Abstract
The prevalence and impact of the overexpression of AmpC and efflux pumps were evaluated with a collection of 190 Pseudomonas aeruginosa isolates recovered from bloodstream infections in a 2008 multicenter study (10 hospitals) in Spain. The MICs of a panel of 13 antipseudomonal agents were determined by microdilution, and the expressions of ampC, mexB, mexY, mexD, and mexF were determined by real-time reverse transcription (RT)-PCR. Up to 39% of the isolates overexpressed at least one of the mechanisms. ampC overexpression (24.2%) was the most prevalent mechanism, followed by mexY (13.2%), mexB (12.6%), mexF (4.2%), and mexD (2.2%). The overexpression of mexB plus mexY, documented for 5.3% of the isolates, was the only combination showing a significantly (P = 0.02) higher prevalence than expected from the frequencies of the individual mechanisms (1.6%). Additionally, all imipenem-resistant isolates studied (25 representative isolates) showed inactivating mutations in oprD. Most of the isolates nonsusceptible to piperacillin-tazobactam (96%) and ceftazidime (84%) overexpressed ampC, while mexB (25%) and mexY (29%) overexpressions gained relevance among cefepime-nonsusceptible isolates. Nevertheless, the prevalence of mexY overexpression was highest among tobramycin-nonsusceptible isolates (37%), and that of mexB was highest among meropenem-nonsusceptible isolates (33%). Regarding ciprofloxacin-resistant isolates, besides the expected increased prevalence of efflux pump overexpression, a highly significant link to ampC overexpression was documented for the first time: up to 52% of ciprofloxacin-nonsusceptible isolates overexpressed ampC, sharply contrasting with the 24% documented for the complete collection (P < 0.001). In summary, mutation-driven resistance was frequent in P. aeruginosa isolates from bloodstream infections, whereas metallo-β-lactamases, detected in 2 isolates (1%) producing VIM-2, although with increasing prevalences, were still uncommon.
INTRODUCTION
The increasing prevalence of nosocomial infections produced by multidrug-resistant (MDR) Pseudomonas aeruginosa strains severely compromises the selection of appropriate treatments and is therefore associated with significant morbidity and mortality (17, 21, 25). The growing threat of antimicrobial resistance in P. aeruginosa results from the extraordinary capacity of this microorganism for developing resistance to almost any available antibiotic by the selection of mutations in chromosomal genes and from the increasing prevalence of transferrable resistance determinants, particularly those encoding class B carbapenemases (or metallo-β-lactamases [MBLs]) or extended-spectrum β-lactamases (ESBLs), frequently cotransferred with genes encoding aminoglycoside-modifying enzymes (18, 19). Among the mutation-mediated resistance mechanisms, particularly noteworthy are those leading to the repression or inactivation of the carbapenem porin OprD, the hyperproduction of the chromosomal cephalosporinase AmpC, or the upregulation of one of the several efflux pumps encoded in the P. aeruginosa genome (11, 13, 22, 32). Furthermore, the accumulation of many of these chromosomal mutations can lead to the emergence of MDR (or even panantibiotic-resistant) strains, which eventually may be responsible for notable outbreaks in the hospital setting (7). Nevertheless, except for some seminal French studies (13), there is still scarce information on the large-scale (suprahospital) epidemiology of P. aeruginosa resistance mechanisms, and there is much less information on the impact of these mechanisms on resistance prevalence. Therefore, a national-scale study was conducted in Spain to determine the prevalence and impact of AmpC and efflux pump overexpression and OprD mutations in P. aeruginosa bloodstream isolates on resistance.
MATERIALS AND METHODS
Strains, molecular typing, and susceptibility testing.
A total of 190 P. aeruginosa isolates recovered from bloodstream infections in a 2008 multicenter study (10 hospitals from different geographic locations) in Spain were studied. Clonal relatedness was evaluated through repetitive extragenic palindromic PCR (REP-PCR) according to previously described protocols (4). The MICs of ceftazidime (Sigma-Aldrich, Madrid, Spain), cefepime (Aventis Pharma, Madrid, Spain), piperacillin (Sigma-Aldrich), piperacillin plus tazobactam (fixed concentration of 4 μg/ml) (Sigma-Aldrich), aztreonam (Sigma-Aldrich), imipenem (Merck, Sharp & Dohme, Madrid, Spain), meropenem (Astra-Zeneca, Madrid, Spain), ciprofloxacin (Sigma-Aldrich), levofloxacin (Roussel Uclaf, Paris, France), gentamicin (Sigma-Aldrich), tobramycin (Sigma-Aldrich), amikacin (Sigma-Aldrich), and colistin (Sigma-Aldrich) were determined by broth microdilution according to CLSI guidelines and breakpoints (5).
Quantification of gene expression by RT-PCR.
