Abstract
The platelet chemokines neutrophil-activating peptide-2 (NAP-2) and thrombocidin-1 (TC-1) differ by only two amino acids at their carboxy-terminal ends. Nevertheless, they display a significant difference in their direct antimicrobial activities, with the longer NAP-2 being inactive and TC-1 being active. In an attempt to rationalize this difference in activity, we studied the structure and the dynamics of both proteins by nuclear magnetic resonance (NMR) spectroscopy. Using 15N isotope-labeled protein, we confirmed that the two monomeric proteins essentially have the same overall structure in aqueous solution. However, NMR relaxation measurements provided evidence that the negatively charged carboxy-terminal residues of NAP-2 experience a restricted motion, whereas the carboxy-terminal end of TC-1 moves in an unrestricted manner. The same behavior was also seen in molecular dynamic simulations of both proteins. Detailed analysis of the protein motions through model-free analysis, as well as a determination of their overall correlation times, provided evidence for the existence of a monomer-dimer equilibrium in solution, which seemed to be more prevalent for TC-1. This finding was supported by diffusion NMR experiments. Dimerization generates a larger cationic surface area that would increase the antimicrobial activities of these chemokines. Moreover, these data also show that the negatively charged carboxy-terminal end of NAP-2 (which is absent in TC-1) folds back over part of the positively charged helical region of the protein and, in doing so, interferes with the direct antimicrobial activity.
INTRODUCTION
The 70-amino-acid residue neutrophil-activating peptide-2 (NAP-2), also known as CXCL-7, is generated through N-terminal proteolytic processing from platelet basic protein (PBP) (52). NAP-2 and its related proteins are released in large amounts from platelets (7, 29) and can be induced in monocytes (44). As a member of the CXC chemokine family which includes interleukin-8 (IL-8) (45) and stromal cell-derived factor 1 (SDF-1) (48), NAP-2 plays an important role in inflammation, blood clotting, and wound healing. It is a potent mediator of neutrophil activation through interactions with the cell surface receptors CXCR-1 and CXCR-2 (38), an activity shared with the homologous chemokines IL-8 and growth-related oncogene-α (GRO-α) (23). PBP-derived products with longer N-terminal regions than NAP-2, namely, connective tissue activating peptide III (CTAP-III) and β-thromboglobulin (β-TG), are typically less active as neutrophil activators; however, these proteins are involved in the chemotaxis of other leukocytes, histamine release, and glycosaminoglycan and proteoglycan metabolism (8, 41).
Recent studies have shown that several chemokines also possess an endogenous antimicrobial activity (56, 60). Detailed structure-function studies have shown that positively charged residues on the surface of these chemokines, especially at their C-terminal α-helices, are largely responsible for their direct antimicrobial activity (4, 9, 28, 39, 56, 63). Peptides of the α-helical portions of some chemokines have been shown to have preferential interactions with anionic lipids over zwitterionic lipids (5, 6), and the full chemokines are believed to act by perturbing bacterial membranes similarly to antimicrobial peptides (10, 17).
In activated platelets, NAP-2 and CTAP-III can become proteolytically truncated at the C terminus by two amino acids to generate thrombocidin-1 (TC-1) and TC-2, respectively (24). This minor change has significant functional consequences because the thrombocidins have direct bactericidal activities against Bacillus subtilis, Escherichia coli, Staphylococcus aureus, and Lactococcus lactis and fungicidal activity against Cryptococcus neoformans, whereas intact NAP-2 and CTAP-III do not. PBP and its derivatives are part of the platelet microbicidal proteins (PMPs) which also include platelet factor-4 (PF-4) (62). These activities have been shown to have important roles for the body's defense against infective endocarditis (13, 34). Proteolysis of the N- and C-terminal ends has been described for numerous chemokines, and it is well known that these events modulate their activity (37, 55). Versions of NAP-2 and CTAP-III with further C-terminal truncations have been described, and many of these have increased activity for neutrophil activation (16). The sequences of NAP-2, TC-1, and the other processed variants of CXCL7 are shown in Fig. 1.
Fig. 1.
Sequence alignment of platelet basic protein and its naturally occurring proteolytically derived variants. Please note that full-length PBP itself is obtained after proteolysis of a 34-residue leader sequence that is required for its secretion. (Adapted with permission from Ehlert et al. [16] and Krijgsveld et al. [24].)
NAP-2 is a homodimer at high concentration in aqueous solution, but it is functionally active as a monomer (7, 43). Its structure was first solved as a tetramer by X-ray crystallography. Subsequently, its solution structure and backbone dynamics were studied by nuclear magnetic resonance (NMR) spectroscopy using a 2-chloroethanol–water cosolvent to dissociate the protein into its monomeric form (61). Similar to other chemokines, the overall fold of NAP-2 consists of an N-terminal region without regular secondary structure that is attached to the remainder of the protein through two disulfide bonds. This is followed by a three-stranded antiparallel β-sheet, a well-defined α-helix, and the protein ends with a short unfolded, solvent-exposed C-terminal tail (61).
Recently, we showed that solution structures of α-helical peptides derived from the corresponding C-terminal portions of NAP-2 and TC-1 determined in an organic cosolvent have a noteworthy difference in their C-terminal tail conformations (39). However, these peptides both have low antimicrobial activities and, as such, the differences in the antimicrobial activities of the full-length NAP-2 and TC-1 proteins could not be adequately explained. Here, NMR relaxation experiments were acquired for samples of 15N isotopically labeled samples of the full NAP-2 and TC-1 chemokines. These data were analyzed using the model-free formalism method (26, 27) to obtain generalized order parameters (S2) and information on chemical exchange rates (Rex) for specific residues. The relaxation information was also used to describe the backbone dynamics of the NAP-2 and TC-1 proteins. Moreover, molecular dynamics simulations were used as well to study fast motions (32), and diffusion NMR experiments (53) were used to characterize the dimerization behavior of the two proteins. These data allowed us to provide a rationale for the functional antimicrobial differences between these two chemokines.
MATERIALS AND METHODS
Purification of recombinant 15N-labeled NAP-2 and TC-1.
