs (molar ratio) | ||
| PC-PG (5:5) | − | Basal LUVs |
| PE-PG (5:5) | − | PE-containing LUVs |
| PC-PG-LPS (5:3:2) | − | LPS-containing LUVs |
| PE-PG-LPS (5:3:2) | − | PE- and LPS-containing LUVs, similar to the outer membrane |
| PE-PG-lipid A (5:3:1) | − | PE- and lipid A-containing LUVs |
| PC-PG-DPG (5:3:2) | − | DPG-containing LUVs |
| PE-PG-DPG (5:3:2) | − | PE- and DPG-containing LUVs |
LUVs, large unilamellar vesicles.
Fig. 3.
Calcein leakage from LUVs containing outer membrane components induced by 500 nM (A) and 250 nM (B) lacticin Q and containing lipid A or DPG induced by 500 nM (C) and 250 nM (D). Lacticin Q in buffer A (10 mM Tris, 75 mM NaCl, 1 mM EDTA, pH 7.0) was incubated at 30°C. EX and EM wavelengths were set at 490 and 520 nm, respectively. As controls, 0% leakage and 100% leakage were obtained by the addition of buffer and 0.1% Triton X-100, respectively. The final lipid concentration was 50 μM. The data are the average results from at least two experiments. It was possible to add 20% LPS or 10% lipid A to LUVs as a maximum ratio.
We analyzed binding between lacticin Q and LUVs by changes in the fluorescence peak of tryptophan residues. Tryptophan residues in hydrophobic environments shift their maximum fluorescent wavelength to a lower level than observed under hydrophilic conditions (blue shift). As shown in Table 3, lacticin Q did not demonstrate any time-dependent changes in the fluorescence peaks of tryptophan in the absence of LUVs (342.3 to 342.9 nm). PC-PG LUVs resulted in the blue shift of the fluorescence peak of lacticin Q, and the fluorescence peaks in the presence of PC-PG LUVs were 6 to 7 nm lower than in the absence of LUVs. Furthermore, PE-PG-LPS LUVs caused a blue shift similar to that observed for PC-PG LUVs. The blue shifts mediated by both LUVs were observed in the first minute of the reaction. These results indicated that the interaction of lacticin Q with PC-PG and PE-PG-LPS LUVs occurred similarly.
Table 3.
LUV-lacticin Q interaction evaluated by blue shift of tryptophan fluorescencea
| Condition | Mean ± SD of fluorescence peak of lacticin Q (nm) at indicated time point (min) |
||
|---|---|---|---|
| 1 | 3 | 10 | |
| Buffer | 342.3 ± 0.3 | 342.9 ± 0.3 | 342.9 ± 0.5 |
| PC-PG | 335.7 ± 0.3 | 336.1 ± 0.3 | 336.0 ± 0.2 |
| PE-PG-LPS | 337.4 ± 1.2 | 336.3 ± 0.7 | 336.2 ± 0.4 |
Lacticin Q and LUVs were mixed in buffer A (as described in the legend to Fig. 3) at 30°C. Excitation and emission wavelengths were set at 290 and 300 to 400 nm, respectively. The fluorescence spectrum of the LUVs was subtracted from that of the LUV-lacticin Q mixture.
Large numbers of peptide bacteriocins from Gram-positive bacteria have been discovered, and many of them are only active against Gram-positive bacteria (9, 22). To the best of our knowledge, this is the first report of a mechanism underlying the selective toxicity of LAB bacteriocins. We demonstrated here that lacticin Q could bind to PE- and LPS-containing membranes (Table 3) but not penetrate them (Fig. 2 and 3). It was previously proposed that toroidal pore formation depended on the physicochemical features of membrane lipids (20, 21). The most important characteristic of the toroidal pore is the rapid transbilayer movement of lipids, the so-called lipid flip-flop (19, 30). To induce lipid flip-flop, it was considered that the inner and outer leaflets of the outer membrane were temporarily connected through the peptide molecules (Fig. 1B). Since lipid flip-flop resulting from pore formation requires the extremely positive curvature of the membrane, the lipid PE, which has a small, hydrophilic head group and, thus, a tendency for negative curvature formation, inhibits the formation of the toroidal pore (19, 21). The previous findings could explain the results presented in Fig. 3A and B for the inhibition of the pore-forming activity of lacticin Q in PE-containing LUVs. In addition, other lipids (lipid A and DPG) that are physicochemically similar to LPS also prevented lacticin Q activity (Fig. 3C and D). A high number of acyl chains may be one of the factors that inhibits the formation of the positive curvature of the toroidal pore. Our findings in this study indicate that the physicochemical features of the outer membrane components are an important factor in the selective toxicity of lacticin Q in eubacteria.
Acknowledgments
This work was partially supported by the Research Project for Utilizing Advanced Technologies in Agriculture, Forestry and Fisheries of the Ministry of Agriculture, Forestry and Fisheries of Japan and by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS).
Footnotes
Published ahead of print on 31 January 2011.