The levels of expression of ampC, mexB, mexD, mexY, and mexF were determined by real-time reverse transcription (RT)-PCR according to previously described protocols (15, 26). Briefly, strains were grown in 10 ml of LB broth at 37°C and 180 rpm to the late log phase (optical density at 600 nm [OD600] of 1) and collected by centrifugation. Total RNA was isolated by using the RNeasy minikit (Qiagen), dissolved in water, and treated with 2 U of Turbo DNase (Ambion) for 30 min at 37°C to remove contaminating DNA. The reaction was stopped by the addition of 5 μl of DNase inactivation reagent to the mixture. A 50-ng sample of purified RNA was then used for one-step reverse transcription and real-time PCR amplification using the QuantiTect SYBR green RT-PCR kit (Qiagen) with a SmartCycler II instrument (Cepheid). Previously described primers (15, 26) were used for the amplification of ampC, mexB, mexD, mexY, mexF, and rpsL (used as a reference to normalize the relative amount of mRNA). Strains were considered positive for ampC, mexD, mexF, or mexY overexpression when the corresponding mRNA level was at least 10-fold higher than that of PAO1, negative if lower than 5-fold, and borderline if between 5- and 10-fold. Strains were considered positive for mexB overexpression when the corresponding mRNA level was at least 3-fold higher than that of PAO1, negative if lower than 2-fold, and borderline if between 2- and 3-fold. All PCRs were performed in duplicate. Strains showing mRNA values of >5-fold for ampC, mexD, mexF, or mexY or >2-fold for mexB in a first experiment were subjected to two additional independent RNA extractions and duplicate PCRs. Mean values (± standard deviations) of mRNA levels obtained in three independent duplicate experiments were considered. Previously obtained PAO1 mutants overexpressing these mechanisms were used as controls (22, 23, 29).
Detection of genes encoding acquired β-lactamases.
Preliminary phenotypic tests included the Etest MBL and double-disk synergy test (DDST) using ceftazidime and amoxicillin-clavulanate (distance from 10 to 30 mm) for the detection of ESBLs. The potential presence of genes encoding acquired β-lactamases was additionally explored by PCR and sequencing for all isolates resistant to imipenem and ceftazidime (MBLs) or ceftazidime (ESBLs). Previously described primers and conditions were used to amplify the genes encoding VIM-, IMP-, PER-, CTX-M-, SHV-, TEM-, and OXA-type β-lactamases (6, 11, 27). After PCR amplification, sequencing reactions were performed with the BigDye Terminator kit (PE Applied Biosystems, Foster City, CA), and sequences were analyzed with an ABI Prism 3100 DNA sequencer (PE Applied Biosystems). The resulting sequences were then compared with those available at the GenBank database (www.ncbi.nih.gov/BLAST).
PCR amplification and sequencing of the oprD gene.
The presence of inactivating mutations in oprD was investigated with 25 representative isolates (representing 22 different clones) resistant to imipenem (MIC > 8 μg/liter) through the PCR amplification of the entire gene with specific primers followed by sequencing, as described previously (31). The nucleotide sequences were transcribed into the amino acid sequence using Vector NTI Advanced 9.0.0 (InforMax; Invitrogen). Nucleotide and amino acid sequences were compared with those of reference strain PAO1.
OMP analysis.
Cultures of P. aeruginosa were grown overnight at 37°C in 5 ml of LB medium and then diluted 100-fold into fresh medium. Bacterial cells were incubated for approximately 5 h with shaking at 37°C to yield late-logarithmic-phase cells. Outer membrane protein (OMP) profiles were examined for the 25 above-mentioned strains using a previously reported method (24). OMPs were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie blue. OprD profiles from clinical isolates were compared with those of reference strain PAO1 and a PAO1-derivative oprD-deficient mutant.
Statistical analysis.
Categorical and quantitative variables were compared by using a χ2 test and a Mann-Whitney U test, respectively. A P value of <0.05 was considered statistically significant.
RESULTS
REP-PCR revealed a remarkable clonal diversity, with up to 116 different patterns identified among the 190 isolates. The activities of the tested antipseudomonal agents against the collection of P. aeruginosa bloodstream isolates from Spanish hospitals are shown in Table 1. Among β-lactams, activity was lowest for cefepime (62% susceptible) and highest for piperacillin-tazobactam (86% susceptible) according to CLSI breakpoints. It should be noted, however, that the application of EUCAST breakpoints (resistant at >16 μg/ml) would drop the susceptibility of the latter antibiotic to 72% (Table 1). Among carbapenems, the activity was higher for meropenem (77% susceptible) than for imipenem (68% susceptible). As for non-β-lactams, nearly 30% of the isolates were resistant to fluoroquinolones (ciprofloxacin and levofloxacin), and 20% were resistant to the aminoglycosides gentamicin and tobramycin. Twelve isolates (6.3%) were resistant to all first-line antibiotics (all β-lactams and fluoroquinolones). The only antibiotics conserving activity in most of the isolates were amikacin (98% susceptible) and colistin (97% susceptible).