15N-labeled recombinant human NAP-2 and TC-1 were expressed as a nonfusion protein in E. coli BL21(DE3) cells grown in 15N-rich growth media (E. coli-OD2 N; Silantes, Munich, Germany). After induction, E. coli cells were pelleted, lysed, and ultracentrifuged, and the supernatant was loaded onto a SP-Sepharose cation-exchange column. SP eluates containing cationic components including NAP-2 or TC-1 were dialyzed and separated by preparative cationic acid-urea polyacrylamide gel electrophoresis. Fractions were analyzed for the presence of NAP-2 or TC-1 on the basis of their respective size by Tris-Tricine SDS-PAGE. The pooled fractions were also tested in radial diffusion and standard broth bactericidal assays with B. subtilis and E. coli to ensure that they had antimicrobial activities comparable to that of unlabeled purified NAP-2 or TC-1. Also, the protein yield recovery was measured by the bicinchoninic acid (BCA) protein assay (49). A typical yield was 5 mg of protein per liter of culture. The purified chemokines were dialyzed against 0.01% acetic acid in Mini Slide-A-Lyzer tubes and lyophilized.
Relaxation experiments.
All NMR experiments were acquired at 310 K. Initial 2D 1H-15N heteronuclear single quantum coherence (HSQC) experiments were run on NAP-2 and TC-1 dissolved in 90%/10% H2O/D2O with the chemical shift standard 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) added to a final concentration of 0.05 mM. Perdeuterated 2-chloroethanol was then added to a concentration of 4% (vol/vol) to dissociate the protein into monomers (33, 61). Judging from the resulting HSQC spectra, a higher concentration of 8% (vol/vol) 2-chloroethanol was required to promote full conversion to the monomeric states of both chemokines. The pH values for the final NAP-2 and TC-1 samples were 4.0 and 3.7, respectively. The concentrations of the samples were 0.4 and 0.6 mM for NAP-2 and TC-1, respectively.
To obtain the backbone assignments, three-dimensional nuclear Overhauser effect spectroscopy (NOESY)-[15N-1H]-HSQC experiments were acquired on a Bruker Avance 700 MHz NMR spectrometer equipped with a triple resonance inverse Cryoprobe with a single axis z-gradient. The mixing time used for both samples was 120 ms.
All backbone dynamics experiments were acquired on a Bruker Avance 500 MHz NMR spectrometer equipped with a triple resonance inverse Cryoprobe. For the longitudinal R1 spin-relaxation experiments, the measured relaxation delays were 42, 98, 154, 196, 294, 406, 602, and 798 ms. For the transverse R2 spin-relaxation experiments, the measured relaxation delays were 6.55, 26.17, 45.79, 65.41, 91.57, 117.73, 137.35, and 163.51 ms. For 1H-15N heteronuclear NOE data acquisition, a 5-s train of 120° proton pulses was used. The data with or without NOE were acquired in an interleaved manner and duplicated for error analysis.
Relaxation data analysis.
NMR spectra were processed using NMRPipe (15), and analysis was performed with NMRView 5.0.4 (21) for spectral visualization. Published assignments for NAP-2 (61) were useful to help make the backbone amide assignments for the HSQC spectra of NAP-2 and TC-1. Three-dimensional NOESY-[15N-1H]-HSQC spectra were used to solve cases of ambiguity and to complete the assignments. R1 and R2 relaxation rates were obtained by fitting the peak intensities to a single exponential decay using the nonlinear least-squares routine of Curvefit in NMRView. The heteronuclear NOE data were also quantified in NMRView, which calculated the intensity ratio between the spectra acquired with or without NOE effect.
The relaxation data were fit to five models as defined in the model-free formalism (26, 27), which assigns each backbone amide spin to one of the five models that describe different spin-relaxation behavior. Depending on the dynamic properties of the particular vector, a specific model is assigned that allows the extraction of the relevant motional parameters. Model selection was performed by using the program FAST-Modelfree (11), which uses ModelFree 4.20 (40) and performs model selection based on the protocol of Mandel et al. (31). The rotational diffusion tensors were estimated based on R2/R1 ratios and pdb structure files for NAP-2 and TC-1 using the programs R2R1_diffusion, pdbinertia, and quadric_diffusion from A. G. Palmer's group (http://www.palmer.hs.columbia.edu/software/diffusion.html). For the model-free calculations, an axially symmetric rotational diffusion tensor was used, the N-H bond length was defined as 1.02 Å, and the chemical shift anisotropy for nitrogen was −172 ppm.
Diffusion NMR spectroscopy.
Pulsed-field gradient diffusion NMR experiments were carried out with lyophilized samples of NAP-2 and TC-1 dissolved in 100% D2O supplemented with 1% dioxane to be used as an internal reference. The concentrations of the chemokines were determined to be 0.1 mM using the BCA protein assay, and the pH values of the samples were 6.3 to 6.4.
The PG-SLED sequence (35, 53) was used to collect pulsed-field-gradient diffusion experiments on a 400-MHz Bruker spectrometer. A total of 64 scans were acquired at each gradient amplitude, with the gradient strength incrementing in 64 steps from 1.25 to 80% of the maximum output of the linear gradient amplifier. Fourier transformation and data analysis were performed using the Bruker XWINNMR software package. Theoretical hydrodynamic radii (RH) were calculated from the empirical equation for folded proteins: RH = 4.75 × N0.29, where N is the number of residues (54). Experimental RH values for the chemokines were calculated as follows: (Dref/Dprotein) × RH(ref), where Dref and Dprotein are the measured diffusion coefficients of dioxane and the protein, respectively, and RH(ref) is the effective hydrodynamic radius of dioxane, taken to be 2.12 Å (22).
MD simulations.
None of the published crystal and NMR structures of NAP-2 contain the coordinates for the last four residues of the protein. As such, we had to construct the last four residues based on the positions of these residues in the structure of the 21 residue peptide recently published (39). To do this, the helical regions of the peptide and NAP-2 (PDB ID 1NAP) were overlaid to provide an optimal fit (residues 55 to 64), and the coordinates were transferred. To minimize steric clashes, the coordinates of the last six residues from the peptide structure were used. For the TC-1 simulations, the same construct was used with the last two residues removed. All of the simulations were conducted with the GROMACS 3.3.3 simulation package (3, 25) and the GROMOS 43a2 force field (47). The use of virtual sites for nonpolar and aromatic hydrogens permits the use of a longer time step of 5 fs (18). The proteins were solvated in a dodecahedral box using explicit simple point charge water molecules, constrained with SETTLE (36). Sodium and chloride counter ions were added to a final concentration of ∼150 mM to represent a physiological concentration and neutralize the overall box charge. Energy minimization was subsequently performed, followed by water and let-go equilibrations. The force constants applied in each of the 50-ps equilibration steps were 1,000, 100, 10, 1, and 0 kJ/mol nm2. The production runs were conducted in triplicate, using randomly generated velocities to sample new trajectories. In total, 3 × 20 ns were accumulated for both the NAP-2 and TC-1 proteins. The neighbor list update and grid searching was performed every four steps, corresponding to a 20-ps periodicity. The simulations were run at 300 K and coupled to an external temperature bath via the Berendsen algorithm, using a coupling constant of 0.1 ps (2). Similarly, the pressure was held constant at 1 bar using isotropic pressure coupling, tp = 1.0 ps, compressibility 4.5 × 10−5 bar−1. Particle Mesh Ewald was used for electrostatics with a cutoff of 0.9 nm (14). Van der Waals interactions were calculated with a twin cutoff of 0.9/1.4 nm. Bond lengths were constrained with LINCS (19).