REFERENCES
- 1. Bartlett G. R. 1959. Phosphorus assay in column chromatography. J. Biol. Chem. 234:466–468 [PubMed] [Google Scholar]
- 2. Bauer R., Dicks L. M. 2005. Mode of action of lipid II-targeting lantibiotics. Int. J. Food Microbiol. 101:201–216 [DOI] [PubMed] [Google Scholar]
- 3. Bonelli R. R., Schneider T., Sahl H. G., Wiedemann I. 2006. Insights into in vivo activities of lantibiotics from gallidermin and epidermin mode-of-action studies. Antimicrob. Agents Chemother. 50:1449–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Breukink E., de Kruijff B. 2006. Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 5:321–332 [DOI] [PubMed] [Google Scholar]
- 5. Breukink E., et al. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361–2364 [DOI] [PubMed] [Google Scholar]
- 6. Brogden K. A. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238–250 [DOI] [PubMed] [Google Scholar]
- 7. Cao-Hoang L., Marechal P. A., Le-Thanh M., Gervais P. 2008. Synergistic action of rapid chilling and nisin on the inactivation of Escherichia coli. Appl. Microbiol. Biotechnol. 79:105–109 [DOI] [PubMed] [Google Scholar]
- 8. Chatterjee C., Paul M., Xie L., van der Donk W. A. 2005. Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105:633–684 [DOI] [PubMed] [Google Scholar]
- 9. Cotter P. D., Hill C., Ross R. P. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777–788 [DOI] [PubMed] [Google Scholar]
- 10. Dalet K., Cenatiempo Y., Cossart P., Hechard Y. 2001. A σ(54)-dependent PTS permease of the mannose family is responsible for sensitivity of Listeria monocytogenes to mesentericin Y105. Microbiology 147:3263–3269 [DOI] [PubMed] [Google Scholar]
- 11. Fimland G., Johnsen L., Dalhus B., Nissen-Meyer J. 2005. Pediocin-like antimicrobial peptides (class IIa bacteriocins) and their immunity proteins: biosynthesis, structure, and mode of action. J. Pept. Sci. 11:688–696 [DOI] [PubMed] [Google Scholar]
- 12. Fujita K., et al. 2007. Structural analysis and characterization of lacticin Q, a novel bacteriocin belonging to a new family of unmodified bacteriocins of gram-positive bacteria. Appl. Environ. Microbiol. 73:2871–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gravesen A., Warthoe P., Knochel S., Thirstrup K. 2000. Restriction fragment differential display of pediocin-resistant Listeria monocytogenes 412 mutants shows consistent overexpression of a putative β-glucoside-specific PTS system. Microbiology 146(Pt. 6):1381–1389 [DOI] [PubMed] [Google Scholar]
- 14. Hechard Y., Pelletier C., Cenatiempo Y., Frere J. 2001. Analysis of σ(54)-dependent genes in Enterococcus faecalis: a mannose PTS permease (EII(Man)) is involved in sensitivity to a bacteriocin, mesentericin Y105. Microbiology 147:1575–1580 [DOI] [PubMed] [Google Scholar]
- 15. Hechard Y., Sahl H.-G. 2002. Mode of action of modified and unmodified bacteriocins from Gram-positive bacteria. Biochimie 84:545–557 [DOI] [PubMed] [Google Scholar]
- 16. Hirakura Y., Kobayashi S., Matsuzaki K. 2002. Specific interactions of the antimicrobial peptide cyclic β-sheet tachyplesin I with lipopolysaccharides. Biochim. Biophys. Acta 1562:32–36 [DOI] [PubMed] [Google Scholar]
- 17. Loh B., Grant C., Hancock R. E. 1984. Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 26:546–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Martinez B., et al. 2008. Specific interaction of the unmodified bacteriocin lactococcin 972 with the cell wall precursor lipid II. Appl. Environ. Microbiol. 74:4666–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matsuzaki K. 1999. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim. Biophys. Acta 1462:1–10 [DOI] [PubMed] [Google Scholar]
- 20. Matsuzaki K., Sugishita K., Fujii N., Miyajima K. 1995. Molecular basis for membrane selectivity of an antimicrobial peptide, magainin 2. Biochemistry 34:3423–3429 [DOI] [PubMed] [Google Scholar]
- 21. Matsuzaki K., et al. 1998. Relationship of membrane curvature to the formation of pores by magainin 2. Biochemistry 37:11856–11863 [DOI] [PubMed] [Google Scholar]
- 22. O'Sullivan L., Ross R. P., Hill C. 2002. Potential of bacteriocin-producing lactic acid bacteria for improvements in food safety and quality. Biochimie 84:593–604 [DOI] [PubMed] [Google Scholar]
- 23. Raetz C. R., Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Twomey D., Ross R. P., Ryan M., Meaney B., Hill C. 2002. Lantibiotics produced by lactic acid bacteria: structure, function and applications. Antonie Van Leeuwenhoek 82:165–185 [PubMed] [Google Scholar]
- 25. Wiedemann I., et al. 2006. Lipid II-based antimicrobial activity of the lantibiotic plantaricin C. Appl. Environ. Microbiol. 72:2809–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wiedemann I., et al. 2006. The mode of action of the lantibiotic lacticin 3147—a complex mechanism involving specific interaction of two peptides and the cell wall precursor lipid II. Mol. Microbiol. 61:285–296 [DOI] [PubMed] [Google Scholar]
- 27. Yoneyama F., Fukao M., Zendo T., Nakayama J., Sonomoto K. 2008. Biosynthetic characterization and biochemical features of the third natural nisin variant, nisin Q, produced by Lactococcus lactis 61-14. J. Appl. Microbiol. 105:1982–1990 [DOI] [PubMed] [Google Scholar]
- 28. Yoneyama F., et al. 2009. Lacticin Q, a lactococcal bacteriocin, causes high-level membrane permeability in the absence of specific receptors. Appl. Environ. Microbiol. 75:538–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yoneyama F., et al. 2009. Peptide-lipid huge toroidal pore, a new antimicrobial mechanism mediated by a lactococcal bacteriocin, lacticin Q. Antimicrob. Agents Chemother. 53:3211–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389–395 [DOI] [PubMed] [Google Scholar]
- 31. Zhang L., Benz R., Hancock R. E. 1999. Influence of proline residues on the antibacterial and synergistic activities of α-helical peptides. Biochemistry 38:8102–8111 [DOI] [PubMed] [Google Scholar]