Table 1.
Activities of antipseudomonal agents against P. aeruginosa bloodstream isolates from Spanish hospitals
| Antibiotica | MIC50 (μg/ml) | MIC90 (μg/ml) | MIC range (μg/ml) | % S (% S according to according to EUCAST breakpoints)b | % I (% I according to according to EUCAST breakpoints)b | % R (% R according to according to EUCAST breakpoints)b |
|---|---|---|---|---|---|---|
| CAZ | 4 | 32 | 0.25–≥128 | 76.3 | 6.8 (0.0) | 16.9 (23.7) |
| FEP | 8 | 32 | 0.25–≥128 | 61.6 | 20.0 (0.0) | 18.4 (38.4) |
| ATM | 8 | 32 | 0.25–≥128 | 67.4 (1.6) | 21.6 (87.4) | 11.0 |
| PIP | 8 | 128 | 1–≥128 | 80.5 (65.8) | 19.5 (34.2) | |
| PIP-Tz | 8 | 128 | 1–≥128 | 86.3 (72.1) | 13.7 (27.9) | |
| IMP | 2 | 32 | 0.12–≥64 | 67.9 | 9.5 | 22.6 |
| MER | 1 | 16 | 0.06–≥64 | 77.4 (70.0) | 7.4 (14.8) | 15.2 |
| CIP | 0.25 | 32 | 0.03–≥32 | 71.6 (65.8) | 1.6 (5.8) | 26.8 (28.4) |
| LEV | 1 | 32 | 0.03–≥32 | 68.4 (62.6) | 2.6 (5.8) | 29.0 (31.6) |
| GEN | 2 | 64 | 0.06–≥64 | 78.9 | 2.1 (0.0) | 19.0 (21.1) |
| TOB | 0.5 | 64 | 0.06–≥64 | 81.6 | 1.1 (0.0) | 17.3 (18.4) |
| AMK | 4 | 8 | 0.12–≥128 | 98.4 (93.7) | 0.5 (4.7) | 1.1 (1.6) |
| COL | 0.5 | 1 | 0.03–8 | 96.8 (98.9) | 2.1 (0.0) | 1.1 |
CAZ, ceftazidime; FEP, cefepime; ATM, aztreonam; PIP, piperacillin; PIP-Tz, piperacillin-tazobactam; IMP, imipenem; MER, meropenem; CIP, ciprofloxacin; LEV, levofloxacin; GEN, gentamicin; TOB, tobramycin; AMK, amikacin; COL, colistin.
Percentage of isolates susceptible (S), intermediate (I), or resistant (R) according to CLSI breakpoints (5). When different, the corresponding percentages of S, I, and R isolates according to EUCAST breakpoints (www.eucast.org) are also indicated in parentheses for comparative purposes.
Two of the isolates were found to produce the MBL VIM-2, whereas ESBLs were not detected in any of the isolates. The prevalence of AmpC and efflux pump overexpression is shown in Table 2. Up to 39% of the isolates overexpressed at least one of the mechanisms. ampC overexpression (24.2%) was the most prevalent mechanism, followed by mexY (13.2%) and mexB (12.6%). The prevalences of mexF (4.2%) and mexD (2.2%) overexpression were considerably lower. Additionally, a certain fraction of the isolates, ranging from 1.6% for mexD to 5.3% for mexY and mexF, were considered to present borderline expression. The means and ranges of values for the overexpression of ampC, mexB, mexD, mexF, and mexY for the studied collection are shown in Table 2.
Table 2.
Prevalences of clinical isolates showing AmpC or efflux pump overexpression
| Resistance gene(s) | No. (%) of isolates with: |
||
|---|---|---|---|
| Overexpressiona | Borderline expression | No overexpression | |
| Any gene | 74 (39.0) | 21 (11.1) | 95 (49.9) |
| ampC | 46 (24.2) | 6 (3.2) | 138 (72.6) |
| mexY | 25 (13.2) | 10 (5.3) | 155 (81.5) |
| mexB | 24 (12.6) | 8 (4.2) | 158 (83.2) |
| mexF | 8 (4.2) | 10 (5.3) | 172 (90.5) |
| mexD | 4 (2.1) | 3 (1.6) | 183 (96.3) |
| Most frequently found combinations | |||
| mexB + mexY | 10 (5.3) | 3 (1.6) | 177 (93.2) |
| ampC + mexY | 10 (5.3) | 2 (1.1) | 178 (93.7) |
| ampC + mexB | 8 (4.2) | 2 (1.1) | 180 (94.7) |
| ampC + mexB + mexY | 3 (1.6) | 2 (1.1) | 185 (97.4) |
Mean values (and ranges) of overexpression (fold increase compared to PAO1) were 419.7 (14.5 to 3,586.1) for ampC, 20.9 (10.2 to 37.4) for mexY, 5.6 (3.1 to 13.3) for mexB, 20.5 (14.2 to 33.2) for mexF, and 20.7 (13.3 to 38.4) for mexD.