RESULTS
Backbone assignments of HSQC spectra.
After acquiring an initial HSQC spectrum of 15N-labeled NAP-2 in H2O/D2O (90%/10%) at 310 K, it was noted that the resonances were rather broad due to oligomerization of the protein. Therefore, 4% perdeuterated 2-chloroethanol was added to reproduce the conditions used previously for NAP-2 in the monomeric form (33, 61). The resulting HSQC spectrum did not completely resemble the previously published data (61), with a few extra low-intensity peaks indicating the presence of a minor conformation from a small proportion of NAP-2 aggregates. The final solutions of both NAP-2 and TC-1 used for the remaining NMR experiments contained 8% 2-chloroethanol to promote full dissociation of the chemokines.
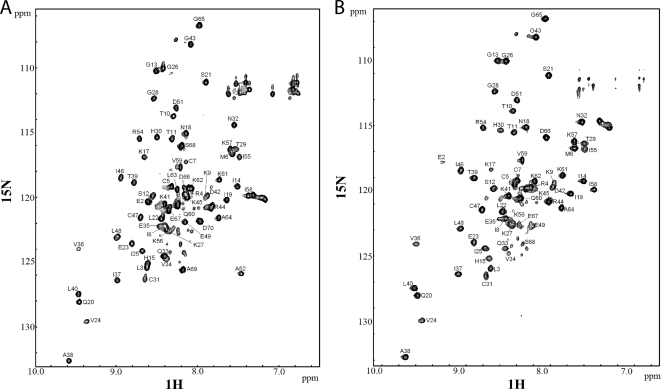
The backbone assignments of the amide peaks in the HSQC spectra were mostly in agreement with the published assignment of NAP-2 (Fig. 2) (61). The three-dimensional NOESY-[15N-1H]-HSQC spectra were essential to determine and/or confirm assignments for peaks that were ambiguous due to peak crowding, slight differences with the previously published HSQC spectra, or the different C-terminal tail of TC-1. Based on the similar spectra obtained for NAP-2 and TC-1, we can conclude that both proteins have very similar structures.
Fig. 2.
1H-15N HSQC spectra of NAP-2 (A) and TC-1 (B) in 8% 2-chloroethanol. Cross-peaks are labeled with resonance assignments. The pH of the samples were 4.0 and 3.7 for NAP-2 and TC-1, respectively, and the spectra were acquired at 310 K.
NMR relaxation experiments.
The resonance assignments were used in the analysis and extraction of heteronuclear NOE values, as well as R1 and R2, the peak intensity decay rates from T1 and T2 relaxation experiments, respectively (Fig. 3). A total of 66 peaks were assigned and used for the analysis out of a total 70 residues in NAP-2. The missing residues include the N terminus at Ala1, where the amino protons undergo fast exchange with the solvent, and Pro16, Pro50, and Pro54, which lack backbone amide protons. For TC-1, the same four residues are missing from the analysis, as well as Ala52 due to line broadening.
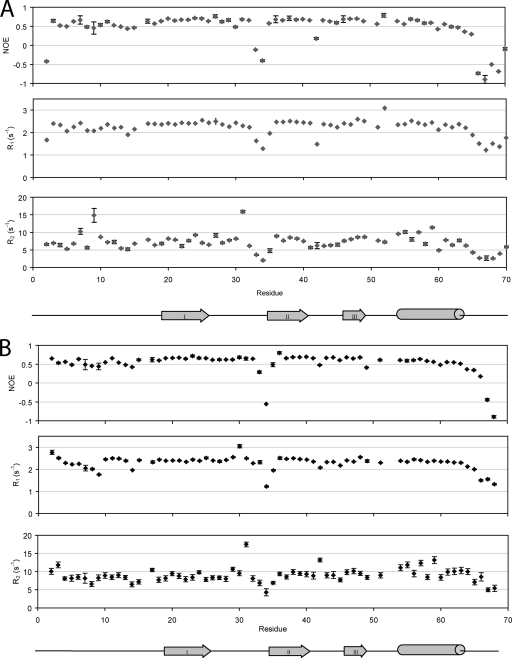
Fig. 3.
1H-15N NMR spin-relaxation data of NAP-2 (A) and TC-1 (B). The plots consist of heteronuclear NOE ratios (top), R1 (middle), and R2 (bottom) relaxation rates. The secondary structure profiles beneath are based on the solution structure of monomeric NAP-2 (61).
The R1, R2, and NOE data are consistent throughout most of the backbones of the NAP-2 and TC-1 proteins. There are two regions in both NAP-2 and TC-1 where these values are significantly lower than the average, indicating fast motion on a ps-ns timescale: the long loop between β-strands I and II; and the C-terminal tail following the α-helix, beginning around Ala64. There are a few residues in NAP-2 and TC-1 with higher than average R2 values, indicating that these residues in particular may undergo conformational exchange on a slower μs-ms timescale.
A few significant differences in the relaxation parameters for NAP-2 and TC-1 were found. For example, the average R2 value for TC-1 (9.09 s−1) is greater than for NAP-2 (7.16 s−1). This reflects slower tumbling for TC-1, which may be due to a higher population of TC-1 dimers than NAP-2. Furthermore, the first few residues of TC-1 have higher R1, R2, and NOE values, indicating a higher order of structural stability, whereas the data for NAP-2 begins with Glu2 being more flexible than the remaining residues. The final major difference between the two chemokines is in their C-terminal tails following the α-helices; this can be attributed to the difference in the sequences of the proteins in that region. The relaxation parameters for both NAP-2 and TC-1 decrease starting at Ala64. The last residues of TC-1 continue with this trend to show increased dynamic behavior. In contrast, the additional residues of NAP-2, specifically Ala69 and Asp70, show a significant increase in motional order more comparable to the folded part of the protein, although those residues still belong to the C-terminal tail that is more flexible. The latter result differs from previously reported 15N dynamics data of NAP-2 (61). In the published data, the C-terminal region becomes increasingly more dynamic and flexible at the end. This result can be attributed to the unnatural Tyr71 residue that was added to the recombinant NAP-2 protein that was used in the earlier studies (33, 61). The purpose for the incorporation of this extraneous C-terminal aromatic residue was for use as an additional NMR probe that had well-resolved resonances (57). Our results here show that the addition presents an artifact where the extra residue disrupts the interactions that the NAP-2 C-terminal tail makes with the rest of the protein.
Model-free analysis.
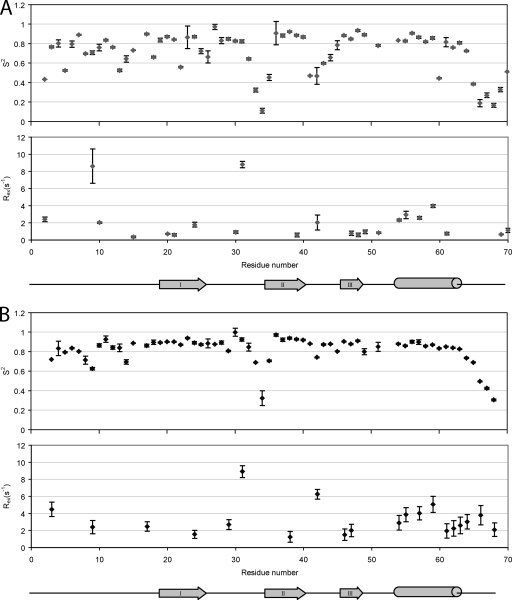
Fast model-free analysis (11) was used to fit the relaxation parameters according to the Lipari-Szabo model-free dynamics formalism (26, 27). From this procedure, the best value for the molecular rotational correlation time, τm, for NAP-2 and TC-1 were 5.5 and 6.0 ns, respectively. These values are greater than those determined for other chemokines of the same size in monomeric form at the same field strength (500 MHz) at temperatures lower than the one used here (310 K), specifically eotaxin (5.09 ns), eotaxin-3 (5.07 ns), and stromal cell-derived factor-1α (5.04 ns) (1, 58, 59). Therefore, a significant amount of monomer-dimer equilibrium is still ongoing in our NAP-2 and TC-1 samples. The higher τm for TC-1 compared to NAP-2 is reflected in the higher R2 relaxation rate values, and this indicates the presence of more extensive dimerization for TC-1.
The S2 order parameters and Rex for residues that could be assigned by the model-free analysis are plotted in Fig. 4. The N-terminal segment of NAP-2 up to Lys17 contains segments of higher flexibility than the protein's regular secondary structure elements. However, this is a much more restrained region than in the related chemokine SDF-1α (1). This is followed by higher S2 values for the first β-strand. β-Strands II and III and the C-terminal α-helix all contain more continuous segments of high motional order, with the flexible loops between the β-strands decreasing in the order parameters. Beyond the NAP-2 α-helix, the order parameters of the C-terminal residues decrease greatly; however, they increase again for the last two residues, Ala69 and Asp70. The first 18 residues of TC-1 together also have somewhat lower values for their order parameters compared to the β-strands and α-helix. However, these values, as well as those for the first β-strand, are higher and more consistent than those of NAP-2. Loop I between the first two β-strands shows high dynamic behavior, specifically for residues Gln33, Val34, and Glu35. Unlike NAP-2, however, loop II of TC-1 between β-strands II and III retains a high motional order. Starting after the α-helix at Leu 63, there is a steady decline in the S2 values to the last residue of TC-1.
Fig. 4.
Model-free data for A) NAP-2 and B) TC-1. The plots consist of order parameters reflecting motions on the ps-ns timescale (top) and exchange rates reflecting motions on the μs-ms timescale (bottom).
Respectively, 22 and 20 residues from NAP-2 and TC-1 required a chemical exchange parameter (Rex) that describes a conformational exchange phenomenon occurring in the μs to ms timescale. These were previously mentioned in regard to residues with higher R2 relaxation rates. In NAP-2, the backbone amides with particularly high Rex values are Lys9 and Cys31, the latter of which is involved in a disulfide bond. There are also four residues belonging to the α-helix of NAP-2 with significant conformational exchange, perhaps due to a slight movement of the α-helix with respect to the remainder of the protein. In TC-1, the spins with particularly high Rex values are Cys31, again involved in a disulfide bond with Cys5, and Lys42. Like NAP-2, four residues from the TC-1 α-helix undergo conformational exchange, probably again reflecting the monomer-dimer equilibrium. TC-1 also has a few residues following the α-helix with increasing Rex values, and therefore it shows movement of the C-terminal tail between different conformations, something that is not observed for NAP-2.
MD simulations.
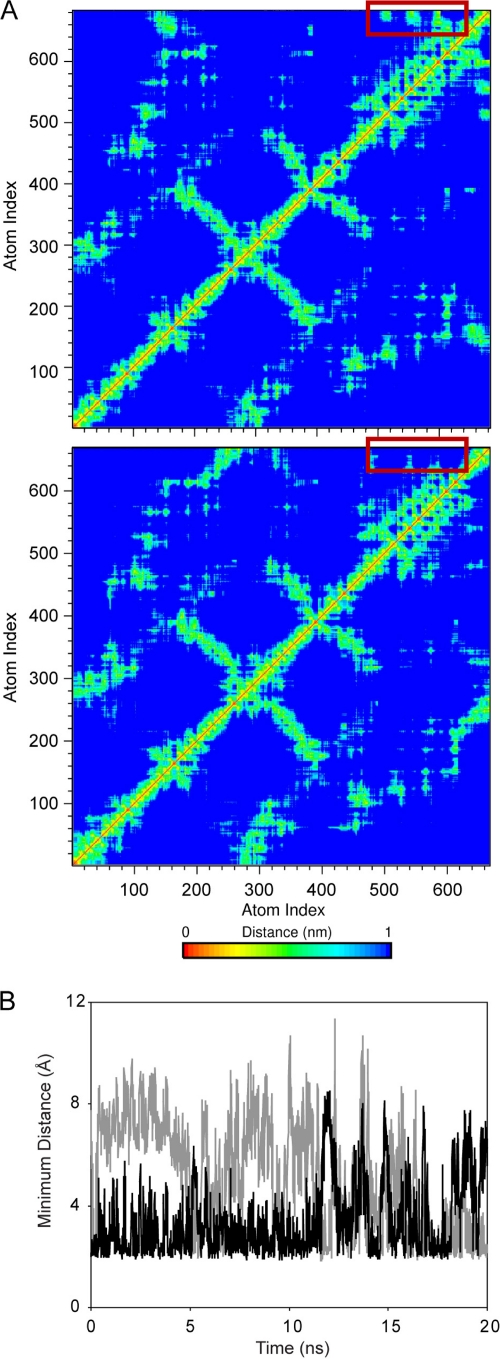
In order to gain further insight into the behavior of the C-terminal regions of NAP-2 and TC-1, we performed molecular dynamics (MD) simulations of the two proteins. Since the positions of the last four residues of NAP-2 are not defined in the available crystal structure of the protein, they had to be added based on the last amino acids as observed in the 21 residue peptide structure recently determined by NMR (39). Because of the observation of several long-range NOEs, in this peptide structure the final Asp70 residue is positioned close to the side chains of two Lys residues in the C-terminal helical portion of NAP-2. This initial starting position is consistent with the dynamic behavior of this residue observed in our present NMR relaxation study. We performed three parallel, unrestrained simulations of monomeric NAP-2 and TC-1 of 20 ns each, to probe fast motions similar to those captured by the 15N NMR relaxation measurements. Analysis of the root mean square deviations showed that both proteins were stable and maintained their overall structure during the MD simulations. The C-terminal tail that is of interest in the present study displays different behavior in the two proteins. In TC-1, the distance from the center of mass of the C terminus to that of the α-helix is consistently higher compared to that of NAP-2 and the number of hydrogen bonds are notably lower. In addition, the standard deviation, which is an indication of the flexibility of this distance, is significantly lower in the NAP-2 trajectories compared to the TC-1 simulations (Table 1). That is, the C-terminal end of the protein is more stably associated with the helix in the NAP-2 simulations compared to those of TC-1. This is evident in distance plots of NAP-2 and TC-1 shown in Fig. 5, which indicate that the last two residues in NAP-2 are crucial for the C terminus to maintain contact with the α-helix. Asp70 is the last residue in NAP-2 and, as such, contains two anionic charges. During the NAP-2 simulations, this residue is observed to interact with a number of positively charged residues such as Lys57 and Lys61 (Fig. 5B). This is consistent with the NOEs observed in the peptide NMR structures (39).