The prevalences of the different combinations of resistance mechanisms are also shown in Table 2. The only combination showing a significantly (P = 0.02 by χ2 test) higher prevalence than expected from the frequencies of the individual mechanisms was mexB plus mexY, documented for 5.3% of the isolates (1.6% expected).
REP-PCR analysis of isolates showing ampC or efflux pump overexpression also revealed a high level of clonal diversity. For instance, a total of 29 different patterns were detected among the 46 isolates showing ampC overexpression. Nevertheless, a certain degree of intra- and interhospital clonal dissemination was evidenced: two of the clones showing ampC overexpression were each detected in 6 isolates, in one case from a single institution and in the other case from two different hospitals in the Barcelona area.
The prevalences of ampC and efflux pump overexpression among the subsets of isolates resistant to different antibiotics are shown in Fig. 1. Most of the isolates nonsusceptible to piperacillin-tazobactam (96%) and ceftazidime (84%) overexpressed ampC. The prevalence of ampC overexpression was also high for isolates nonsusceptible to other β-lactams, such as cefepime (52%) or meropenem (60%), although efflux pump overexpression gained protagonism among isolates nonsusceptible to these antibiotics, with prevalences of mexB and mexY overexpression reaching values close to 30%. Regarding ciprofloxacin-resistant isolates, besides the expected increased prevalence of efflux pump overexpression, a highly significant (unexpected) link to ampC overexpression was documented: up to 52% of ciprofloxacin-nonsusceptible isolates overexpressed ampC, in sharp contrast with the 24% documented for the complete collection (P < 0.001 by χ2 test). Interestingly, the same link to ampC overexpression was observed for the aminoglycosides (Fig. 1). As for isolates resistant to all first-line agents (all β-lactams and aminoglycosides), 100% overexpressed ampC, 33% overexpressed mexY, and 25% overexpressed mexB. Finally, it is worth noting that the prevalence of mexY overexpression was highest among tobramycin-nonsusceptible isolates (37%), and that of mexB overexpression was highest among meropenem-nonsusceptible isolates (33%).
Fig. 1.
Prevalences of ampC, mexB, or mexY overexpression in several subsets of isolates nonsusceptible to different antibiotics. * indicates a P value of <0.05 and ** indicates a P value of <0.001 compared to the corresponding subsets of susceptible isolates.
Table 3 shows the activities of the tested antibiotics in the subsets of isolates overexpressing ampC, mexB, or mexY. As could be anticipated, the levels of activity of all β-lactams were low among isolates overexpressing ampC, but again, a remarkable unexpected link between ampC overexpression and fluoroquinolone and aminoglycoside resistance was noted. For example, up to 60% of isolates overexpressing ampC were nonsusceptible to ciprofloxacin (28% for the complete collection), and up to 49% were nonsusceptible to tobramycin (18% for the complete collection). Regarding mexB-overexpressing isolates, activity was particularly low for aztreonam, cefepime, the carbapenems, and the fluoroquinolones, and for mexY, activity was low for the same antibiotics plus the aminoglycosides gentamicin and tobramycin.
Table 3.
Activities of antipseudomonal agents against P. aeruginosa bloodstream isolates with diverse resistance mechanisms
| Antibiotica | All isolates (n = 190) |
Isolates overexpressing ampC (n = 45)b |
Isolates overexpressing mexB (n = 24) |
Isolates overexpressing mexY (n = 25) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % S | MIC50 | MIC90 | % S | MIC50 | MIC90 | % S | MIC50 | MIC90 | % S | MIC50 | MIC90 | |
| CAZ | 76.3 | 4 | 32 | 17.8 | 32 | 128 | 58.7 | 8 | 64 | 68.0 | 8 | 32 |
| FEP | 61.6 | 8 | 32 | 17.8 | 32 | 64 | 25.0 | 16 | 32 | 16.0 | 16 | 32 |
| ATM | 67.4 | 8 | 32 | 22.2 | 16 | 64 | 33.3 | 16 | 32 | 40.0 | 16 | 16 |
| PIP | 80.5 | 8 | 128 | 28.9 | 128 | 128 | 79.2 | 32 | 128 | 68.0 | 16 | 128 |
| PIP-Tz | 86.3 | 8 | 128 | 44.4 | 128 | 256 | 79.2 | 16 | 256 | 80.0 | 16 | 128 |
| IMP | 67.9 | 2 | 32 | 37.8 | 16 | 32 | 41.7 | 8 | 32 | 32.0 | 16 | 32 |
| MER | 77.4 | 1 | 16 | 44.4 | 8 | 16 | 41.7 | 8 | 32 | 44.0 | 8 | 16 |
| CIP | 71.6 | 0.25 | 32 | 40.0 | 32 | 32 | 45.8 | 2 | 32 | 24.0 | 32 | 32 |
| LEV | 68.4 | 1 | 32 | 35.6 | 32 | 32 | 33.3 | 8 | 32 | 16.0 | 32 | 32 |
| GEN | 78.9 | 2 | 64 | 46.7 | 32 | 64 | 66.7 | 2 | 64 | 40.0 | 32 | 64 |
| TOB | 81.6 | 0.5 | 64 | 51.1 | 4 | 64 | 66.7 | 1 | 64 | 48.0 | 8 | 64 |
| AMK | 98.4 | 4 | 8 | 100 | 4 | 8 | 100 | 4 | 8 | 100 | 4 | 16 |
| COL | 96.8 | 0.5 | 1 | 93.3 | 0.5 | 1 | 95.8 | 0.5 | 1 | 100 | 0.5 | 1 |
CAZ, ceftazidime; FEP, cefepime; ATM, aztreonam; PIP, piperacillin; PIP-Tz, piperacillin-tazobactam; IMP, imipenem; MER, meropenem; CIP, ciprofloxacin; LEV, levofloxacin; GEN, gentamicin; TOB, tobramycin; AMK, amikacin; COL, colistin.
One of the isolates showing ampC overexpression additionally produced the MBL VIM-2 and was therefore excluded from this analysis.
Additionally, the involvement of oprD mutations in imipenem resistance was investigated with 25 representative isolates (representing 22 different clones) resistant to this antibiotic. Sequencing of the entire oprD gene was carried out, and the expression of OprD was also assessed by an analysis of the OMP profiles in order to correlate the presence of inactivating mutations to the absence or decreasing expression of OprD. Consistent with previously reported observations (11), nearly all isolates presented inactivating mutations in oprD. Nine of the isolates (36%) showed point mutations leading to premature stop codons, 8 (32%) showed frameshift mutations originated by the insertion or deletion of 1 bp, 4 (16%) showed larger deletions (10 to 100 bp), and 2 (8%) showed an interruption of the gene by insertion sequences (IS1471-like and ISPa1328). One further isolate had a deletion of 5 nucleotides starting at position 1132 and a 1-bp deletion at position 1149, which generated an in-frame deletion resulting in the loss of Gly-378 and Tyr-379 and amino acid substitutions (K380Y, N381A, Y382G, and G383L). This isolate also displayed a point mutation in the stop codon leading to a larger (13-amino-acid) predicted protein. Finally, another isolate did not show apparent inactivating mutations but presented several polymorphisms compared to PAO1 and lacked OprD expression according to OMP profiles (data not shown).
OprD inactivation is also known to confer reduced susceptibility to meropenem, although clinical resistance to this carbapenem is thought to require additional mechanisms, such as AmpC or MexAB-OprM hyperproduction (10, 11, 30). Consistent with this observation, as shown in Table 4, a high proportion of imipenem-nonsusceptible and meropenem-nonsusceptible isolates overexpressed ampC (62.5%) or mexB (30%), in contrast to isolates nonsusceptible only to imipenem.
Table 4.
Prevalences of ampC, mexB, and mexY overexpression among isolates showing particular resistance phenotypes
| No. (%) of isolates | Resistance phenotypea |
No. (%) of isolates showing overexpression of: |
|||||
|---|---|---|---|---|---|---|---|
| IMP | MER | CAZ | FEP | ampC | mexB | mexY | |
| 190 (100) | All | All | All | All | 46 (24.2) | 24 (12.6) | 25 (13.2) |
| 95 (50) | S | S | S | S | 4 (4.2) | 5 (5.3) | 2 (2.1) |
| 21 (11.1) | R | S | All | All | 4 (19.0) | 1 (4.8) | 4 (19.0) |
| 40 (21.1) | R | R | All | All | 25 (62.5) | 12 (30.0) | 13 (32.5) |
| 30 (15.8) | All | All | S | R | 1 (3.3) | 8 (26.7) | 13 (43.3) |
| 43 (22.6) | All | All | R | R | 37 (86.0) | 10 (23.3) | 8 (18.6) |
| 27 (14.2) | R | R | R | R | 24 (88.9) | 8 (29.6) | 8 (29.6) |
IMP, imipenem; MER, meropenem; CAZ, ceftazidime; FEP, cefepime. “R” phenotypes include CLSI intermediate and resistant categories. Imipenem-sensitive–meropenem-resistant (n = 3) and ceftazidime-susceptible–cefepime-resistant (n = 2) phenotypes were not included in the table due to the nonsignificant number of isolates. “All” indicates all possible phenotypes (S/I/R).