Table 1.
Distance and hydrogen bonds of C-terminal residues to the α-helix in the MD simulations
| Chemokine | Avg ± SD |
|
|---|---|---|
| C-terminal distance to helix (nm)a | No. of hydrogen bondsb | |
| NAP-2 | 0.84 ± 0.10 | 2.60 ± 1.55 |
| 0.84 ± 0.06 | 3.74 ± 2.28 | |
| 0.72 ± 0.05 | 2.42 ± 1.34 | |
| TC-1 | 1.54 ± 0.23 | 2.49 ± 1.35 |
| 1.17 ± 0.31 | 1.48 ± 1.19 | |
| 0.93 ± 0.34 | 1.88 ± 1.23 | |
Minimum distance of center of mass of the last four residues to the α-helix.
Averaged for the last four residues to the α-helix over the last 15 ns of the trajectories.
Fig. 5.
MD simulation results for the NAP-2 and TC-1 chemokines. (A) Representative distance plots averaged using the NMR 1/r6 distance dependence for NAP-2 (top panel) and TC-1 lower panel. The additional two residues in NAP-2 form considerable contacts with the α-helix during the protein (red boxes). These contacts are absent in the TC1 trajectory due to the lack of Asp70. (B) The minimum distance between Asp70 of NAP-2 and Lys57 (black) and Lys61 (gray) illustrating the pronounced electrostatic interactions that take place between the far C terminus and the α-helix.
Diffusion NMR experiments.
To investigate further the monomer-dimer properties of NAP-2 and TC-1, NMR diffusion studies were conducted using the internal standard dioxane, as has been done for other chemokines (9, 20, 51). For these experiments, the concentrations and conditions of the chemokine samples were carefully maintained to be consistent with each other for direct comparison, and a dilute protein concentration of 0.1 mM was used to obtain a more observable difference in the dimerization properties of the chemokines. The results are shown in Table 2. The hydrodynamic radius calculated for NAP-2 is close to that of a monomer for a theoretical folded, globular protein of the same size. The higher experimental RH value for TC-1 indicates that there is a higher proportion of dimers than monomers present in solution for this truncated protein.
Table 2.
Hydrodynamic radii of NAP-2 and TC-1 theoretically calculated for globular proteins and determined experimentally from diffusion NMR experiments
| Chemokine | Theoretical RH (Å) |
Experimental RH (avg Å ± SD) | |
|---|---|---|---|
| Monomer | Dimer | ||
| NAP-2 | 16.4 | 20.1 | 16.3 ± 0.8 |
| TC-1 | 16.3 | 20.0 | 19.0 ± 0.8 |
DISCUSSION
Platelets play a major role in blood clot formation, wound healing, and cytokine signaling. In addition, microbicidal proteins can be released from activated platelets (50, 62, 64). TC-1 has been characterized as a C-terminal deletion product of NAP-2 (24). The loss of the two carboxy-terminal amino acid residues, presumably through the activity of a carboxypeptidase, represents a functional switch that turns TC-1 into an active microbicidal and fungicidal protein (24). To examine the structural basis for this phenomenon, backbone dynamics experiments by NMR spectroscopy were carried out and analyzed using the Lipari-Szabo model-free formalism.
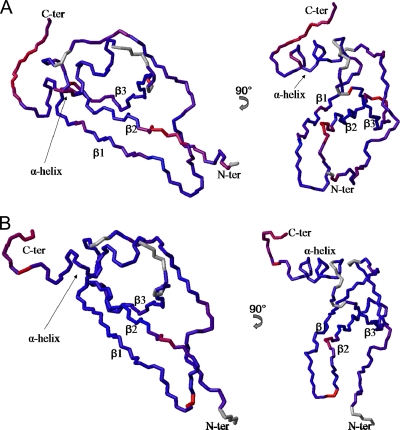
In Fig. 6, the main chains of the NAP-2 and TC-1 structural models are colored according to S2. These derived order parameters are sensitive to local motions on the ps-ns timescale. Overall, both proteins exhibit dynamic behavior typical of other chemokines (1, 58, 59, 61), with an aperiodic flexible N-terminal segment encompassing the first 15 or so residues that are most important for chemokine receptor binding, as has been found for other CXC chemokines (12, 42). This is followed by ordered regular secondary structure elements, with the loop between the first two β-strands showing some flexibility, and a flexible C-terminal segment past the α-helix.
Fig. 6.
Backbone chains of NAP-2 (A) and TC-1 (B) with residues colored along a gradient according to their S2 order parameters. Red and blue residues represent high and low extremes in flexibility, respectively, with purple representing intermediate flexibility. Residues for which no S2 values were assigned are colored in gray. The structure models are based on a combination of the NAP-2 crystal structure (PDB ID 1NAP) and the solution structures of the corresponding C-terminal α-helix peptides solved previously (39).