Table 4 also shows the prevalences of ampC, mexB, and mexY overexpression according to the ceftazidime-cefepime resistance phenotype. Consistent with data from previously reported studies (12), a very high prevalence (43%) of mexY overexpression was observed for isolates nonsusceptible only to cefepime, whereas ampC overexpression was documented for most (86%) of the isolates nonsusceptible to both antibiotics.
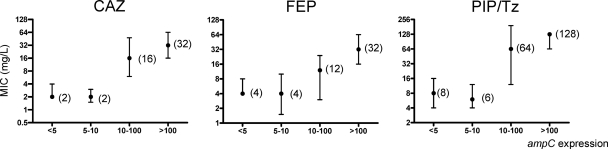
Finally, Fig. 2 shows the correlation between the level of ampC expression and the level of resistance (MICs) to ceftazidime, cefepime, and piperacillin-tazobactam. While MICs for isolates showing borderline expression (5- to 10-fold higher than that of reference strain PAO1) were not significantly higher than those for negative (<5-fold) isolates, the MICs of the three antibiotics sharply increased at higher expression levels (Fig. 2). As found for ampC, the median MICs for isolates with borderline mexB overexpression (2- to 3-fold) were nearly identical to those for negative (<2-fold) isolates, but a tendency (not reaching statistical significance) toward higher MICs of cefepime was noted for isolates showing borderline mexY expression (not shown).
Fig. 2.
Effect of the level of ampC expression on the MICs of ceftazidime (CAZ), cefepime (FEP), and piperacillin-tazobactam (PIP-Tz). Median MIC values are shown; error bars represent the interquartile ranges.
DISCUSSION
The prevalence and impact of the overexpression of the genes encoding the chromosomal cephalosporinase AmpC (ampC) and the four major efflux pumps (mexB, mexD, mexF, and mexY) on resistance were investigated with a large collection of P. aeruginosa bloodstream isolates from a Spanish multicenter study, providing national-scale epidemiologic information on these relevant resistance mechanisms, which was previously available only from preceding studies in France (13). In our study, up to 39% of the isolates overexpressed at least one of the mechanisms, with ampC (24.2%), mexY (13.2%), or mexB (12.6%) being the gene more frequently overexpressed. While the prevalences of ampC (20%) and mexB (11%) overexpression obtained by the French study were similar, that of mexY (36%) was much higher. One factor possibly contributing to this difference is the breakpoints used for defining AmpC and efflux pump overexpression, which vary substantially in the literature (8, 11, 13, 26, 34) and still need to be harmonized. In any case, the lower breakpoints used for mexY in the French study are comparable to our breakpoints for borderline expression, which would increase the prevalence to 18.5%, still half of that documented by the above-mentioned study. This apparent discrepancy could well reflect differences in antibiotic use in both countries, since the incidence of isolates hyperproducing MexXY-OprM has been shown to be very much influenced by the use of aminoglycosides, fluoroquinolones, and cefepime, as opposed to carbapenems (14). It should be noted, however, that the first large outbreak of a P. aeruginosa strain hyperproducing MexXY-OprM was recently reported for a Spanish hospital (28). Indeed, an understanding of the interplay between the specific antibiotic use policies, the local clonal epidemiology, and the dynamics of P. aeruginosa resistance is an issue of major relevance that should be further investigated.
Another important issue to consider for an understanding of resistance dynamics is the interconnections between the different resistance mechanisms. A statistically significant link between mexB and mexY overexpressions was documented for the first time in this study. Our results are in agreement with previously reported data showing that the production of both resistance mechanisms simultaneously is frequently found for P. aeruginosa isolates, despite the finding that independent mutations are apparently responsible for the overexpression of each efflux pump (20). Thus, the epidemiological, biological, or mechanistic factors responsible for this link remain to be elucidated. Perhaps, what is more surprising and concerning is the strong statistical link between ampC overexpression and fluoroquinolone resistance documented. Indeed, this study adds AmpC hyperproduction in P. aeruginosa to the growing and concerning examples of the strong linkage of fluoroquinolone resistance to β-lactam resistance mechanisms, particularly the extended-spectrum β-lactamases in members of the Enterobacteriaceae or methicillin resistance in Staphylococcus aureus. It still needs to be experimentally addressed whether this link is favored by the mutagenic effects of fluoroquinolones (3) and/or genetic capitalism (a strain resistant to one antibiotic is more likely to acquire resistance to a second antibiotic) (2).