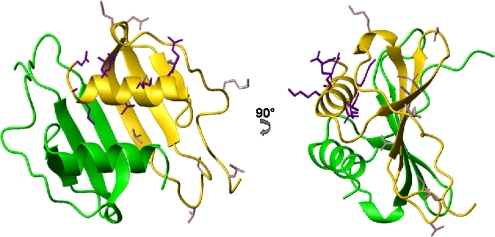
The larger τm for TC-1 compared to NAP-2 in the same solvent and temperature with similar pH values points to TC-1 forming dimers more readily than NAP-2. This is confirmed by the results from the diffusion NMR experiments. The NAP-2 dimer, as part of the tetrameric (dimer of dimers) complex solved by X-ray crystallography (30), is shown in Fig. 7. The dimer interface of NAP-2 involves hydrogen bonding between the two β-I strands which run antiparallel to each other as well as contacts between the two α-helices. The side chains of the α-helix residues of both chemokines with conformational exchange Rex terms, as determined by model-free analysis seem to be involved in establishing intra- and intermolecular contacts. The α-helix therefore seems to move as a unit and play a role in dimerization, with TC-1 having more residues with Rex terms in that segment. Such movements have also been observed for other chemokines such as MIP-3α and SDF-1α, and they often accompany the monomer-dimer dissociation process (1, 9). In vivo, NAP-2 and presumably TC-1 are in a monomer-dimer equilibrium and may even form higher-order oligomers; however, NAP-2 is thought to bind as a monomer to its chemokine receptors (43, 46).
Fig. 7.
Two views of the NAP-2 dimer structure solved by X-ray crystallography (PDB ID 1NAP) (30) showing the dimer interface consisting of β-strand I and the α-helix. The side chains of TC-1 residues with Rex terms are shown in dark violet for the α-helix region and light pink for the remainder TC-1.
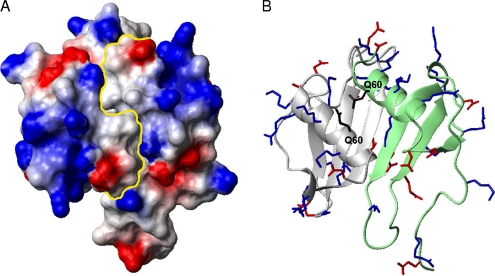
The role of dimerization to promote antimicrobial activity may be due to the formation of a large cationic solvent-exposed surface area across the antiparallel α-helices that is slightly interrupted by the polar side chain of Gln60 in both monomer subunits (Fig. 8). This is a feature common to all antimicrobial chemokines, which also contain hydrophobic surfaces (56). For the NAP-2 or TC-1 dimer, there are hydrophobic patches on either side of the large cationic surface; however, the surface directly behind it contains some negatively charged and polar side chains. Exactly how the two C-terminal residues of NAP-2 shift the monomer-dimer equilibrium in solution compared to TC-1 is unclear at present and would require further work in the future.
Fig. 8.
(A) Electrostatic potential surface plot of the NAP-2 dimer (PDB ID 1NAP) with a yellow line showing the divide between the monomer subunits and the large, positively charged patch encircled. (B) Ribbon diagram of the dimer in the same orientation with positively charged side chains in blue, negatively charged side chains in red, and the bridging surface Gln60 residues in black, which slightly interrupt the large cationic surface.
Another major difference between the 15N relaxation data of the chemokines occurs exactly at the region of the amino acid sequence difference, the C-terminal tail. The last two residues of NAP-2 are not as flexible as the residues that precede them, whereas the last residues of TC-1 are clearly the most flexible. This supports the structures determined for the α-helix peptides that show that the last Asp residue of the NAP-2 peptide can fold back and interact with the positively charged region of the α-helix (39). This does not occur with the C-terminal end of the TC-1 peptide. Overall, the MD simulation and NMR relaxation results are consistent with the notion that the double negatively charged Asp70 residue of NAP-2 effectively shields part of the positive surface of the protein. Such an effect is not seen for the C-terminal Ser68 residue of TC-1. This would therefore decrease the possibility of NAP-2 interactions with the negatively charged bacterial membranes to lead to reduced antimicrobial activity. This restrained C-terminal flexibility was not observed in earlier backbone dynamics experiments of NAP-2. This is presumably due to the extra C-terminal Tyr residue that was introduced in that study, which would cause steric hindrance and prevent the preceding residues to have any interactions with the rest of the protein. In contrast to NAP-2, the shortened C-terminal end of TC-1, while still negatively charged with its sequence ending in Asp66, Glu67, and Ser68, is fully flexible and solvent exposed. This would allow for exposure of a large, positively charged surface on TC-1, particularly in a dimer form of the protein.
The 15N backbone dynamics work and MD simulations presented here have revealed a basis for the emergence of the TC-1 antimicrobial activities after it has been produced from NAP-2. The two-amino-acid extension of NAP-2 is less mobile and seems to mask a cationic surface on the protein, interfering with the direct bactericidal mechanism. The shortened tail of TC-1 is highly mobile and not restrained by any other areas of the chemokine. TC-1 is also better able to form dimers than NAP-2. Through this dimerization, a large, mostly continuous cationic surface is assembled, and this promotes interactions with the negatively charged bacterial membranes, resulting in bacterial killing.
ACKNOWLEDGMENTS
We thank Deane McIntyre for the management and upkeep of the NMR facilities.
Operating support for this research was provided by the Institute for Infection and Immunity of the Canadian Institutes of Health Research. L.T.N. was supported by a studentship award from the Alberta Heritage Foundation for Medical Research (AHFMR). H.J.V. holds a Scientist Award from the AHFMR.
Footnotes
Published ahead of print on 14 February 2011.
REFERENCES
- 1. Baryshnikova O. K., Sykes B. D. 2006. Backbone dynamics of SDF-1α determined by NMR: interpretation in the presence of monomer-dimer equilibrium. Protein Sci. 15:2568–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., Dinola A., Haak J. R. 1984. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690 [Google Scholar]
- 3. Berendsen H. J. C., van der Spoel D., van Drunen R. 1995. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 91:43–56 [Google Scholar]
- 4. Bhattacharjya S., Ramamoorthy A. 2009. Multifunctional host defense peptides: functional and mechanistic insights from NMR structures of potent antimicrobial peptides. FEBS J. 276:6465–6473 [DOI] [PubMed] [Google Scholar]
- 5. Bourbigot S., et al. 2009. Antimicrobial peptide RP-1 structure and interactions with anionic versus zwitterionic micelles. Biopolymers 91:1–13 [DOI] [PubMed] [Google Scholar]
- 6. Bourbigot S., Fardy L., Waring A. J., Yeaman M. R., Booth V. 2009. Structure of chemokine-derived antimicrobial peptide interleukin-8α and interaction with detergent micelles and oriented lipid bilayers. Biochemistry 48:10509–10521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brandt E., Ludwig A., Petersen F., Flad H. D. 2000. Platelet-derived CXC chemokines: old players in new games. Immunol. Rev. 177:204–216 [DOI] [PubMed] [Google Scholar]
- 8. Castor C. W., Miller J. W., Walz D. A. 1983. Structural and biological characteristics of connective tissue activating peptide (CTAP-III), a major human platelet-derived growth factor. Proc. Natl. Acad. Sci. U. S. A. 80:765–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chan D. I., Hunter H. N., Tack B. F., Vogel H. J. 2008. Human macrophage inflammatory protein 3α: protein and peptide nuclear magnetic resonance solution structures, dimerization, dynamics, and anti-infective properties. Antimicrob. Agents Chemother. 52:883–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan D. I., Prenner E. J., Vogel H. J. 2006. Tryptophan- and arginine-rich antimicrobial peptides: structures and mechanisms of action. Biochim. Biophys. Acta 1758:1184–1202 [DOI] [PubMed] [Google Scholar]
- 11. Cole R., Loria J. P. 2003. FAST-Modelfree: a program for rapid automated analysis of solution NMR spin-relaxation data. J. Biomol. NMR. 26:203–213 [DOI] [PubMed] [Google Scholar]
- 12. Crump M. P., et al. 1997. Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 16:6996–7007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dankert J., Krijgsveld J., van Der Werff J., Joldersma W., Zaat S. A. 2001. Platelet microbicidal activity is an important defense factor against viridans streptococcal endocarditis. J. Infect. Dis. 184:597–605 [DOI] [PubMed] [Google Scholar]
- 14. Darden T., York D., Pedersen L. 1993. Particle mesh Ewald: an N log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092 [Google Scholar]
- 15. Delaglio F., et al. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277–293 [DOI] [PubMed] [Google Scholar]
- 16. Ehlert J. E., Gerdes J., Flad H. D., Brandt E. 1998. Novel C-terminally truncated isoforms of the CXC chemokine beta-thromboglobulin and their impact on neutrophil functions. J. Immunol. 161:4975–4982 [PubMed] [Google Scholar]
- 17. Epand R. M., Vogel H. J. 1999. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta 1462:11–28 [DOI] [PubMed] [Google Scholar]
- 18. Feenstra K. A., Hess B., Berendsen H. J. C. 1999. Improving efficiency of large time-scale molecular dynamics simulations of hydrogen-rich systems. J. Comput. Chem. 20:786–798 [DOI] [PubMed] [Google Scholar]
- 19. Hess B., Bekker H., Herman J. C., Berendsen H. J. C., Fraaije J. G. E. M. 1997. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463–1472 [Google Scholar]
- 20. Jansma A. L., Kirkpatrick J. P., Hsu A. R., Handel T. M., Nietlispach D. 2010. NMR analysis of the structure, dynamics, and unique oligomerization properties of the chemokine CCL27. J. Biol. Chem. 285:14424–14437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson B. A. 2004. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278:313–352 [DOI] [PubMed] [Google Scholar]
- 22. Jones J. A., Wilkins D. K., Smith L. J., Dobson C. M. 1997. Characterisation of protein unfolding by NMR diffusion measurements. J. Biomol. NMR 10:199–203 [Google Scholar]
- 23. Katancik J. A., Sharma A., de Nardin E. 2000. Interleukin 8, neutrophil-activating peptide-2 and GRO-alpha bind to and elicit cell activation via specific and different amino acid residues of CXCR2. Cytokine 12:1480–1488 [DOI] [PubMed] [Google Scholar]
- 24. Krijgsveld J., et al. 2000. Thrombocidins, microbicidal proteins from human blood platelets, are C-terminal deletion products of CXC chemokines. J. Biol. Chem. 275:20374–20381 [DOI] [PubMed] [Google Scholar]
- 25. Lindahl E., Hess B., van der Spoel D. 2001. GROMACS 3.0: a package for molecular simulation and trajectory analysis J. Mol. Model. 7:306–317 [Google Scholar]
- 26. Lipari G., Szabo A. 1982. Model-free approach to the interpretation of nuclear magnetic-resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 104:4546–4559 [Google Scholar]
- 27. Lipari G., Szabo A. 1982. Model-free approach to the interpretation of nuclear magnetic-resonance relaxation in macromolecules. 2. Analysis of experimental results. J. Am. Chem. Soc. 104:4559–4570 [Google Scholar]
- 28. Liu B., Wilson E. 2010. The antimicrobial activity of CCL28 is dependent on C-terminal positively charged amino acids. Eur. J. Immunol. 40:186–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Majumdar S., Gonder D., Koutsis B., Poncz M. 1991. Characterization of the human beta-thromboglobulin gene: comparison with the gene for platelet factor 4. J. Biol. Chem. 266:5785–5789 [PubMed] [Google Scholar]
- 30. Malkowski M. G., Wu J. Y., Lazar J. B., Johnson P. H., Edwards B. F. 1995. The crystal structure of recombinant human neutrophil-activating peptide-2 (M6L) at 1.9-Å resolution. J. Biol. Chem. 270:7077–7087 [DOI] [PubMed] [Google Scholar]
- 31. Mandel A. M., Akke M., Palmer A. G., III 1995. Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J. Mol. Biol. 246:144–163 [DOI] [PubMed] [Google Scholar]