While most isolates nonsusceptible to ceftazidime or piperacillin-tazobactam were found to hyperproduce AmpC, MexAB-OprM, and (particularly) MexX-OprM, overexpression gained protagonism among cefepime-nonsusceptible isolates. In agreement with data from previous works (11, 32, 35), all imipenem-resistant isolates studied were found to be deficient in the carbapenem porin OprD, due mainly to the presence of inactivating mutations in oprD. For those isolates additionally resistant to meropenem, a strong link with AmpC and MexAB-OprM overexpression was evidenced, consistent with previously reported findings (10, 11, 30). Moreover, the highest prevalence of mexB overexpression (33%) was found among meropenem-nonsusceptible isolates. On the other hand, the highest prevalence of mexY overexpression (37%) was found among tobramycin-nonsusceptible isolates. However, it should be noted that the overexpression of MexXY-OprM is known to confer low-level resistance to aminoglycosides (1). Our results indeed indicate that the overexpression of this efflux system is a frequent coadjuvant of aminoglycoside-modifying enzymes in aminoglycoside resistance, as evidenced by its strong linkage to gentamicin and tobramycin resistance, despite most of the isolates being susceptible to amikacin according to CLSI breakpoints.
Finally, 1% of the isolates (4% of those resistant to imipenem) were found to be MBL producers. While this prevalence of MBLs was still much lower than those reported previously for South America (9), the Far East (16), or certain European countries (33), it denotes an approximately 10-fold increase compared to that reported by a multicenter study performed in Spain 5 years earlier (11).
In summary, the overexpression of AmpC and efflux pumps as well as the mutational inactivation of OprD are frequent among P. aeruginosa isolates from bloodstream infections in Spanish hospitals, whereas MBL production, although increasing, is still infrequent.
ACKNOWLEDGMENTS
We are grateful to all clinical microbiologists and infectious disease specialists involved in this project from the Hospital Universitario de Bellvitge (Barcelona), Consorsi Sanitari Parc Tauli (Barcelona), Hospital Universitario Virgen de la Macarena (Sevilla), Hospital Vall de Hebrón (Barcelona), Hospital Universitari Son Dureta (Palma de Mallorca), Hospital de Sant Pau (Barcelona), Hospital Universitario Marqués de Valdecilla (Santander), Hospital Virgen del Rocío (Sevilla), Hospital Reina Sofía (Córdoba), and Hospital Mutua de Tarrasa (Barcelona).
This work was supported by the Ministerio de Ciencia e Innovación of Spain and the Instituto de Salud Carlos III, through the Spanish Network for the Research in Infectious Diseases (REIPI C03/14 and RD06/0008), and grants PI08/0802 and PS09/00033.
Footnotes
Published ahead of print on 28 February 2011.
REFERENCES
- 1. Aires J. R., Köhler T., Nikaido H., Plésiat P. P. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baquero F. 2004. From pieces to patterns: evolutionary engineering in bacterial pathogens. Nat. Rev. Microbiol. 2:510–518 [DOI] [PubMed] [Google Scholar]
- 3. Blázquez J., Oliver A., Gómez-Gómez J. M. 2002. Mutation and evolution of antibiotic resistance: antibiotics as promoters of antibiotic resistance? Curr. Drug Targets 3:345–349 [DOI] [PubMed] [Google Scholar]
- 4. Cano M. E., et al. 2009. Detection of plasmid-mediated quinolone resistance genes in clinical isolates of Enterobacter spp. in Spain. J. Clin. Microbiol. 47:2033–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2008. Performance standards for antimicrobial susceptibility testing, vol. 28, no. 3, 18th informational supplement. M100-S18 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6. Coque T. M., Oliver A., Perez-Diaz J. C., Baquero F., Canton R. 2002. Genes encoding TEM-4, SHV-2, and CTX-M-10 extended-spectrum beta-lactamases are carried by multiple Klebsiella pneumoniae clones in a single hospital (Madrid, 1989 to 2000). Antimicrob. Agents Chemother. 46:500–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deplano A., et al. 2005. Molecular characterization of an epidemic clone of panantibiotic-resistant Pseudomonas aeruginosa. J. Clin. Microbiol. 43:1198–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dumas J. L., van Delden C., Perron K., Köhler T. 2006. Analysis of antibiotic resistance gene expression in Pseudomonas aeruginosa by quantitative real-time-PCR. FEMS Microbiol. Lett. 254:217–225 [DOI] [PubMed] [Google Scholar]
- 9. Fritsche T. R., Sader H. S., Toleman M. A., Walsh T. R., Jones R. N. 2005. Emerging metallo-beta-lactamase-mediated resistances: a summary report from the worldwide SENTRY antimicrobial surveillance program. Clin. Infect. Dis. 41:S276–S278 [DOI] [PubMed] [Google Scholar]
- 10. Giske C. G., Buarø L., Sundsfjord A., Wretlind B. 2008. Alterations of porin, pumps, and penicillin-binding proteins in carbapenem resistant clinical isolates of Pseudomonas aeruginosa. Microb. Drug Resist. 14:23–30 [DOI] [PubMed] [Google Scholar]
- 11. Gutiérrez O., et al. 2007. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa isolates from Spanish hospitals. Antimicrob. Agents Chemother. 51:4329–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hocquet D., Nordmann P., El Garch F., Cabanne L., Plésiat P. 2006. Involvement of the MexXY-OprM efflux system in emergence of cefepime resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 50:1347–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hocquet D., et al. 2007. Pseudomonas aeruginosa may accumulate drug resistance mechanisms without losing its ability to cause bloodstream infections. Antimicrob. Agents Chemother. 51:3531–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hocquet D., et al. 2008. Relationship between antibiotic use and incidence of MexXY-OprM overproducers among clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52:1173–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Juan C., Moyá B., Pérez J. L., Oliver A. 2006. Stepwise upregulation of the Pseudomonas aeruginosa chromosomal cephalosporinase conferring high-level beta-lactam resistance involves three AmpD homologues. Antimicrob. Agents Chemother. 50:1780–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee K., et al. 2003. VIM- and IMP-type metallo-beta-lactamase-producing Pseudomonas spp. and Acinetobacter spp. in Korean hospitals. Emerg. Infect. Dis. 9:868–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leibovici L., et al. 1998. The benefit of appropriate empirical antibiotic treatment in patients with bloodstream infection. J. Intern. Med. 244:379–386 [DOI] [PubMed] [Google Scholar]
- 18. Lister P. D., Wolter D. J., Hanson N. D. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 22:582–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Livermore D. M. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34:634–640 [DOI] [PubMed] [Google Scholar]
- 20. Llanes C., et al. 2004. Clinical strains of Pseudomonas aeruginosa overexpressing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob. Agents Chemother. 48:1797–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mesaros N., et al. 2007. Pseudomonas aeruginosa: resistance and therapeutics options in the turn of the new millennium. Clin. Microbiol. Infect. 13:560–578 [DOI] [PubMed] [Google Scholar]
- 22. Moya B., et al. 2009. Beta-lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog. 5:e1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mulet X., et al. 2009. Azithromycin in Pseudomonas aeruginosa biofilms: bactericidal activity and selection of nfxB mutants. Antimicrob. Agents Chemother. 53:1552–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mushtaq S., Ge Y., Livermore D. M. 2004. Doripenem versus Pseudomonas aeruginosa in vitro: activity against characterized isolates, mutants, and transconjugants and resistance selection potential. Antimicrob. Agents Chemother. 48:3086–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Obritsch M. D., Fish D. N., MacLaren R., Jung R. 2004. National surveillance of antimicrobial resistance in Pseudomonas aeruginosa isolates obtained from intensive care unit patients from 1993 to 2002. Antimicrob. Agents Chemother. 48:4606–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oh H., Stenhoff S., Jalal S., Wretlind B. 2003. Role of efflux pumps and mutations in genes for topoisomerases II and IV in fluoroquinolone-resistant Pseudomonas aeruginosa strains. Microb. Drug Resist. 8:323–328 [DOI] [PubMed] [Google Scholar]
- 27. Oliver A., et al. 2002. Mechanisms of decreased susceptibility to cefpodoxime in Escherichia coli. Antimicrob. Agents Chemother. 46:3829–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peña C., et al. 2009. Nosocomial outbreak of a non-cefepime-susceptible ceftazidime-susceptible Pseudomonas aeruginosa strain overexpressing MexXY-OprM and producing an integron-borne PSE-1 beta-lactamase. J. Clin. Microbiol. 47:2381–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Plasencia V., et al. 2007. Influence of high mutation rates on the mechanisms and dynamics of in vitro and in vivo resistance development to single or combined antipseudomonal agents. Antimicrob. Agents Chemother. 51:2574–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Quale J., Bratu S., Gupta J., Landman D. 2006. Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 50:1633–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rodríguez M. C., et al. 2010. Molecular characterization of Pseudomonas aeruginosa isolates in Cantabria, Spain, producing VIM-2 metallo-beta-lactamase. Enferm. Infecc. Microbiol. Clin. 28(2):99–103 [DOI] [PubMed] [Google Scholar]
- 32. Rodríguez-Martínez J. M., Poirel L., Nordmann P. 2009. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53:4783–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rossolini G. M., et al. 2008. First countrywide survey of acquired metallo-beta-lactamases in Gram-negative pathogens in Italy. Antimicrob. Agents Chemother. 52:4023–4029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tomás M., et al. 2010. Efflux pumps, OprD porin, AmpC beta-lactamase, and multiresistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 54:2219–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trias J., Nikaido H. 1990. Outer membrane protein D2 catalyzes facilitated diffusion of carbapenems and penems through the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 34:52–57 [DOI] [PMC free article] [PubMed] [Google Scholar]