- 32. Markwick P. R., et al. 2009. Toward a unified representation of protein structural dynamics in solution. J. Am. Chem. Soc. 131:16968–16975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mayo K. H., Yang Y., Daly T. J., Barry J. K., La Rosa G. J. 1994. Secondary structure of neutrophil-activating peptide-2 determined by 1H-nuclear magnetic resonance spectroscopy. Biochem. J. 304(Pt. 2):371–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mercier R. C., Rybak M. J., Bayer A. S., Yeaman M. R. 2000. Influence of platelets and platelet microbicidal protein susceptibility on the fate of Staphylococcus aureus in an in vitro model of infective endocarditis. Infect. Immun. 68:4699–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Merrill M. R. 1993. NMR diffusion measurements using a composite gradient PGSE sequence. J. Magn. Reson. A 103:223–225 [Google Scholar]
- 36. Miyamoto S., Kollman P. A. 1992. SETTLE: an analytical version of the SHAKE and RATTLE algorithms for rigid water models. J. Comput. Chem. 13:952–962 [Google Scholar]
- 37. Mortier A., Van Damme J., Proost P. 2008. Regulation of chemokine activity by posttranslational modification. Pharmacol. Ther. 120:197–217 [DOI] [PubMed] [Google Scholar]
- 38. Moser B., Schumacher C., von Tscharner V., Clark-Lewis I., Baggiolini M. 1991. Neutrophil-activating peptide 2 and gro/melanoma growth-stimulatory activity interact with neutrophil-activating peptide 1/interleukin 8 receptors on human neutrophils. J. Biol. Chem. 266:10666–10671 [PubMed] [Google Scholar]
- 39. Nguyen L. T., Chan D. I., Boszhard L., Zaat S. A., Vogel H. J. 2010. Structure-function studies of chemokine-derived carboxy-terminal antimicrobial peptides. Biochim. Biophys. Acta 1798:1062–1072 [DOI] [PubMed] [Google Scholar]
- 40. Palmer A. G., Rance M., Wright P. E. 1991. Intramolecular motions of a zinc finger DNA-binding domain from Xfin characterized by proton-detected natural abundance C-12 heteronuclear NMR-spectroscopy. J. Am. Chem. Soc. 113:4371–4380 [Google Scholar]
- 41. Proudfoot A. E., et al. 1997. Structure and bioactivity of recombinant human CTAP-III and NAP-2. J. Protein Chem. 16:37–49 [DOI] [PubMed] [Google Scholar]
- 42. Rajagopalan L., Rajarathnam K. 2004. Ligand selectivity and affinity of chemokine receptor CXCR1: role of N-terminal domain. J. Biol. Chem. 279:30000–30008 [DOI] [PubMed] [Google Scholar]
- 43. Rajarathnam K., et al. 1997. Neutrophil-activating peptide-2 and melanoma growth-stimulatory activity are functional as monomers for neutrophil activation. J. Biol. Chem. 272:1725–1729 [DOI] [PubMed] [Google Scholar]
- 44. Schaffner A., King C. C., Schaer D., Guiney D. G. 2004. Induction and antimicrobial activity of platelet basic protein derivatives in human monocytes. J. Leukoc. Biol. 76:1010–1018 [DOI] [PubMed] [Google Scholar]
- 45. Schmid J., Weissman C. 1987. Induction of mRNA for a serine protease and a beta-thromboglobulin-like protein in mitogen-stimulated human leukocytes. J. Immunol. 139:250–256 [PubMed] [Google Scholar]
- 46. Schnitzel W., Monschein U., Besemer J. 1994. Monomer-dimer equilibria of interleukin-8 and neutrophil-activating peptide 2. Evidence for IL-8 binding as a dimer and oligomer to IL-8 receptor B. J. Leukoc. Biol. 55:763–770 [DOI] [PubMed] [Google Scholar]
- 47. Scott W. R. P., et al. 1999. The GROMOS biomolecular simulation program package. J. Phys. Chem. A 103:3596–3607 [Google Scholar]
- 48. Shirozu M., et al. 1995. Structure and chromosomal localization of the human stromal cell-derived factor 1 (SDF1) gene. Genomics 28:495–500 [DOI] [PubMed] [Google Scholar]
- 49. Stoscheck C. M. 1990. Quantitation of protein. Methods Enzymol. 182:50–68 [DOI] [PubMed] [Google Scholar]
- 50. Tang Y. Q., Yeaman M. R., Selsted M. E. 2002. Antimicrobial peptides from human platelets. Infect. Immun. 70:6524–6533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Veldkamp C. T., Peterson F. C., Pelzek A. J., Volkman B. F. 2005. The monomer-dimer equilibrium of stromal cell-derived factor-1 (CXCL 12) is altered by pH, phosphate, sulfate, and heparin. Protein Sci. 14:1071–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Walz A., Baggiolini M. 1990. Generation of the neutrophil-activating peptide NAP-2 from platelet basic protein or connective tissue-activating peptide III through monocyte proteases. J. Exp. Med. 171:449–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weljie A. M., Yamniuk A. P., Yoshino H., Izumi Y., Vogel H. J. 2003. Protein conformational changes studied by diffusion NMR spectroscopy: application to helix-loop-helix calcium binding proteins. Protein Sci. 12:228–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wilkins D. K., et al. 1999. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry 38:16424–16431 [DOI] [PubMed] [Google Scholar]
- 55. Wolf M., Albrecht S., Marki C. 2008. Proteolytic processing of chemokines: implications in physiological and pathological conditions. Int. J. Biochem. Cell Biol. 40:1185–1198 [DOI] [PubMed] [Google Scholar]
- 56. Yang D., et al. 2003. Many chemokines including CCL20/MIP-3α display antimicrobial activity. J. Leukoc. Biol. 74:448–455 [DOI] [PubMed] [Google Scholar]
- 57. Yang Y., Mayo K. H., Daly T. J., Barry J. K., La Rosa G. J. 1994. Subunit association and structural analysis of platelet basic protein and related proteins investigated by 1H NMR spectroscopy and circular dichroism. J. Biol. Chem. 269:20110–20118 [PubMed] [Google Scholar]
- 58. Ye J., Mayer K. L., Mayer M. R., Stone M. J. 2001. NMR solution structure and backbone dynamics of the CC chemokine eotaxin-3. Biochemistry 40:7820–7831 [DOI] [PubMed] [Google Scholar]
- 59. Ye J., Mayer K. L., Stone M. J. 1999. Backbone dynamics of the human CC-chemokine eotaxin. J. Biomol. NMR 15:115–124 [DOI] [PubMed] [Google Scholar]
- 60. Yeaman M. R., Yount N. Y. 2007. Unifying themes in host defense effector polypeptides. Nat. Rev. Microbiol. 5:727–740 [DOI] [PubMed] [Google Scholar]
- 61. Young H., Roongta V., Daly T. J., Mayo K. H. 1999. NMR structure and dynamics of monomeric neutrophil-activating peptide 2. Biochem. J. 338(Pt. 3):591–598 [PMC free article] [PubMed] [Google Scholar]
- 62. Yount N. Y., et al. 2004. Platelet microbicidal protein 1: structural themes of a multifunctional antimicrobial peptide. Antimicrob. Agents Chemother. 48:4395–4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yount N. Y., et al. 2007. Structural correlates of antimicrobial efficacy in IL-8 and related human kinocidins. Biochim. Biophys. Acta 1768:598–608 [DOI] [PubMed] [Google Scholar]
- 64. Zaat S. A., et al. 1997. Cell-adherent glucan does not protect endocarditis-causing viridans streptococci against bactericidal proteins from human blood platelets. Adv. Exp. Med. Biol. 418:709–712 [DOI] [PubMed] [Google Scholar]